Abstract
Spirolaxine is a natural product isolated from Sporotrichum laxum ATCC 15155, which has shown a variety of biological activities including promising anti-Helicobacter pylori property. To understand how this compound is biosynthesized, the genome of S. laxum was sequenced. Analysis of the genome sequence revealed two putative type III polyketide synthase (PKS) genes in this strain, Sl-pks1 and Sl-pks2, which are located adjacent to each other (~2.0 kb apart) in a tail-to-tail arrangement. Disruption of these two genes revealed that Sl-PKS2 is the dedicated PKS involved in the biosynthesis of spirolaxine. The intron-free Sl-pks2 gene was amplified from the cDNA of S. laxum and ligated into the expression vector pET28a for expression in Escherichia coli BL21-CodonPlus (DE3)-RIL. The major products of Sl-PKS2 in E. coli were characterized as alkylresorcinols that contain a C13–C17 saturated or unsaturated hydrocarbon side chain based on the spectral data. This enzyme was purified and reacted with malonyl-CoA and a series of fatty acyl-SNACs (C6–C10). Corresponding alkylresorcinols were formed from the decarboxylation of the synthesized tetraketide resorcylic acids, together with fatty acyl-primed triketide and tetraketide pyrones as byproducts. This work provides important information about the PKS involved in the biosynthesis of spirolaxine, which will facilitate further understanding and engineering of the biosynthetic pathway of this medicinally important molecule.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
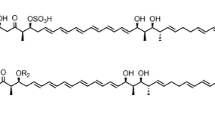
Fungi produce numerous structurally and functionally diverse natural products. Some fungal metabolites have been developed into therapeutics such as penicillin (antibacterial) and lovastatin (anti-cholesterol), while others have shown interesting biological activities and may represent promising lead compounds for new drug development. Spirolaxine (1, Fig. 1) is a natural product originally discovered from Sporotrichum laxum ATCC 15155 (Basidiomycetes) as a plant growth inhibitor (Arnone et al. 1990). It was later found to possess significant anti-Helicobacter pylori activity, representing a potential drug candidate for the treatment of H. pylori-related diseases (Blaser 1992). 1 was also reported to exhibit cholesterol-lowering activity (Robinson and Brimble 2007). A more recent research reported that 1 has inhibitory activities toward endothelial cells (BMEC and HUVEC) and a variety of tumor cell lines (i.e., LoVO and HL60) (Dimitrov et al. 2012; Giannini et al. 2004; Tsukamoto et al. 1998). Besides 1, two other 5,5-spiroacetal phthalides, CJ-12,954 (2, Fig. 1) and CJ-13,014 (3, Fig. 1), were also isolated from the cultures of the Basidiomycetae Phanerochaete velutina. All these molecules showed anti-H. pylori activity (Dekker et al. 1997), while 1 showed twice as much potency as the other two spiroacetal compounds 2 and 3.
1 belongs to a group of interesting secondary metabolites that feature a unique 3-methoxy-5-hydroxy-phthalide nucleus linked to a 6,5-spiroacetal group through a five-membered methylene chain. Owing to the interesting biological activities and structural characteristics, the structures of these compounds were determined and many efforts were devoted to the development of a chiral synthetic route of 1 and its derivatives (Bava et al. 2005; Wang et al. 2013). However, it remains unknown how 1 is assembled by particular biosynthetic enzymes in the producing host. In our previous research, we proposed that a linear polyketide intermediate synthesized by a highly reducing polyketide synthase (hrPKS) is taken by a non-reducing polyketide synthase (nrPKS) as the starter unit and extended through three rounds of decarboxylative condensation of malonyl-CoA. The polyketide backbone undergoes subsequent C-2/C-7 and C-1/C-8 cyclizations. The formed phthalide core structure is then released from the enzyme and further modified by a series of tailoring enzymes to yield 1 (Wang et al. 2013). This proposed route is similar to the formation of the family of fungal benzenediol lactone (BDL) polyketides such as radicicol and dehydrocurvularin (Xu et al. 2014a; Xu et al. 2014b; Xu et al. 2013a; Xu et al. 2013b).
On the other hand, fungal type III PKSs are found to synthesize akylresorcinols (Seshime et al. 2005), which may also serve as the biosynthetic precursor of 1 for a series of tailoring reactions. Filamentous fungi are a known source of natural products with wide applications in agriculture, medicine, industry, and nutrition. Since the discovery of the first fungal type III PKS ORAS from Neurospora crassa (Funa et al. 2007), several fungal type III PKSs have been reported. For example, Aspergillus niger AnPKS (Li et al. 2011) catalyzes the synthesis of pentaketide alkylresorcylic acids, while BPKS from Botrytis cinerea accepts C4–C18 aliphatic acyl-CoAs and benzoyl-CoA as the starters to form pyrones, resorcylic acids, and resorcinols through sequential condensation with malonyl-CoA (Jeya et al. 2012). CsyA from Aspergillus oryzae synthesizes triketide and tetraketide pyrones (Yu et al. 2010), and CsyB from the same strain takes a starter fatty acyl-CoA, malonyl-CoA, and acetoacetyl-CoA to synthesize 3-acetyl-4-hydroxy-6-alkyl-α-pyrone (Hashimoto et al. 2014).
To understand the biosynthetic process of 1, the genomic DNA of S. laxum ATCC 15155 was sequenced in this work. However, analysis of the genomic sequence did not yield any pair of hrPKS and nrPKS that are proposed to synthesize 1. Instead, we found two putative type III PKS genes, Sl-pks1 (GenBank accession no. KU560626) and Sl-pks2 (GenBank accession no. KU560627), which are located adjacent to each other in a tail-to-tail arrangement. Knockout of Sl-pks1 has no effects on spirolaxine biosynthesis, whereas disruption of Sl-pks2 abolished the production of 1, which thus revealed that Sl-PKS2 is a dedicated PKS involved in the biosynthesis of 1. This enzyme was functionally characterized as an alkylresorcinol synthase through in vitro enzymatic reactions and isolation of in vivo products from the heterologous host Escherichia coli.
