Abstract
Although the specific function of SCO2127 remains elusive, it has been assumed that this hypothetical protein plays an important role in carbon catabolite regulation and therefore in antibiotic biosynthesis in Streptomyces coelicolor. To shed light on the functional relationship of SCO2127 to the biosynthesis of actinorhodin, a detailed analysis of the proteins differentially produced between the strain M145 and the Δsco2127 mutant of S. coelicolor was performed. The delayed morphological differentiation and impaired production of actinorhodin showed by the deletion strain were accompanied by increased abundance of gluconeogenic enzymes, as well as downregulation of both glycolysis and acetyl-CoA carboxylase. Repression of mycothiol biosynthetic enzymes was further observed in the absence of SCO2127, in addition to upregulation of hydroxyectoine biosynthetic enzymes and SCO0204, which controls nitrite formation. The data generated in this study reveal that the response regulator SCO0204 greatly contributes to prevent the formation of actinorhodin in the ∆sco2127 mutant, likely through the activation of some proteins associated with oxidative stress that include the nitrite producer SCO0216.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptomycetes are well-known producers of natural products, including over half of the antibiotics currently used in medical practice (Demain 2015). Production of these metabolites is tightly related to the development cycle. While the synthesis of natural products occurs after the emergence of aerial mycelium in semisolid media (Chater 2006), in liquid cultures takes place at the well-defined transition phase between the compartmentalized and multinucleated mycelium, which corresponds with the switch from the exponential to stationary growth (Manteca et al. 2008). In the model actinomycete S. coelicolor, is particularly easy to identify this event because of the production of prodiginine and actinorhodin (ACT), which have a characteristic red and blue appearance, respectively (Tsao et al. 1985; Bystrykh et al. 1996). ACT is probably the most studied antibiotic produced by S. coelicolor. Its biosynthetic pathway has been thoroughly described (Fernández-Moreno et al. 1992; Fernández-Moreno et al. 1994) and currently, a large number of either specific or regulatory proteins are recognized (Arias et al. 1999; Kang et al. 2007; Taguchi et al. 2007; Lee et al. 2009; Feng et al. 2011; Rico et al. 2014). Despite this, little is known about more complex regulatory networks that inhibit its production in S. coelicolor.
Carbon catabolite regulation (CCR) affects the utilization of carbon sources and the synthesis of secondary metabolites by reducing either the expression or the activities of the corresponding enzymes when a favored carbon source is present in the culture medium (Ruiz et al. 2010; Görke and Stülke 2008). Pioneer studies on Streptomyces CCR include the identification of S. coelicolor mutants resistant to grow in cultures with the glucose analog 2-deoxyglucose (DogR mutants) (Hodgson 1982). S. coelicolor DogR mutants started to be studied in the early 80s because their phenotype includes the well-known ‘symptoms’ of carbon catabolite resistance. These are characterized by a significant reduction of glucose kinase activity (Seno and Chater 1983), which reduces levels of glycolytic intermediates (Jault et al. 2000; Ramos et al. 2004). Genetic complementation of S. coelicolor DogR mutants with a 2.9-kb DNA fragment containing the gene glkA (SCO2126) resulted in a reversion of the DogR phenotype (Ikeda et al. 1984) and supported glucose kinase (GlkA) as the top regulator of the CCR in S. coelicolor. However, such 2.9 kb DNA fragment also contained the sco2127 gene (just upstream of glkA) (Angell et al. 1992). Further studies on the anthracyclines producer Streptomyces peucetius var. caesius DogR mutants even show that complementation with sco2127 completely restores wild type levels of antibiotics (Guzman et al. 2005). Despite sco2127 codes for a soluble protein (Chávez et al. 2009), it has been difficult to determine its activity due to the lack of sequence homology to any known protein. Whatever the role of SCO2127 in CCR, we are interested in the identification of new factors functionally related to this protein that may affect the production of ACT in S. coelicolor.
In this study, a comparison between the wild type and the Δsco2127 mutant was performed at the translational level. The deletion strain displayed a dramatic reduction in the production of ACT. This phenotype was accompanied by an altered expression of mycothiol and hydroxyectoine biosynthetic enzymes, as well as the unexpected accumulation of nitrite. An in silico analysis, followed by deletion of the response regulator encoded by sco0204, enabled us to identify that this gene controls the formation of nitrite in the Δsco2127 strain. The results presented here illustrate how genome-wide biological data sets can be used to study tightly regulated complex phenotypes and expand our understanding of novel regulatory networks that influence the synthesis of ACT in S. coelicolor M145.
Materials and methods
Bacterial strains and growth conditions
S. coelicolor M145, a plasmid-free derivative of S. coelicolor A3(2), was used as the wild-type strain. The markerless Δsco2127 mutant was obtained as described elsewhere (Gust et al. 2002) from the previously reported apramycin resistant mutant Δsco2127:aac(3)IV (Chávez et al. 2011). This technique allowed us to use the apramycin resistant cassette to replace the gene sco0204, generating the double mutant Δsco2127 Δsco0204:aac(3)IV. For spore harvesting, the strains were routinely grown as confluent lawns on MS plates (Kieser et al. 2000). For fermentation studies, 50 mL of modified R5 liquid medium (Fernández et al. 1998) was utilized. The medium lacked sucrose but was supplemented with 55 mM arabinose as the sole carbon source. The medium was inoculated into 250-mL baffled flasks (Sigma-Aldrich; St. Louis, Missouri, US) with 4 × 106 spores/mL and incubated at 30 °C under agitation (200 rpm).
Actinorhodin and nitrite determination
Actinorhodin production during the stationary phase was determined according to Bystrykh et al. (1996). Colorimetric determination of nitrite was performed according to the Griess-Ilosvay principle, using adjusted quantities of suitable Api 20 E™ reagents (bioMérieux, Inc.; Marcy-I’Étoile, France).
Protein extraction
Cells from two separate biological replicates of 50 mL-flask cultures of M145 and the sco2127 deficient strain were harvested at mid exponential and stationary growth phase by centrifugation (11,000×g at 4 °C). The pellets were washed with PBS buffer (8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4 per liter, pH 7.5 and then resuspended in lysis buffer 50 mM Tris–HCl, pH 7.5, Thermo protease inhibitor as described elsewhere (Licona-Cassani et al. 2014). Homogeneous cell disruption was achieved by sonication and cellular debris removed by centrifugation 10,000×g, 5 min at 4 °C. To further remove impurities, treatment with DNase I and RNase A (Thermo Fisher Scientific Inc.; Waltham, Massachusetts, USA) was performed. The crude protein extract was dialyzed against Milli-Q water, quantified using the 2D Quant Kit (GE Healthcare; Little Chalfont, United Kingdom) and lyophilized.
Protein digestion
SWATH samples were prepared as specified in Wiśniewski et al. (2009). Briefly, 50 μg of protein from each strain were diluted in 800 μL of 8 M urea/50 mM AMBIC and loaded into four Amicon Ultra-0.5 mL centrifugal filters with a nominal cut-off of 30 kDa (Millipore; Billerica, Massachusetts, USA). Proteins were washed twice with 200 μL of 8 M urea/50 mM AMBIC by centrifugation (14, 000×g for 15 min), followed by two washes in 100 μL of 50 mM AMBIC. In-filter trypsin digestion was performed (overnight at 37 °C, 50 rpm) using 5.0 μg of MS-grade trypsin (Promega; Madison, Wisconsin, USA) in 30 μL of AMBIC (50 mM). Digested proteins were collected by three rounds of centrifugation with 30 μL of AMBIC (50 mM). Final pH of the samples was adjusted to 3.0 using formic acid. Protein samples were concentrated using a vacuum centrifuge and finally concentrated using a C-18 ZipTip (Millipore) to assure that quantities injected in each LC injection were equal.
SWATH library reference and sample preparation
Information-dependent acquisition (IDA) was used to generate a reference library from a pooled sample that included 50 μg of proteins from each of the strains tested in two biological replicates. To avoid column overloading, C-18 ZipTips were used. Before MS analysis, samples were concentrated using a vacuum centrifuge to remove residual acetonitrile and resuspended in 99.5 μL of 0.1 % formic acid. Synthetic peptide mixture (HRM calibration kit from Biognosys; Zurich, Switzerland) was spiked to each sample for retention time shift correction according to Bernhardt et al. (2012) and Escher et al. (2012). IDA and SWATH injections were performed in duplicate from 4 to 0.5 μg of peptides, respectively.
Statistical analysis
PeakView 1.2 (ABSciex) was used for SWATH MS/MS data processing. Data was log2-transformed and quantile-normalized (Bolstad et al. 2003) and technical replicates were averaged. Only proteins with at least two peptides with >95 % confidence were used for statistical analysis. Sample statistical analyses of two biological replicates were performed by fitting the data for each protein to a linear model using the R Limma package (Smyth 2005). Limma’s empirical Bayes method was used to calculate a moderated t-statistic test for the contrast, and proteins were classified as differentially expressed when their adjusted p value was lower than 0.05 (Smyth 2004).
Liquid chromatography coupled to mass spectrometry (LC-MS) analysis
LC-MS/MS was used for proteomic and metabolomics analysis. Central carbon metabolites were measured at mid-exponential phase using an ion-pairing HPLC-MS/MS method, as described in Dietmair et al. (2012) and modified in McDonald et al. (2014). For proteomic discovery (IDA) and quantification (SWATH), samples were initially separated using a Shimadzu Prominence nano-LC system as previously described in Kappler and Nouwens (2013) using the modifications reported by Orellana et al. (2015).
Results
The ∆sco2127 mutant showed impairment of actinorhodin biosynthesis and morphological differentiation.