Materials and methods
General method
Polyketide products were detected and isolated on an Agilent 1200 high performance liquid chromatography (HPLC) instrument. Electrospray ionization mass spectrometry (ESI-MS) spectra were collected on an Agilent 6130 Single quadrupole LC-MS. Nuclear magnetic resonance (NMR) spectra were acquired in CDCl3 on a JEOL NMR instrument (300 MHz for 1H NMR and 75 MHz for 13C NMR). The chemical shift (δ) values were given in parts per million (ppm). The coupling constants (J values) were reported in Hertz (Hz).
Strains and materials
DNA manipulations were performed using standard techniques with Escherichia coli XL1-Blue as a host organism. Expression vector pET28a was purchased from Novagen (Madison, WI, USA). E. coli BL21-CodonPlus (DE3)-RIL was purchased from Stratagene (La Jolla, CA, USA) as the expression host. S. laxum strain ATCC 15155 was obtained from the American Type Culture Collection. Malonyl-CoA was purchased from Sigma–Aldrich (St. Louis, MO, USA). Straight-chain saturated fatty acyl-SNACs (N-acetylcysteamine thioesters) were synthesized as previously described (Yu et al. 2010).
Extraction and sequencing of genomic DNA
S. laxum was cultured in 50 mL of potato dextrose broth (PDB) at 28 °C for 4 days, and the genomic DNA was extracted using the previously reported method (Yu et al. 2010). The genome was then sequenced using an Illumina MiSeq desktop sequencer and assembled with the short read de novo assembler Velvet. Augustus was used for gene prediction (Stanke and Morgenstern 2005), and BLASTX was used for annotation. FramePlot (Ishikawa and Hotta 1999) was used to analyze the predicted PKS genes, with Phanerochaete chrysosporium transcriptome as the reference. Two putative type III PKS genes, Sl-pks1 (GenBank accession no. KU560626) and Sl-pks2 (GenBank accession no. KU560627), were found in the sequence data.
Knockout of Sl-pks2 and Sl-pks1
The genomic DNA of S. laxum was used as the template to amplify a 2.7-kb fragment (left arm) upstream of Sl-pks2 with a set of specific primers Sl-PKS2-ko-la-F and Sl-PKS2-ko-la-R (Table 1). Similarly, a 3.0-kb downstream fragment (right arm) was amplified with another set of primers Sl-PKS2-ko-ra-F and Sl-PKS2-ko-ra-R (Table 1). The PCR products were first ligated to the cloning vector pJET1.2 to yield pSUN56 and pSUN55, respectively (Table 2). After the plasmids were sequenced, the inserts were subsequently cloned into the pAg1-H3 vector to yield pSUN63 (Table 2).
To disrupt Sl-pks1, the two arms (3.0 and 3.5 kb) were amplified with two sets of primers including Sl-PKS1-ko-la-F, Sl-PKS1-ko-la-R, Sl-PKS1-ko-ra-F, and Sl-PKS1-ko-ra-R (Table 1). Ligation of the two arms into pJET1.2 yielded pSUN106 and pSUN107, respectively (Table 2). The disruption plasmid pSUN110 (Table 2) was obtained after the two arms were cloned into pAg1-H3.
The two disruption plasmids, pSUN63 and pSUN110, were respectively introduced into Agrobacterium tumefaciens LBA4404 strain by electroporation, which carries the Ti plasmid pAL4404. This plasmid only has the vir and ori regions of the Ti plasmid. Electroporation conditions were 25 μF, 200 Ω, and 2.5 kV (0.2 cm cuvettes) on a Gene Pulser® electroporator (Bio-Rad, USA).
A. tumefaciens LBA4404 cells containing the T-DNA binary vectors were grown overnight at 28 °C in YM broth (yeast extract 0.4 g L−1, mannitol 10 g L−1, NaCl 0.1 g L−1, MgSO4 0.1 g L−1, K2HPO4·3H2O 0.5 g L−1, pH 7.0) containing 50 μg mL−1 kanamycin. Subsequently, 1 mL of the culture was centrifuged at 3500 rpm for 5 min. The pelleted cells were washed twice with 1 mL of induction medium (IM) (K2HPO4 3 g L−1, NaH2PO4 1 g L−1, (NH4)2SO4 0.5 g L−1, MgSO4·7H2O 0.3 g L−1, KCl 0.15 g L−1, CaCl2 0.005 g L−1, FeSO4·7H2O 0.0025 g L−1, glucose 10 mM, 2-[N-morpholino]ethanesulphonic acid (MES) 40 mM, glycerol 0.5 % (w/v), pH 5.3), inoculated in 50 mL of fresh IM, and grown for an additional 5 h at 28 °C. The final OD600 value of the cultures should be around 0.5. Before co-cultivation, the cells were washed twice as described above with an equal volume of sterile 0.85 % NaCl (w/v) solution and re-suspended in 1 mL of 0.85 % NaCl.
S. laxum was incubated on potato dextrose agar plates at 28 °C for 5 days to allow spore germination. Spores were washed with sterile 0.85 % NaCl (w/v) solution and re-suspended with 1 mL of above-prepared A. tumefasciens cells. The mixture was inoculated onto sterile cellulose films and applied over IM-agar plates containing 500 nM acetosyringone (AS) and incubated at 25 °C for 24 h. After co-cultivation, the films that have both S. laxum and A. tumefasciens were transferred to a new batch of IM-agar plates containing 250 μg mL−1 hygromycin B and 100 μg mL−1 ampicillin for colony selection and inhibition of A. tumefaciens LBA4404. The plates were incubated at 28 °C for 7 days until single colonies appeared.
To confirm the knockout of gene Sl-pks2 in S. laxum, we extracted the genomic DNA of S. laxum mutants with ZR Fungal/Bacterial DNA MiniPrep™ (ZYMO Research, CA, USA). Two pairs of check primers, including the genome-specific primers Sl-PKS2-ko-la-check-F and Sl-PKS2-ko-ra-check-R as well as the vector-specific primers HYG-check-F and HYG-check-R (from the HYG gene) (Table 1), were used to amplify two specific fragments from the mutant. For analyzing the deletion of Sl-pks1, the genome-specific primers Sl-PKS1-la-check-F and Sl-PKS1-ra-check-R were used with the two vector-specific primers (Table 1).