Phenotypic characterization of S. coelicolor M145 and its derivative mutant Δsco2127 was performed in 250-mL baffled flasks containing 50 mL of modified R5 liquid media supplemented with arabinose 55 mM as the carbon source. In these conditions, the SCO2127 deficient strain showed a clear reduction of biomass along the fermentation (Fig. 1a). However, during the exponential growth phase, the two strains showed similar specific growth rates, i.e., 0.11 ± 0.01 and 0.09 ± 0.02 h−1, respectively. Although the biosynthesis of actinorhodin in both cultures started at 48 h (8 h after entering the stationary phase), pigment production in the deletion mutant decreased more than 90 % at stationary phase (Fig. 1b). According to this, the ∆sco2127 strain also showed a severe damage of mycelial development when streaked on modified R5 plates (Fig. 2).
Deletion of sco2127 alters the abundance of several proteins
SCO2127 is one of 3694 hypothetical proteins (47 % of the total ORFs) encoded in S. coelicolor M145 (Bentley et al. 2002). A lack of sequence homology to any known protein makes difficult to formulate a hypothesis explaining the phenotypic differences between M145 and the SCO2127 deficient strain. Therefore, we used proteomics to generate possible explanations for the virtual lack of actinorhodin biosynthesis observed in the absence of SCO2127. Of the 346 proteins differentially produced between the M145 strain and the deletion mutant at stationary growth phase (Online Resource 1), those involved in the biosynthesis of actinorhodin showed the lowest abundance in the ∆sco2127 strain (Table 1). Other proteins that showed less abundance in the absence of SCO2127 included enolase (SCO3096), and pyruvate kinase (SCO5423), which are involved in glycolysis. Moreover, downregulation of acetyl-CoA carboxylase (SCO5535), which catalyzes the formation malonyl-CoA, was observed. In contrast, the abundance of the gluconeogenic enzymes glyceraldehyde-3-phosphate dehydrogenase (SCO7040) and fructose 1, 6-bisphosphatase (SCO5047) increased in the ∆sco2127 strain.
The Δsco2127 strain shows altered abundance of stress-related proteins
Mycothiol (MSH) helps to maintain the redox balance and counteracts the effects of reactive oxygen species in S. coelicolor M145 and Mycobacterium tuberculosis (Buchmeier et al. 2003). The abundance of two out (SCO1663 and SCO4967) of five enzymes required for the biosynthesis of MSH was reduced 1.56- and 2.02-fold, respectively, in the Δsco2127 strain at stationary phase. In contrast, high abundance of SCO1865 (1.99-fold) and SCO1867 (2.3-fold), which are involved in the biosynthesis of the compatible solute hydroxyectoine (ECT), was observed in the deletion mutant (Table S1). Similar to these enzymes, deletion of sco2127 also resulted in upregulation of different proteins associated with redox stress including a zinc-containing alcohol dehydrogenase SCO0179 (6.53-fold), a universal stress protein SCO0200 (1.99-fold), a luxR family two-component response regulator SCO0204 (2.5-fold) and two subunits of nitrate reductase SCO0216-17 (2.8-fold each).
An additional comparison between the proteomic profiles of M145 and ∆sco2127 strains was performed at exponential phase, just to ensure that the differential abundance of these enzymes was not simply due to the growth phase. Interestingly, similar abundance pattern (either up- or downregulated) for SCO1663, SCO4967, SCO1865, SCO1867, SCO0179, SCO0204, and SCO0216 was noticed. However, the abundance of both SCO0200 and SCO0217 did not show statistically significant change between the two strains at exponential phase. Despite this, low abundance of SCO4151 (3.14-fold), which is additionally involved in the biosynthesis of MSH, was detected in the ∆sco2127 mutant (Fig. 3).
Abundance profile of ten proteins differentially produced between the ∆sco2127 mutant and the parent strain M145 at both, exponential and stationary growth phase. According to the color bar on top, protein abundance in the deletion strain is indicated as red (high) and green (low), respectively. Gray denotes the absence of data
Regarding the enzymes encoded by the act gene cluster, it is noteworthy that, unlike what was observed in the stationary phase, only the ACT transporter (SCO5083) and the ketoacyl reductase (SCO5086) decreased their abundance (1.75- and 1.41-fold, respectively) in the ∆sco2127 mutant at exponential phase.
The SCO2127 deficient strain accumulates nitrite
In S. coelicolor M145, nitrite is produced by the nitrate reductase enzyme complex SCO0216-19. Moreover, nitrite induces delay of growth and development, and negatively affects the formation of ACT (Fischer et al. 2014), which resembles the phenotype of the ∆sco2127 mutant (Fig. 2). Since the nitrate reductase SCO0216 showed higher abundance in the SCO2127 deficient strain, compared to M145 at both exponential and stationary growth phases (Fig. 3), a colorimetric assay was subsequently performed to determine quantitatively whether the mutant accumulated nitrite. Due to interference caused by the presence of ACT, nitrite could not be determined in any of the two strains at stationary phase (data not shown). However, more than a threefold accumulation of nitrite was observed in the ∆sco2127 mutant (10.34 ± 0.51 nmol*mgDCW−1), about the parent M145 strain (3.08 ± 1.39 nmol*mgDCW−1) at exponential phase (Fig. 4).
The response regulator SCO0204 controls nitrite formation in the Δsco2127 mutant
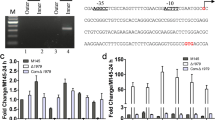
Differentially expressed DNA-binding proteins points to relevant sub-regulatory networks of the Δsco2127 mutant, thus, a phylogenetic footprint analysis was conducted to identify the putative regulatory targets of SCO0204. The analysis consists in retrieve the promoters of all orthologous of SCO0204 in the class Actinobacteria to find the conserved elements using the footprint-discovery tool (Janky and van Helden 2008a). The resulting motif, i.e., a G + C rich inverted repeat 5′-cAGGGCCGAtCGGCCCtg-3′, was identified as the binding site for regulatory gene sco0204 and was subsequently searched in the promoter of genes corresponding to our differentially expressed protein data set according to Turatsinze et al. (2008). This bioinformatic approach showed the motif to be located upstream of multiple genes, including sco0179, sco0200, sco0204, and sco0216 (Table 2). Particularly, the putative binding sites of SCO0204 discovered upstream of the nitrate reductase-encoding gene sco0216 suggest that the transcriptional regulator SCO0204 controls its expression and ultimately the accumulation of nitrite in the ∆sco2127 mutant.
To validate our bioinformatic predictions, we generated the double mutant Δsco2127 Δsco0204:aac(3)IV and evaluated its nitrite production. As expected, the double mutant displayed a 60 % decrease in nitrite accumulation compared to the SCO2127 deficient strain (Fig. 4). As nitrite concentration in the double mutant was reduced, compared to the Δsco2127 single mutant, ACT production was improved over 10-fold in the Δsco2127 Δsco0204:aac(3)IV strain (Fig. 4). Consistently, morphological differentiation of the double mutant clearly enhanced over the ∆sco2127 strain (Fig. 2).
Discussion
The idea that SCO2127 plays an important role in Streptomyces CCR is derived from the study of spontaneous mutants resistant to grow in the presence of the glucose analog 2-deoxyglucose (DogR mutants) (Angell et al. 1992; Angell et al. 1994; Guzman et al. 2005; Guzmán et al. 2005). However, such mutants exhibit, by their very nature, a variety of genotypes (Ikeda et al. 1984), a situation that has been practically ignored but that certainly has hindered the participation of the hypothetical protein SCO2127 in CCR and therefore its influence in the production of antibiotics. Whatever the role of SCO2127 in this regulatory mechanism, in this work, we have analyzed for the first time the markerless ∆sco2127 mutant to avoid conclusions obtained from polar effects. Also, this allows a better insight of the impact that the protein encoded by SCO2127 has in the production of actinorhodin specifically. The results obtained in this work reveal that SCO2127 is necessary for the biosynthesis of ACT in S. coelicolor. Further proteome profile analysis of the ∆sco2127 mutant showed a differential abundance of several proteins, including upregulation of the nitrate reductase SCO0216, which has been confirmed to produce nitrite in S. coelicolor (Fischer et al. 2014). Nitrite leads to the spontaneous formation of nitric oxide, a highly reactive, cytotoxic and lipophilic radical that reacts with DNA and proteins (Cooper 1999; Weiss 2006). Moreover, it was recently found that nitrite prevented the formation of actinorhodin and induced a delay of growth and development in S. coelicolor M145 cultures (Fischer et al. 2014). Thus, it is likely that the fall in actinorhodin formation and probably the impairment of morphological differentiation showed by the ∆sco2127 strain can be due to a stress condition caused by an accumulation of nitrite.
In the M145 strain, it has been observed that the lack of mshA, mshC, and mshD produce an undetectable level of MSH and a negative effect on morphological differentiation (Park et al. 2006). Therefore, we hypothesize that delayed morphological differentiation observed in the deletion mutant is additionally linked to downregulation of SCO1663 (MshC), SCO4151 (MshD) and probably SCO4967 (Mca), a key enzyme involved in MSH-dependent detoxification (Park et al. 2006). As MSH is required to sustain a redox balance (Buchmeier et al. 2003; Newton et al. 2008; Hesketh et al. 2015), it is possible that the ∆sco2127 mutant is defenseless against the effects of reactive oxygen species likely produced from nitrite. As a result of either nitrite accumulation or downregulation of some MSH biosynthetic enzymes, it seems that the oxidative stress exhibited by the deletion mutant further increased the abundance of SCO1865 and SCO1867, which are required for the biosynthesis of ECT (Bursy et al. 2008). Although the presence of ECT is yet to be determined, upregulation of these enzymes strongly suggests that the ∆sco2127 strain likely produces ECT to stabilize its proteins, nucleic acids or the cell membrane (Roychoudhury et al. 2013). Consistent with this, it has been observed a higher abundance of diverse enzymes involved in the biosynthesis of ECT when S. coelicolor is exposed to stress originated by vancomycin (Hesketh et al. 2015).