Amplification of intron-free Sl-pks2 from the cDNA of S. laxum and construction of pET28a-Sl-pks2
Wild-type S. laxum was grown in potato dextrose broth at 28 °C for 3 days. The cells were harvested by filtration, from which the RNA was extracted with TRIzol Reagent (Invitrogen, USA). The extracted RNA was purified with a Direct-zol RNA MiniPrep kit (Zymo Research, CA, USA). The cDNA was synthesized from total RNA with M-MuLV Reverse Transcriptase (NEB, MA, USA). With the cDNA as a template, a 1227-bp DNA fragment of the intron-free Sl-pks2 gene was amplified by PCR with a set of specific primers including Sl-PKS2-F (5′-CGGGATCCATGTCTCCCGCAAAGCTC-3′, with a BamHI site shown by underlining) and Sl-PKS2-R (5′-CGGAATTCTTACCCCGAGTCAGAGGAACC-3′, with an EcoRI site shown by underlining). The resulting intron-free fragment was cloned into the pJET1.2 cloning vector and subsequently cloned into pET28a vector between the BamHI and EcoRI sites, resulting in pSUN26.
In vivo polyketide biosynthesis
E. coli BL21-CodonPlus (DE3)-RIL cells harboring pET28a-Sl-pks2 were grown in Luria–Bertani (LB) broth containing 50 μg mL−1 kanamycin and 50 μg mL−1 chloramphenicol at 37 °C and 250 rpm. Until the OD600 reached 0.6, 200 μM isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to induce the overexpression of Sl-PKS2. The induced broth was maintained at 28 °C and 250 rpm for an additional 24 h. The cells were harvested by centrifugation at 3500 rpm for 5 min. Polyketide products were extracted from both the supernatant with ethyl acetate and the pellet with methanol. The organic extracts were combined and concentrated in rotator vapor to an oily residue. The extract was analyzed on an Agilent 1200 HPLC instrument (Agilent Eclipse Plus C18, 5 μm, 4.6 mm × 250 mm), eluted with a linear gradient of 75 to 90 % (v/v) acetonitrile-water (containing 0.1 % trifluoroacetic acid) over 45 min at a flow rate of 1 mL min−1. The products were detected at 280 nm.
Purification and structural characterization of alkylresorcinols 5–9
To isolate compounds 5–9 for structural characterization, the culture was scaled up to 1 L and grown for 72 h. Briefly, 1 mL of seed culture (OD600 = 1.0) of E. coli BL21-CodonPlus (DE3)-RIL/pET28a-Sl-pks2 was inoculated into 1 L of LB medium containing 50 μg mL−1 kanamycin and chloramphenicol and grown to OD600 of 0.6. Two hundred micromolars IPTG was then added to induce protein expression. After fermentation, the culture was extracted as described above. To isolate these compounds, a typical purification procedure was given below. The crude extract was fractionated on a silica gel-60 column, eluted with a stepwise gradient of ethyl acetate-hexane (0:100, 1:99, 2:98, 3:97, 5:95, 10:90, 15:85, 20:80 v/v, each 250 mL) to afford 8 fractions. The fractions containing the target compounds were further separated by reverse-phased HPLC (Agilent Eclipse Plus C18, 5 μm, 4.6 mm × 250 mm), eluted with acetonitrile-water at a flow rate of 1 mL min−1. Compound 5 was found to be in fraction 6 after silica gel column chromatography. This fraction was further separated on HPLC with 67 % acetonitrile-water. The peak at 23.7 min was collected to yield 5.1 mg of 5 in pure form. Compounds 6 and 7 were found to be in fraction 5 after silica gel column chromatography. This fraction was further separated on HPLC with 74 % acetonitrile-water. The peaks at 32.4 min and 35.6 min were collected to yield 6.4 mg of 6 and 8.1 mg of 7, respectively, in pure form. Compounds 8 and 9 were found to be in fraction 3 after silica gel column chromatography. This fraction was further separated on HPLC with 82 % acetonitrile-water. The peak at 47.9 and 50.5 min were collected to yield 10.1 mg of 8 and 9.4 mg of 9, respectively.
Compound 5
ESI-MS (+): [M + H]+ m/z 291.2; lH NMR (300 MHz, CDC13): δ 6.24 (2H, d, J = 1.8 Hz, H-4 and H-6), 6.19 (1H, t, J = 1.8 Hz, H-2), 5.34–5.36 (2H, m, H-6′ and H-7′), 2.49 (2H, t, J = 7.5 Hz, H-1′), 2.01 (4H, m, H-5′ and H-8′), 1.57 (2H, m, H-2′), 1.31 (approx. 12H, m), 0.89 (3H. t, J = 6.9 Hz, H-13′). 13C NMR (75 MHz, CDCl3): δ 156.8 (C-1 and C-3), 146.3 (C-5), 130.0 and 130.2 (C-6′ and C-7′), 108.2 (C-4 ad C-6), 100.4 (C-2), 36.0 (C-l′), 32.0, 31.3 (C-2′), 30.0–29.2 (m), 27.4, 22.9, 14.3 (C-13′).
Compound 6
ESI-MS (+): [M + H]+ m/z 293.2; lH NMR (300 MHz, CDC13): δ 6.25 (2H, d, J = 1.8 Hz, H-4 and H-6), 6.20 (1H, t, J = 1.8 Hz, H-2), 2.47 (2H, t, J = 7.5 Hz, H-1′), 1.55 (2H, m, H-2′), 1.31 (approx. 20H, brs), 0.89 (3H. t, J = 6.9 Hz, H-15′). 13C NMR (75 MHz, CDCl3): δ 156.8 (C-1 and C-3), 146.4 (C-5), 108.2 (C-4 and 6), 100.3 (C-2), 36.0 (C-l′), 32.1, 31.3 (C-2′), 29.9–29.5 (m), 27.4, 22.9 (s), 14.3 (C-13′).