In addition to SCO0216, the proteins encoded by sco0179, sco0200, sco0204, and sco0217 showed more abundance in the deletion mutant. These genes are distributed along what appears to be the same genetic loci. Hence, they can be commonly regulated. Upregulation of several genes located in virtually the same region, as well as their corresponding proteins, including the transcriptional regulator SCO0204, had been previously identified in S. coelicolor under distinct genetic backgrounds (Gubbens et al. 2012; Daigle et al. 2015). Interestingly, sco0204 is the only gene within this locus that codes for a DNA-binding protein and therefore is a strong candidate to carry out this regulation. SCO0204 shares 65 % of sequence identity with SCO3818 (Wang et al. 2009), which suggests that expression of the mentioned locus in the ∆sco2127 strain may also be under the influence of SCO3818. However, the absence of SCO3818 in our datasets discards this possibility.
Identification of putative binding sites for SCO0204 upstream of several genes located mainly between sco0167 and sco0219, including the nitrate reductase SCO0216 encoding gene, reinforces the hypothesis that the expression of these genes is under the control of SCO0204. In fact, the consensus sequence, as well as the target genes identified in this work for SCO0204, is strikingly similar to those recently obtained using a different bioinformatic pipeline (Daigle et al. 2015).
Although most of the proteins belonging to the putative regulatory network of SCO0204 were not differentially produced in the present study, the hypothetical involvement of this regulon in response to stress (Daigle et al. 2015) fits well with the observed accumulation of nitrite in the ∆sco2127 strain. Therefore, it can be concluded that in the deleted mutant, the response regulator SCO0204 induces the expression of nitrate reductase and thus the formation of nitrite, which has a negative impact on ACT production. Further deletion of sco0204 in the ∆sco2127 strain resulted in a decreased amount of nitrite since SCO0204 induces nitrite production in the absence of SCO2127. In contrast, a significant increment in ACT concentration was observed in the ∆sco2127 ∆sco0204:aac(3)IV strain, accompanying nitrite shortage. Although the specific production of ACT improved significantly in the double mutant about the sco2127 deficient strain, the ACT produced by the ∆sco2127 ∆sco0204:aac(3)IV mutant did not reach the level shown by the M145 strain. A possible explanation of this event may result from the differential abundance of other proteins associated with the absence of SCO2127, for example, the low abundance of SCO5535. In S. coelicolor, sco5535 codes for the carboxyl transferase subunit of the acetyl-CoA carboxylase (Rodríguez et al. 2001), whose main physiological role is the synthesis of malonyl-CoA. Malonyl-CoA is an essential precursor for the production of many polyketide-based antibiotics, such as ACT (Hopwood and Sherman 1990). Therefore, we assume that downregulation of SCO5535 diminishes the activity of acetyl-CoA carboxylase and thus the biosynthesis of malonyl-CoA, preventing the full recovery of ACT in the ∆sco2127 ∆sco0204:aac(3)IV mutant.
Regarding the central carbon metabolism, deletion of sco2127 also resulted in a differential abundance of diverse enzymes involved in gluconeogenesis and glycolysis. A previous study showed that the flux of carbon through glycolysis and the tricarboxylic acid cycle decreased in a S. coelicolor mutant that does not produce the four endogenous antibiotics including ACT (Coze et al. 2013). Hence, in addition to downregulation of SCO5535, the inability of the ∆sco2127 ∆sco0204:aac(3)IV mutant to recuperate the level of ACT observed in the M145 strain is consistent with additional changes originally caused by deletion of sco2127, specifically the high and low abundance of gluconeogenic and glycolytic enzymes, respectively.
Although an exhaustive analysis of the ∆sco2127 mutant as well as complementary studies, such as crystallization of the SCO2127, is necessary to understand its physiological role. This study highlights the use of genome-wide proteome analysis to describe the phenotypes caused by deletion of uncharacterized proteins and specifically provides new insights into the role of SCO0204 in the S. coelicolor M145 response to stress and its impact on the production of antibiotics.
Change history
09 March 2020
The published online version of this paper contains mistake. The authors first and last names have been interchanged. The correct version is given above.