Compound 7
ESI-MS (+): [M + H]+ m/z 319.2, [M + Na]+ m/z 341.2; lH NMR (300 MHz, CDC13): δ 6.25 (2H, d, J = 1.8 Hz, H-4 and H-6), 6.20 (1H, t, J = 1.8 Hz, H-2), 5.34–5.36 (2H, m, H-8′ and H-9′), 2.48 (2H, t, J = 7.5 Hz, H-1′), 2.03 (4H, m, H-7′ and H-10′), 1.56 (2H, m, H-2′), 1.29 (approx. 16H, brs), 0.88 (3H, t, J = 6.9 Hz, H-15′). 13C NMR (75 MHz, CDCl3): δ 156.9 (C-1 and C-3), 146.4 (C-5), 130.2 (C-8′ and C-9′), 108.3 (C-4 and C-6), 100.4 (C-2), 36.1 (C-l′), 32.2, 32.3 (C-2′), 30.0–29.3 (m), 27.4, 22.9, 14.3 (C-15′).
Compound 8
ESI-MS (+): [M + H]+ m/z 321.2, [M + Na]+ m/z 343.2; lH NMR (300 MHz, CDC13): δ 6.25 (2H, d, J = 1.8 Hz, H-4 and H-6), 6.20 (1H, t, J = 1.8 Hz, H-2), 2.48 (2H, t, J = 7.5 Hz, H-1′), 1.58 (2H, m, H-2′), 1.29 (approx. 24H, brs), 0.89 (3H, t, J = 6.9 Hz, H-15′). 13C NMR (75 MHz, CDCl3): δ 157.0 (C-1 and C-3), 146.3 (C-5), 108.1 (C-4 and C-6), 100.3 (C-2), 36.1 (C-l′), 32.1, 32.0 (C-2′), 29.9–29.5 (m), 27.4, 22.9, 14.3 (C-15′).
Compound 9
ESI-MS (+): [M + H]+ m/z 347.0; lH NMR (300 MHz, CDC13): δ 6.25 (2H, d, J = 1.8 Hz, H-4 and H-6), 6.20 (1H, t, J = 1.8 Hz, H-2), 5.35–5.36 (2H, m, H-10′ and H-11′), 2.49 (2H, t, J = 7.5 Hz, H-1′), 2.03 (4H, m, H-9′ and H-12′), 1.57 (2H, m, H-2′), 1.29 (approx. 20H, brs), 0.88 (3H, t, J = 6.9 Hz, H-17′). 13C NMR (75 MHz, CDCl3): δ 156.9 (C-1 and C-3), 146.3 (C-5), 130.2 and 130.0 (C-10′ and C-11′), 108.2 (C-4 and C-6), 100.4 (C-2), 36.0 (C-l′), 32.0, 31.3 (C-2′), 30.0–29.2 (m), 27.4, 22.9, 14.3 (C-17′).
Purification of His6-tagged Sl-PKS2
For production of His6-tagged Sl-PKS2, E. coli BL21-CodonPlus (DE3)-RIL harboring pET28a-Sl-pks2 was grown at 18 °C in Luria–Bertani (LB) broth containing 50 μg mL−1 kanamycin and 50 μg mL−1 chloramphenicol. When the OD600 reached 0.6, 200 μM IPTG was added to induce the expression of Sl-PKS2. The induced culture was maintained at 28 °C for an additional 16 h. Cells were harvested by centrifugation, resuspended in the lysis buffer composed of 20 mM Tris–HCl (pH 7.9) and 0.5 M NaCl, and disrupted by sonication. A crude cell-lysate was prepared by centrifugation at 13,000 rpm at 4 °C for 15 min to remove cell debris. N-His6-tagged Sl-PKS2 was purified using nickel–nitrilotriacetic acid (Ni–NTA) agarose (Qiagen, Valencia, CA, USA) according to the manual from the manufacturer. The purified His6-tagged protein was concentrated and desalted against buffer A [50 mM Tris–HCl (pH 7.9), 2 mM ethylenediaminetetraacetic acid (EDTA) and 1 mM dithiothreitol (DTT)] with a centrifugal filter device (Millipore, Billerica, MA, USA).
PKS assay
A typical reaction mixture contained 200 μM malonyl-CoA, 100 μM acyl-SNAC, 100 mM phosphate buffer (pH 7.0), and 10 μg of Sl-PKS2 in a total volume of 100 μL. Reaction was incubated at 30 °C for 60 min before being quenched with 50 μL of methanol. The samples were centrifuged at 15,000 rpm for 5 min and the products in the supernatant were analyzed on an Agilent 1200 HPLC instrument (Agilent Eclipse Plus C18, 5 μm, 4.6 mm × 250 mm). The samples were eluted with a linear gradient of 30 to 90 % (v/v) acetonitrile-water (containing 0.1 % trifluoroacetic acid) over 45 min at a flow rate of 1 mL min−1, and the compounds were detected at 280 nm. The identity of the synthesized products was determined by the comparison of the retention times and UV absorptions with those of the products previously reported (Yu et al. 2010), which was further confirmed by the mass spectra measured on an Agilent 6130 quadrupole LC-MS.
Results
Discovery and analysis of two putative type III PKS genes from S. laxum
S. laxum ATCC 15155 is a producer of 1. The genome of this strain was sequenced. A total of 11,347 proteins were predicted by Augustus, which were compared with the NCBI nonredundant (NR) protein databases by BLASTX to predict their biological functions. The sequenced data were further analyzed using a gene annotation tool FramePlot with P. chrysosporium transcriptome as the reference. Two putative chalcone synthase (CHS)-like type III PKS genes, Sl-pks1 and Sl-pks2, are arranged tail to tail on the genome, separated only by a region of ~2.0 kb. BLAST analysis of the primary amino acid sequences of the deduced proteins yielded a lot of homologs from the CHS superfamily enzymes, although the closest homologs to Sl-PKS1 and Sl-PKS2 are two unidentified proteins (XP_007391993.1, 85 % identity and XP_007391992.1, 72 % identity) from Phanerochaete carnosa HHB-10118-sp. Sequence alignment of Sl-PKSs with N. crassa ORAS (Funa et al. 2007) and A. oryzae CsyA revealed that both S. laxum Sl-PKSs maintain an almost identical CoA binding site with the reported type III PKSs and a conserved catalytic triad of Cys-158, His-312, and Asn-345 (numbering in Sl-PKS2) (Fig. S1).
Knockout of Sl-pks1 and Sl-pks2 from S. laxum ATCC 15155
To determine whether Sl-PKS1 and Sl-PKS2 are involved in spirolaxine biosynthesis, targeted gene knockout was performed, respectively. A disruption vector pAg1-H3 (Fig. 2a) containing a hygromycin phosphotransferase B gene (HYG) cassette flanked with the 3.0-kb left arm and 3.5-kb right arm for Sl-pks1 knockout. The left and right arms were gene fragments upstream and downstream of the target gene on the genome of S. laxum. Similarly, a 2.7-kb left arm and a 3.0-kb right arm are used for the knockout of Sl-pks2. The products of wild-type S. laxum and the two mutants were analyzed by HPLC. As shown in Fig. 2b, the wild type (trace I) produced two major compounds on WSH plates including 1 at 34.1 min and its analog sporotricale (4, Fig. 1) at 24.2 min. Knockout of Sl-pks1 did not affect the product profile and the two major products were still generated (trace II), suggesting that this gene is not involved in the biosynthesis of these compounds. In contrast, the Sl-pks2 knockout mutant of S. laxum failed to produce these compounds (trace III). Genomic DNA of the knockout mutants was extracted and used as the template for PCR amplification to check whether the target genes were deleted. Genome- and vector-specific primers (Table 1) were used. For the S. laxum-ΔSl-pks2 mutant, a 3.0-kb fragment (lane 1, Fig. 2c) was amplified from the upstream region of the disruption site using the genome-specific primer Sl-PKS2-la-check-F and the vector-specific primer HYG-check-R. Similarly, a downstream fragment (4.2 kb, lane 2, Fig. 2c) was obtained using HYG-check-F and Sl-PKS2-ra-check-R. No products were amplified from the genome of the wild type (lanes 3 and 4, Fig. 2c), which confirmed that the loss of spirolaxine production was a consequence of the targeted gene replacement of Sl-pks2. Knockout of Sl-pks1 was verified in the same way. Two fragments (4.4 kb, lane 5, and 3.3 kb, lane 6, Fig. 2c) were amplified from the mutant, but not the wild type, further confirming that Sl-PKS1 is not involved in spirolaxine biosynthesis. Thus, Sl-PKS2 was characterized as a dedicated PKS involved in the biosynthesis of 1.
Gene disruption of Sl-pks1 and Sl-pks2. a Double crossover strategy to knock out Sl-pks1 and Sl-pks2. pAG-H3 and hygromycin B were used as the vector and selection marker, respectively. b HPLC analysis of the products of wild type S. laxum (I), S. laxum-ΔSl-pks1 (II) and S. laxum-ΔSl-pks2 (III). c Verification of the correct knockout mutants. Genome-specific check primers (Sl-PKS1-ko-la-check-F, Sl-PKS1-ko-ra-check-R, Sl-PKS2-ko-la-check-F and Sl-PKS2-ko-ra-check-R) and vector-specific primers HYG-check-F and HYG-check-R were used to amplify particular fragments from the mutants, while no products were obtained from the wild type using the same sets of primers. M: 1 kb Plus DNA ladder; 1: a 3.0-kb fragment upstream of the disruption site amplified from S. laxum-ΔSl-pks2 with Sl-PKS2-ko-la-check-F and HYG-check-R; 2: a 4.2-kb fragment downstream of the disruption site amplified from S. laxum-ΔSl-pks2 with HYG-check-F and Sl-PKS2-ko-ra-check-R; 3: no product from wild-type S. laxum with Sl-PKS2-ko-la-check-F and HYG-check-R; 4: no product from wild-type S. laxum with HYG-check-F and Sl-PKS2-ko-ra-check-R; 5: a 4.4-kb fragment upstream of the disruption site amplified from S. laxum-ΔSl-pks1 with Sl-PKS1-ko-la-check-F and HYG-check-R; 6: a 3.3-kb fragment downstream the disruption site amplified from S. laxum-ΔSl-pks1 with HYG-check-F and Sl-PKS1-ko-ra-check-R; 7: no product from wild-type S. laxum with Sl-PKS1-ko-la-check-F and HYG-check-R; 8: no product from wild type S. laxum with HYG-check-F and Sl-PKS1-ko-ra-check-R
Characterization of the products synthesized by Sl-PKS2 in E. coli
Having investigated the Sl-pks2 knockout strain, we further examined whether Sl-PKS2 can be expressed in a heterologous host such as E. coli to produce corresponding polyketide products. Fatty acyl-CoAs or acyl-ACPs, naturally present in E. coli, are the intermediates in fatty acid biosynthesis or metabolism. In our previous research, A. oryzae type III PKS CsyA was found to take these starter units to synthesize alkylpyrones in E. coli (Yu et al. 2010). In E. coli, fatty acids are assembled by fatty acid synthases from acetyl-CoA and malonyl-CoA. These fatty acyl-ACPs or acyl-CoAs could be accepted as starters for condensation reactions catalyzed by PKSs with abroad starter flexibility, such as the type I PKS Gibberella zeae PKS13 (Zhou et al. 2008) and type III PKS CsyB (Hashimoto et al. 2014). We have previously isolated 1 and analogs as well as alkylresorcylic acids from S. laxum. These compounds feature a C16 or C18 fatty acyl side chain attached to a benzene ring (Wang et al. 2013). Thus, we hypothesized that Sl-PKS2 can accept various acyl-ACPs or acyl-CoAs as starter units that are available in E. coli (White et al. 2005). We amplified the intron-free Sl-pks2 (1227 bp) gene from the cDNA of S. laxum (Fig. 3a), and ligated the gene with the expression vector pET28a to yield pSUN26, which was used to transform E. coli BL21-CodonPlus (DE3)-RIL. The IPTG-induced broth was extracted with an equal volume of ethyl acetate. As shown in Fig. 3b, HPLC analysis revealed that five major products (5–9, trace I) were synthesized by Sl-PKS2, which were not present in E. coli BL21-CodonPlus (DE3)-RIL-pET28a (trace II) that was used as negative control.