References
Angell S, Schwarz E, Bibb M (1992) The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol Microbiol 6:2833–2844
Angell S, Lewis C, Buttner M, Bibb M (1994) Glucose repression in Streptomyces coelicolor A3(2): a likely regulatory role for glucose kinase. Mol Gen Genet 244:135–143
Arias P, Fernández-Moreno MA, Malpartida F (1999) Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3 ( 2 ) as a DNA-binding protein. J Bacteriol 181:6958–6968. doi:10.1038/ja.2015.13
Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang C-H, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream M-A, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2. Nature 417:141–147. doi:10.1038/417141a
Bernhardt OM, Selevsek N, Gillet LC, Rinner O, Picotti P, Aebersold R, Reiter L (2012) Spectronaut: a fast and efficient algorithm for MRM-like processing of data independent acquisition (SWATH-MS) data. In: Proceedings of the 60th ASMS Conference on Mass Spectrometry and Allied Topics. Vancouver, Canada,
Bolstad B, Irizarry R, Astrand M, Speed T (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193
Buchmeier NA, Newton GL, Koledin T, Fahey RC (2003) Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol Microbiol 47:1723–1732. doi:10.1046/j.1365-2958.2003.03416.x
Bursy J, Kuhlmann AU, Pittelkow M, Hartmann H, Jebbar M, Pierik AJ, Bremer E (2008) Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl Environ Microbiol 74:7286–7296. doi:10.1128/AEM.00768-08
Bystrykh LV, Fernández-Moreno MA, Herrema JK, Malpartida F, Hopwood DA, Dijkhuizen L (1996) Production of actinorhodin-related “blue pigments” by Streptomyces coelicolor A3 ( 2 ). J Bacteriol 178:2238–2244
Chater K (2006) Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos Trans R Soc Lond Ser B Biol Sci 361:761–768. doi:10.1098/rstb.2005.1758
Chávez A, García-Huante Y, Ruiz B, Langley E, Rodríguez-Sanoja R, Sanchez S (2009) Cloning and expression of the sco2127 gene from Streptomyces coelicolor M145. J Ind Microbiol Biotechnol 36:649–654. doi:10.1007/s10295-009-0533-z
Chávez A, Forero A, Sánchez M, Rodríguez-Sanoja R, Mendoza-Hernández G, Servín-Gonzalez L, Sánchez B, García-Huante Y, Rocha D, Langley E, Ruiz B, Sánchez S (2011) Interaction of SCO2127 with BldKB and its possible connection to carbon catabolite regulation of morphological differentiation in Streptomyces coelicolor. Appl Microbiol Biotechnol 89:799–806. doi:10.1007/s00253-010-2905-8
Cooper CE (1999) Nitric oxide and iron proteins. Biochim Biophys Acta - Bioenerg 1411:290–309. doi:10.1016/S0005-2728(99)00021-3
Coze F, Gilard F, Tcherkez G, Virolle M-J, Guyonvarch A (2013) Carbon-flux distribution within Streptomyces coelicolor metabolism: a comparison between the actinorhodin-producing strain M145 and its non-producing derivative M1146. PLoS One 8:e84151. doi:10.1371/journal.pone.0084151
Daigle F, Lerat S, Bucca G, Sanssouci É, Smith CP, Malouin F, Beaulieu C (2015) A terD domain-encoding gene (SCO2368) is involved in calcium homeostasis and participates in calcium regulation of a DosR-like regulon in Streptomyces coelicolor. J Bacteriol 197:913–923. doi:10.1128/JB.02278-14
Demain, A.L Sanchez, S (2015) The need for new antibiotics. In: Sanchez, S., Demain, A.L. (Eds.), Antibiotics: current innovations and future trends, Caister Academic Press,, pp. 65–82. ISBN: 978-1-908230-54-6
Dietmair S, Hodson MP, Quek L-E, Timmins NE, Gray P, Nielsen LK (2012) A multi-omics analysis of recombinant protein production in Hek293 cells. PLoS One 7:e43394. doi:10.1371/journal.pone.0043394
Escher C, Reiter L, MacLean B, Ossola R, Herzog F, Chilton J, MacCoss MJ, Rinner O (2012) Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics 12:1111–1121. doi:10.1002/pmic.201100463
Feng WH, Mao XM, Liu ZH, Li YQ (2011) The ECF sigma factor SigT regulates actinorhodin production in response to nitrogen stress in Streptomyces coelicolor. Appl Microbiol Biotechnol 92:1009–1021. doi:10.1007/s00253-011-3619-2
Fernández E, Weißbach U, Reillo CS, Braña AF, Méndez C, Rohr J, Salas JA (1998) Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J Bacteriol 180:4929–4937
Fernández-Moreno MA, Martínez E, Boto L, Hopwood DA, Malpartida F (1992) Nucleotide sequence and deduced functions of a set of cotranscribed genes of Streptomyces coelicolor A3 (2) including the polyketide synthase for the antibiotic actinorhodin. J Biol Chem 267:19278–19290
Fernández-Moreno MA, Martínez E, Caballero JL, Ichinose K, Hopwood DA, Malpartida F (1994) DNA sequence and functions of the actVI region of the actinorhodin biosynthetic gene cluster of Streptomyces coelicolor A3(2). J Biol Chem 269:24854–24863
Fischer M, Falke D, Pawlik T, Sawers RG (2014) Oxygen-dependent control of respiratory nitrate reduction in mycelium of Streptomyces coelicolor A3(2). J Bacteriol 196:4152–4162. doi:10.1128/JB.02202-14
Görke B, Stülke J (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi:10.1038/nrmicro1932
Gubbens J, Janus M, Florea BI, Overkleeft HS, van Wezel GP (2012) Identification of glucose kinase-dependent and -independent pathways for carbon control of primary metabolism, development and antibiotic production in Streptomyces coelicolor by quantitative proteomics. Mol Microbiol 86:1490–1507. doi:10.1111/mmi.12072
Gust B, Kieser T, Chater KF (2002) REDIRECT technology: PCR-targeting system in Streptomyces coelicolor. Norwich: John Innes Centre.