Expression of intron-free Sl-pks2 in E. coli BL21-CodonPlus (DE3)-RIL and analysis of the in vivo polyketide products. a Amplification of the Sl-pks2 gene from the cDNA and genomic DNA. M: 1 kb Plus DNA ladder, 1: intron-free Sl-pks2 (1227 bp); 2: intron-containing Sl-pks2 (1348 bp). b HPLC analysis of the products of ethyl acetate extracts of the cultures of E. coli BL21-CodonPlus (DE3)-RIL-pSUN26 (trace I) and E. coli BL21-CodonPlus (DE3)-RIL-pET28a (trace II). Five major products 5–9 were detected. c The structures of alkylresorcinols 5–9
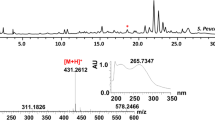
These five products were isolated from the scaled-up culture of E. coli BL21-CodonPlus (DE3)-RIL-pSUN26. Compounds 5–9 were obtained as yellowish oil. They were identified as alkylresorcinols with either an unsaturated or saturated aliphatic side chain (Fig. 3c) based on their UV (maximum UV absorptions at 203, 232 and 278 nm), MS, and NMR spectra and a comparison with previously published data (Suzuki et al. 1997). The molecular weights of 5–9 were determined to be 290, 292, 318, 320, and 346 based on their [M + H]+ ion peaks at m/z 291, 293, 319, 321, and 347 in the ESI-MS, respectively (Fig. S2 in the Supplementary Material). Accordingly, compounds 6 and 8 were deduced to be alkylresorcinols with a saturated side chain, while 5, 7, and 9 have an unsaturated side chain. The 1H and 13C NMR spectra of 6 revealed two aromatic proton signals at δH 6.25 (2H, d, J = 1.8 Hz, H-4 and H-6) and δH 6.20 (1H, t, J = 1.8 Hz, H-2), and four carbon signals at δC 100.3 (C-2), 108.2 (C-4 and C-6), 146.4 (C-5), and 156.8 (C-1 and C-3), suggesting that a resorcinol moiety is present in the structure. In addition, proton signals typical for a long aliphatic chain including the benzylic methylene at δH 2.47 (2H, t, J = 7.5 Hz) and terminal methyl group at δH 0.89 (3H, t, J = 6.9 Hz) were also found. Thus, 6 was identified as 1,3-dihydroxyphenyl-5-tridecane, a resorcinol derivative with a saturated C13 side chain. 8 has highly similar NMR data to those of 6, but has an additional CH2-CH2 unit. Thus, it was characterized as 1,3-dihydroxyphenyl-5-pentadecane that has a saturated C15 side chain. The NMR spectra of 5 also showed the aromatic proton and carbon signals which are similar to 6. However, two overlapping olefinic protons at δH 5.34–5.36 and carbon signals at δC 130.0 indicated that there is a double bond in the side chain. Thus, 5 was characterized as 1,3-dihydroxyphenyl-5-tridecene. Similarly, 7 and 9 were identified as resorcinol derivatives containing an unsaturated C15 and C17 side chain, named 1,3-dihydroxyphenyl-5-pentadecene and 1,3-dihydroxyphenyl-5-heptadecene, respectively. These data were in good agreement with the data reported previously for 5-alk(en)ylresorcinols (Knodler et al. 2008; Suzuki et al. 1997). The thioesters of common fatty acids in E. coli (Mejía et al. 1999; Shaw and Ingraham 1965) are proposed to be used by Sl-PKS2 as the starter units to synthesize 5–9. Specifically, the thioesters of unsaturated fatty acids including tetradecenoic acid (cis-7), hexadecenoic acid (cis-9), and octadecenoic acid (cis-11) are used by Sl-PKS2 to afford 5, 7, and 9, while the thioesters of myristic acid (C14:0) and palmitic acid (C16:0) were used to synthesize 6 and 8, respectively. The position of the double bond in 5, 7, and 9 was proposed to be the same as that in tetradecenoic acid (cis-7), hexadecenoic acid (cis-9), and octadecenoic acid (cis-11) which served as the starter substrates. Accordingly, their structures were deduced to be 1,3-dihydroxyphenyl-5-cis-6′-tridecene, 1,3-dihydroxyphenyl-5-cis-8′-pentadecene and 1,3-dihydroxyphenyl-5-cis-10′-heptadecene, respectively.
Functional characterization of Sl-PKS2 through in vitro reactions
His6-tagged Sl-PKS2 was purified (final yield 15 mg L−1) through Ni–NTA chromatography for the in vitro reactions. The purified protein showed a single protein band at ∼46 kDa on SDS-PAGE (Fig. 4a). In order to test whether fatty acyl-CoAs are appropriate starter units for Sl-PKS2, we synthesized the SNAC esters of C6–C10 fatty acids (C6, C7, C8, C9, and C10) to mimic the corresponding CoA esters. These starter units were respectively reacted with Sl-PKS2, with malonyl-CoA as the extender substrate. HPLC analysis of the reactions revealed that Sl-PKS2 has broad substrate specificity toward SNAC starters containing fatty acyl groups ranging from C6 to C10 (Fig. 4b). For these starter units, Sl-PKS2 yielded three groups of product, and the retention times of the products were consistent with the chain length. As observed in Fig. 4b, the shorter the chain length of the starters, the shorter the retention times of the resulting products on reversed-phase HPLC. The peaks (groups a and b) have maximum UV absorptions at 204 and 286 nm, which is characteristic UV absorbance pattern for pyrones. Through ESI-MS analysis (Fig. S3 in the Supplementary Material) and a comparison with the pyrone compounds we previously reported (Yu et al. 2010), we characterized the products as fatty acyl-primed triketide (group a, 10a-14a, Fig. 4c) and tetraketide pyrones (group b, 10b-14b, Fig. 4c). In contrast, group c compounds (10c-14c, Fig. 4c) showed the same UV spectra as 5–9 we obtained from E. coli, suggesting that they are alkylresorcinols. Further, LC-MS allowed us to determine the length of the side chains in these 5-alkylresorcinols. The pathways leading to the formation of these in vitro products are summarized in Fig. 4c.