Guzman S, Carmona A, Escalante L, Imriskova I, López R, Rodrígues-Sanoja R, Ruiz B, Servín-González L, Sánchez S, Langley E (2005) Pleiotropic effect of the SCO2127 gene on the glucose uptake, glucose kinase activity and carbon catabolite repression in Streptomyces peucetius var. caesius. Microbiology 151:1717–1723. doi:10.1099/mic.0.27557-0
Guzmán S, Ramos I, Moreno E, Ruiz B, Rodríguez-Sanoja R, Escalante L, Langley E, Sanchez S (2005) Sugar uptake and sensitivity to carbon catabolite regulation in Streptomyces peucetius var. caesius. Appl Microbiol Biotechnol 69:200–206. doi:10.1007/s00253-005-1965-7
Hesketh A, Deery MJ, Hong H-J (2015) High-resolution mass spectrometry based proteomic analysis of the response to vancomycin-induced cell wall stress in Streptomyces coelicolor A3(2. J Proteome Res 14:2915–2928. doi:10.1021/acs.jproteome.5b00242
Hodgson DA (1982) Glucose repression of carbon source uptake and metabolism in Streptomyces coelicolor A3(2) and its perturbation in mutants resistant to 2-deoxyglucose. J Gen Microbiol 128:2417–2430
Hopwood DA, Sherman DH (1990) Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet 24:37–62
Ikeda H, Seno ET, Bruton CJ, Chater KF (1984) Genetic mapping, cloning and physiological aspects of the glucose kinase gene of Streptomyces coelicolor. Mol Gen Genet 196:501–507
Janky R, van Helden J (2008a) RSAT-footprint-discovery. In: Regul. Seq. Anal. Tools. http://embnet.ccg.unam.mx/rsa-tools/. Accessed 1 Jan 2014
Janky R, van Helden J (2008b) Evaluation of phylogenetic footprint discovery for predicting bacterial cis-regulatory elements and revealing their evolution. BMC Bioinformatics 9:37. doi:10.1186/1471-2105-9-37
Jault JM, Fieulaine S, Nessler S, Gonzalo P, Di Pietro A, Deutscher J, Galinier A (2000) The HPr kinase from Bacillus subtilis is a homo-oligomeric enzyme which exhibits strong positive cooperativity for nucleotide and fructose 1,6-bisphosphate binding. J Biol Chem 275:1773–1780. doi:10.1074/jbc.275.3.1773
Kang SH, Huang J, Lee HN, Hur YA, Cohen SN, Kim ES (2007) Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces. J Bacteriol 189:4315–4319. doi:10.1128/JB.01789-06
Kappler U, Nouwens AS (2013) The molybdoproteome of Starkeya novella—insights into the diversity and functions of molybdenum containing proteins in response to changing growth conditions. Metallomics 5:325–334. doi:10.1039/c2mt20230a
Kieser T, Bibb M, Buttner M, Chater KF, Hopwood D (2000) Practical Streptomyces Genetics. The John Innes Foundation, Norwich
Lee HN, Im JH, Lee MJ, Lee SY, Kim ES (2009) A putative secreted solute binding protein, SCO6569 is a possible AfsR2-dependent down-regulator of actinorhodin biosynthesis in Streptomyces coelicolor. Process Biochem 44:373–377. doi:10.1016/j.procbio.2008.12.002
Licona-Cassani C, Lim S, Marcellin E, Nielsen LK (2014) Temporal dynamics of the Saccharopolyspora erythraea phosphoproteome. Mol Cell Proteomics 13:1219–1230. doi:10.1074/mcp.M113.033951
Manteca A, Alvarez R, Salazar N, Yagüe P, Sanchez J (2008) Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor. Appl Environ Microbiol 74:3877–3886. doi:10.1128/AEM.02715-07
McDonald TS, Tan KN, Hodson MP, Borges K (2014) Alterations of hippocampal glucose metabolism by even versus uneven medium chain triglycerides. J Cereb Blood Flow Metab 34:153–160. doi:10.1038/jcbfm.2013.184
Newton GL, Buchmeier N, Fahey RC (2008) Biosynthesis and functions of mycothiol, the unique protective thiol of actinobacteria. Microbiol Mol Biol Rev 72:471–494. doi:10.1128/MMBR.00008-08
Orellana CA, Marcellin E, Schulz BL, Nouwens AS, Gray PP, Nielsen LK (2015) High-antibody-producing chinese hamster ovary cells up-regulate intracellular protein transport and glutathione synthesis. J Proteome Res 14:609–618. doi:10.1021/pr501027c
Park J-H, Cha C-J, Roe J-H (2006) Identification of genes for mycothiol biosynthesis in Streptomyces coelicolor A3(2. J Microbiol 44:121–125
Ramos I, Guzman S, Escalante L, Imriskova I, Rodriguez-Sanoja R, Sanchez S, Langley E (2004) Glucose kinase alone cannot be responsible for carbon source regulation in Streptomyces peucetius var. caesius. Res Microbiol 155:267–274. doi:10.1016/j.resmic.2004.01.004