Purification of Sl-PKS2 from E. coli BL21-CodonPlus (DE3)-RIL and in vitro reactions. a SDS-PAGE analysis of the purification of Sl-PKS2. M: Protein ladder; 1: cell lysate; 2: purified Sl-PKS2. b HPLC analysis of in vitro reactions of Sl-PKS2 with various acyl-SNACs (C6–C10) and malonyl-CoA. I: hexanoyl-SNAC (C6); II: heptanoyl-SNAC (C7); III: octanoyl-SNAC (C8); IV: nonanoyl-SNAC (C9); V: decanoyl-SNAC (C10). Three groups of product were formed, including triketide pyrones 10a-14a, tetraketide pyrones 10b-14b, and alkylresorcinols 10c-14c. c Summary of the in vitro reactions of Sl-PKS2
Discussion
Type III PKSs are involved in the biosynthesis of many fungal metabolites, such as csypyrone B1 (Hashimoto et al. 2014) and resorcinols (Funa et al. 2007). Based on the functional characterization of Sl-PKS2 and the metabolites we previously isolated from the host (Wang et al. 2013), a type III polyketide biosynthetic pathway was proposed to synthesize 1 (Fig. 5). Palmitoyl-CoA (15) or palmitoyl-ACP (16) was adopted as the starter substrate by a type III PKS (Sl-PKS2), which elongates the chain using three molecules of malonyl-CoA to yield a nascent polyketide chain 17. This intermediate is cyclized via an intramolecular C-2/C-7 aldol condensation. The regioselective C-2/C-7 cyclization by the dedicated type III PKS is observed in the biosynthesis of various natural products (Funa et al. 2007). The polyketide intermediate can be hydrolyzed and released from the enzyme to yield the resorcylic acid derivative 18. Hydroxylation of C-21 yielded 19. A ring is formed between C-1 carboxyl and C-8 to yield the phthalide structure 20, which can be further methylated at 3-OH to afford 21. 21 is subsequently hydroxylated at C-18 to yield 22, which is oxidized and then cyclized to form the methyltetrahydrofuran ring to give rise to 23. A hydroxyl group at C-14 was introduced to afford 24, which is then used to form the tetrahydropyran ring and yield the final product spirolaxine (1). In our previous research, we isolated (R)-3,5-dihydroxy-7-(16-hydroxyheptadecyl)benzoic acid and (R)-3,5-dihydroxy-7-(15-hydroxyheptadecyl)benzoic acid from S. laxum (Wang et al. 2013), which indicated that Sl-PKS2 also can accept stearoyl-CoA (C18:0) as a starter unit. This, together with the in vivo and in vitro results in this work, revealed that Sl-PKS2 has broad substrate specificity toward acyl starter units.
We identified Sl-PKS2 from S. laxum as a new fungal type III PKS which synthesizes fatty acyl-primed resorcylic acid compounds. Our work demonstrated that Sl-PKS2 activities can be completely reconstituted both in vitro and in E. coli. The products of Sl-PKS2 in E. coli are alkylresorcinols, which resulted from decarboxylation of corresponding alkylresorcylic acids (Funa et al. 2007). This supports the proposed biosynthetic pathway of 1 in Fig. 5. Consistently, in vitro reaction of Sl-PKS2 with various fatty acyl-SNACs yielded corresponding alkylresorcinols as major products. Interestingly, triketide and tetraketide pyrones were also found in these in vitro reactions. Pyrones are common byproducts of type III PKSs. The triketide pyrones are formed from the pre-mature intermediates that undergo spontaneous cyclization. Once the nascent polyketide chain reached the full length after three rounds of condensation, it will be cyclized by the enzyme to yield alkylresorcylic acids and subsequently alkylresorcinols. Spontaneous cyclization can lead to the synthesis of tetraketide alkylpyrones. Formation of these pyrone byproducts is likely due to the starter units used in the in vitro reactions. They are shorter than the abundant C16 and C18 fatty acyl-CoAs (or -ACPs) in E. coli and S. laxum that are natural substrates for Sl-PKS2. In addition, protein interactions are common in cells. The environment difference between the E. coli cells and the in vitro reaction buffer may cause the different performance of Sl-PKS2.
5-Alkylresorcinols belong to the large family of natural compounds referred to as phenolic lipids, which are widely distributed in many organisms, such as plants, fungi, and bacteria. They have shown functions in cellular biochemistry, membrane structure, and physiology (Kozubek and Tyman 1999). In this work, we confirmed that Sl-PKS2 is involved in the biosynthesis of the medicinally important compound spirolaxine, which for the first time revealed that this molecule is assembled via a type III, instead of type I biosynthetic pathway. Although Sl-pks1 sits adjacent to Sl-pks2 on the genome, it is not involved in spirolaxine biosynthesis. It was also found that the proposed oxygenase genes are not present in proximity to Sl-pks2 (Table S1), suggesting that the alkylresorcinols synthesized by this enzyme are shared by multiple pathways. In fact, phenolic lipids produced by type III PKSs play a critical role as minor components in the biological membrane in both prokaryotes and eukaryotes (Baerson et al. 2010; Miyanaga and Horinouchi 2010). It is proposed that alkylresorcinols synthesized by Sl-PKS2 is used as phenolic lipids by other biological processes in S. laxum. This explains why Sl-pks2 is not clustered with other spirolaxine biosynthetic genes. Additional work is thus needed to characterize the tailoring enzymes for complete elucidation of the biosynthetic pathway of this anti-H. pylori natural product.