Rico S, Santamaria RI, Yepes a, Rodriguez H, Laing E, Bucca G, CP S, Diaz M (2014) Deciphering the regulon of Streptomyces coelicolor AbrC3, a positive response regulator of antibiotic production. Appl Environ Microbiol 80:2417–2428. doi:10.1128/AEM.03378-13
Rodríguez E, Banchio C, Diacovich L, Bibb MJ, Gramajo H (2001) Role of an essential acyl coenzyme A carboxylase in the primary and secondary metabolism of Streptomyces coelicolor A3 (2). Appl Environ Microbiol 67:4166–4176. doi:10.1128/AEM.67.9.4166
Roychoudhury A, Bieker A, Häussinger D, Oesterhelt F (2013) Membrane protein stability depends on the concentration of compatible solutes—a single molecule force spectroscopic study. Biol Chem 394:1465–1474. doi:10.1515/hsz-2013-0173
Ruiz B, Chávez A, Forero A, García-Huante Y, Romero A, Sánchez M, Rocha D, Sánchez B, Rodríguez-Sanoja R, Langley E, Sánchez S (2010) Production of secondary metabolites: regulation by the carbon source. Crit Rev Microbiol 36:146–167
Sánchez B, Rodríguez-Sanoja R, Langley E, Sánchez S (2010) Production of secondary metabolites: regulation by the carbon source. Crit Rev Microbiol 36:146–167
Seno ET, Chater KF (1983) Glycerol catabolic enzymes and their regulation in wild-type and mutant strains of Streptomyces coelicolor A3(2). J Gen Microbiol 129:1403–1413
Smyth G (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:1–25
Smyth G (2005) Bioinformatics and computational biology solutions using R and Bioconductor. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S (eds) LIMMA: linear models for microarray data. Springer, New york, pp. 397–420
Taguchi T, Okamoto S, Lezhava A, Li A, Ochi K, Ebizuka Y, Ichinose K (2007) Possible involvement of ActVI-ORFA in transcriptional regulation of actVI tailoring-step genes for actinorhodin biosynthesis. FEMS Microbiol Lett 269:234–239. doi:10.1111/j.1574-6968.2007.00627.x
Tsao S-W, Rudd BAM, He X-G, Chang C-J, Floss HG (1985) Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J Antibiot (Tokyo) 38:128–131. doi:10.7164/antibiotics.38.128
Turatsinze J-V, Thomas-Chollier M, Defrance M, van Helden J (2008) Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat Protoc 3:1578–1588. doi:10.1038/nprot.2008.97
Wang W, Shu D, Chen L, Jiang W, Lu Y (2009) Cross-talk between an orphan response regulator and a noncognate histidine kinase in Streptomyces coelicolor. FEMS Microbiol Lett 294:150–156. doi:10.1111/j.1574-6968.2009.01563.x
Weiss B (2006) Evidence for mutagenesis by nitric oxide during nitrate metabolism in Escherichia coli. J Bacteriol 188:829–833. doi:10.1128/JB.188.3.829-833.2006
Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. doi:10.1038/nmeth.1322
Acknowledgments
The authors thank Dr. Rosa M. Gutiérrez-Ríos and Dr. Luis Servín-González for critical reading of the manuscript. We thank Jesús Villegas, Abel Blancas, Marco A. Ortíz, and Daniel Vazquez for technical and bioinformatic support. Finally, we would like to acknowledge Dr. Mark Hudson and Dr. Amanda S. Nouwens for technical support in the proteomic experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This work was funded by grants from DGAPA, PAPIIT, UNAM IN201413, and CONACYT CB 219686 to Sergio Sánchez. Funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication. This work did not involve experiments on humans or animals. The authors declare the absence of financial or non-financial competing interest. The authors alone are responsible for the content of this article. All authors have read and approved the submission.
Electronic supplementary material
Table S1
(PDF 464 kb)
Rights and permissions
About this article
Cite this article
H., T.V., Cuauhtemoc, LC., Nidia, MC. et al. Deletion of the hypothetical protein SCO2127 of Streptomyces coelicolor allowed identification of a new regulator of actinorhodin production. Appl Microbiol Biotechnol 100, 9229–9237 (2016). https://doi.org/10.1007/s00253-016-7811-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7811-2