References
Arnone A, Assante G, Nasini G, Vajna de Pava O (1990) Secondary mold metabolites. Part 28. Spirolaxine and sporotricale: two long-chain phthalides produced by Sporotrichum laxum. Phytochemistry 29:613–616
Baerson SR, Schröder J, Cook D, Rimando AM, Pan Z, Dayan FE, Noonan BP, Duke SO (2010) Alkylresorcinol biosynthesis in plants: new insights from an ancient enzyme family? Plant Signal Behav 5:1286–1289
Bava A, Clericuzio M, Giannini G, Malpezzi L, Meille SV, Nasini G (2005) Absolute configuration of the fungal metabolite spirolaxine. Eur J Org Chem 2005(11):2292–2296
Blaser MJ (1992) Helicobacter pylori: its role in disease. Clin Infect Dis 15(3):386–391
Dekker KA, Inagaki T, Gootz TD, Kaneda K, Nomura E, Sakakibara T, Sakemi S, Sugie Y, Yamauchi Y, Yoshikawa N, Kojima N (1997) CJ-12,954 and its congeners, new anti-Helicobacter pylori compounds produced by Phanerochaete velutina: fermentation, isolation, structural elucidation and biological activities. J Antibiot 50(10):833–839
Dimitrov I, Furkert DP, Fraser JD, Radcliff FJ, Finch O, Brimble MA (2012) Synthesis and anti-Helicobacter pylori activity of analogues of spirolaxine methyl ether. MedChemComm 3(8):938–943
Funa N, Awakawa T, Horinouchi S (2007) Pentaketide resorcylic acid synthesis by type III polyketide synthase from Neurospora crassa. J Biol Chem 282(19):14476–14481
Giannini G, Pisano C, Riccioni T, Marcellini M, Chiarucci I, Bava A, Nasini G (2004) Spirolaxine, a fungal metabolite from Sporotrichum laxum: determination of relative stereochemistry and biological activity in vitro. Proc Amer Assoc Cancer Res 45:1011
Hashimoto M, Koen T, Takahashi H, Suda C, Kitamoto K, Fujii I (2014) Aspergillus oryzae CsyB catalyzes the condensation of two beta-ketoacyl-CoAs to form 3-acetyl-4-hydroxy-6-alkyl-alpha-pyrone. J Biol Chem 289(29):19976–19984
Ishikawa J, Hotta K (1999) FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol Lett 174(2):251–253
Jeya M, Kim TS, Tiwari MK, Li J, Zhao H, Lee JK (2012) The Botrytis cinerea type III polyketide synthase shows unprecedented high catalytic efficiency toward long chain acyl-CoAs. Mol BioSyst 8(11):2864–2867
Knodler M, Conrad J, Wenzig EM, Bauer R, Lacorn M, Beifuss U, Carle R, Schieber A (2008) Anti-inflammatory 5-(11’Z-heptadecenyl)- and 5-(8’Z,11’Z-heptadecadienyl)-resorcinols from mango (Mangifera indica L.) peels. Phytochemistry 69(4):988–993
Kozubek A, Tyman JH (1999) Resorcinolic lipids, the natural non-isoprenoid phenolic amphiphiles and their biological activity. Chem Rev 99(1):1–26
Li J, Luo Y, Lee JK, Zhao H (2011) Cloning and characterization of a type III polyketide synthase from Aspergillus niger. Bioorg Med Chem Lett 21(20):6085–6089
Mejía R, Gómez-Eichelmann MC, Fernández MS (1999) Fatty acid profile of Escherichia coli during the heat-shock response. IUBMB Life 47(5):835–844
Miyanaga A, Horinouchi S (2010) Type III polyketide synthases responsible for phenolic lipid synthesis. In: Timmis KN (ed) Handbook of hydrocarbon and lipid microbiology. Springer-Verlag, Berlin, Heidelberg, pp. 519–525
Robinson JE, Brimble MA (2007) Synthesis of the anti-Helicobacter pylori agent (+)-spirolaxine methyl ether and the unnatural (2″S)-diastereomer. Org Biomol Chem 5(16):2572–2582
Seshime Y, Juvvadi PR, Fujii I, Kitamoto K (2005) Discovery of a novel superfamily of type III polyketide synthases in Aspergillus oryzae. Biochem Biophys Res Commun 331(1):253–260
Shaw MK, Ingraham JL (1965) Fatty acid composition of Escherichia coli as a possible controlling factor of the minimal growth temperature. J Bacteriol 90(1):141–146
Stanke M, Morgenstern B (2005) AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33(Web Server):W465-W467
Suzuki Y, Esumi Y, Uramoto M, Kono Y, Sakurai A (1997) Structural analyses of carbon chains in 5-alk(en)ylresorcinols of rye and wheat whole flour by tandem mass spectrometry. Biosci Biotechnol Biochem 61(3):480–486
Tsukamoto M, Sano H, Kadota Y, Ojiri K, Suda H (1998) Anticancer spirolaxine and its fermentative manufacture. JP10007557A,
Wang S, Zhang S, Zhou T, Zhan J (2013) Three new resorcylic acid derivatives from Sporotrichum laxum. Bioorg Med Chem Lett 23(21):5806–5809
White SW, Zheng J, Zhang Y-M, Rock CO (2005) The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem 74:791–831
Xu Y, Zhou T, Espinosa-Artiles P, Tang Y, Zhan J, Molnar I (2014a) Insights into the biosynthesis of 12-membered resorcylic acid lactones from heterologous production in Saccharomyces cerevisiae. ACS Chem Biol 9(5):1119–1127
Xu Y, Zhou T, Zhang S, Espinosa-Artiles P, Wang L, Zhang W, Lin M, Gunatilaka AAL, Zhan J, Molnar I (2014b) Diversity-oriented combinatorial biosynthesis of benzenediol lactone scaffolds by subunit shuffling of fungal polyketide synthases. Proc Natl Acad Sci U S A 111(34):12354–12359
Xu Y, Zhou T, Zhang S, Xuan L-J, Zhan J, Molnar I (2013a) Thioesterase domains of fungal nonreducing polyketide synthases act as decision gates during combinatorial biosynthesis. J Am Chem Soc 135(29):10783–10791
Xu Y, Zhou T, Zhou Z, Su S, Roberts SA, Montfort WR, Zeng J, Chen M, Zhang W, Lin M, Zhan J, Molnar I (2013b) Rational reprogramming of fungal polyketide first-ring cyclization. Proc Natl Acad Sci U S A 110(14):5398–5403
Yu D, Zeng J, Chen D, Zhan J (2010) Characterization and reconstitution of a new fungal type III polyketide synthase from Aspergillus oryzae. Enzyme Microb Tech 46(7):575–580
Zhou H, Zhan J, Watanabe K, Xie X, Tang Y (2008) A polyketide macrolactone synthase from the filamentous fungus Gibberella zeae. Proc Natl Acad Sci U S A 105(17):6249–6254
Acknowledgments
This work was supported by a Grant-In-Aid from the American Heart Association (16GRNT26430067), grants from the National Natural Science Foundation of China (31170763, 31470787), a Hunan Science and Technology Key Project (2014FJ1007), and the Hundred Talents Program of Hunan Province, China.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
ESM 1
(PDF 473 kb)
Rights and permissions
About this article
Cite this article
Sun, L., Wang, S., Zhang, S. et al. Identification of a type III polyketide synthase involved in the biosynthesis of spirolaxine. Appl Microbiol Biotechnol 100, 7103–7113 (2016). https://doi.org/10.1007/s00253-016-7444-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7444-5