Abstract
Enantioselective hydrolysis of racemic epoxides mediated by epoxide hydrolases (EHs) is one of the most promising approaches to obtain enantiopure epoxides. In this study, we identified and characterized a novel EH (TpEH1) from Tsukamurella paurometabola by analyzing the conserved catalytic residues of EH. TpEH1 was overexpressed and purified, and its catalytic properties were studied using racemic phenyl glycidyl ether (PGE) and its derivatives as substrates. TpEH1 showed excellent enantioselectivity to the substrates PGE, 3-methylPGE, and 3-nitroPGE. The highest enantioselectivity (E > 100) was achieved when 3-nitroPGE was used as the substrate. The recombinant Escherichia coli TpEH1 demonstrated high substrate tolerance toward PGE and could hydrolyze PGE at concentrations of up to 400 mM (60 g/L) with high enantioselectivity (E = 65), giving (R)-PGE with enantiomeric excess of more than 99 % ee and 45 % yield within 1 h. This concentration of PGE is the highest reported concentration catalyzed by native EHs to date. Thus, the easily available and highly active E. coli TpEH1 showed great potential for the practical preparation of optically pure (R)-PGE.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epoxides are recognized as important synthons for fine organic synthesis due to their chemical versatility (Kotik et al. 2012). Among known epoxides, phenyl glycidyl ether (PGE) is an important building block widely used for the production of bioactive compounds, such as β-blockers (Bisi et al. 2003), neuroprotective molecules (Pieper et al. 2010), β-secretase-cleaving enzyme (BACE) inhibitors (John et al. 2003), and modulators of toll-like receptor 7 (Basith et al. 2011). The enantioselective hydrolysis of racemic epoxides mediated by epoxide hydrolases (EHs, E.C.3.3.2.3) is a promising approach to obtaining enantiopure epoxides (Lin et al. 2011). Enantioselective EHs selectively hydrolyze one enantiomer of racemic epoxide into the corresponding vicinal diol by adding one molecule of water, leaving the slow reacting enantiomer. Therefore, effective production of optically pure epoxides depends on the substrate building blocks and appropriate EH.
Most EHs are ubiquitously found in nature and have been identified in many organisms, including mammals, plants, insects, and various microorganisms (Bala and Chimni 2010). EHs from Aspergillus niger (Deregnaucourt et al. 2007), Solanum tuberosum (Monterde et al. 2004), Agrobacterium radiobacter AD1 (Rui et al. 2005), Vigna radiata (Zhu et al. 2013), Bacillus megaterium (Zhao et al. 2011), and Sphingomonas sp. HXN-200 (Wu et al. 2013) have been shown to demonstrate great industrial potentiality to produce valuable enantiopure epoxides and vicinal diols. However, the development of highly active, highly enantioselective EHs for the practical production of optically desired epoxides in high enantiomeric excess (ee), high concentration, and high yield is still a substantial challenge. To develop an industrial-scale process, mining of novel EHs with high enantioselectivity in the presence of high substrate concentrations is required.
With the increased availability of public genome information, many putative EHs can be discovered from GenBank. The potential EH activities of these putative EHs could be further confirmed based on the presence of conserved motifs with multiple alignments. Most EHs belong to the α/β hydrolase fold family and have a conserved catalytic unit composed of an α/β hydrolase fold core domain and a variable lid domain at the top. The catalytic center is situated between the core domain and the lid domain with a catalytic triad consisting of a carboxylate nucleophile Asp, a general-base His, and a charge-relay carboxylate (Asp or Glu). In addition, two Tyr residues that provide hydrogen bonding to the epoxide oxygen are always present from the lid pointing toward the catalytic triad. The other two motifs, HGXP and G-X-Sm-X-S/T (Sm = small residue, X = any residue) constituting the oxyanion hole, are also conserved in the α/β hydrolase fold family. Since many species have been sequenced, novel EHs can be identified by performing a search of the genomic databases (Barth et al. 2004; van Loo et al. 2006; Widersten et al. 2010).
In this study, a novel EH (TpEH1) was discovered from Tsukamurella paurometabola DSM20162 based on the analysis of conserved catalytic residues of epoxide hydrolase, and its biochemical properties were studied in detail. The ability of TpEH1 to hydrolyze racemic PGE and its derivatives was analyzed, and the recombinant Escherichia coli TpEH1 was used to produce (R)-PGE at high concentrations and with high enantioselectivity.
Materials and methods
Materials
The genomic DNA of T. paurometabola DSM20162 was stored in our lab. All enzymes used for molecular cloning were from Takara (Dalian, China). The expression vector pET-28a (+) and E. coli strains DH5α and BL21 (DE3) were used for cloning and expressing TpEH1. Racemic PGE, (S)-PGE, (R)-PGE, cyclohexene oxide, styrene oxide, cyclohexanediol, 3-phenoxy-1, 2-propanediol, and 4-chlorophenyl glycidyl ether were purchased from Sigma Aldrich (Milwaukee, WI, USA). The other substrates were synthesized as previously described (Zhang et al. 2010).
Database mining and sequence analysis
Sequences of putative EHs or α/β hydrolases from different bacterial genomes were selected from NCBI (http://www.ncbi.nlm.nih.gov). The candidates were further confirmed for the presence of the conserved motifs and residues of the putative EH. One predicted that α/β hydrolase sequence (accession number YP_003645475) of T. paurometabola DSM20162 was selected and confirmed by multisequence alignment with other reported EHs using DNAMAN.
Cloning of the TpEH1 gene from T. paurometabola DSM20162
The full-length TpEH1 gene flanked by BamHI and HindIII sites was amplified by polymerase chain reaction (PCR) with forward (TpEH1-F, 5′-CGCGGATCCATGACGATCACCCCGCACACCGTG-3′) and reverse primers (TpEH1-R, 5′-CCCAAGCTTTCAGCCCGCGGGGTGGTTCTCC-3′). The obtained DNA fragment was digested and ligated to the corresponding digested plasmid pET-28a (+). The resulting recombinant plasmid was then transformed into BL21 (DE3) for the expression of EH.
Expression and purification of TpEH1
A single transformant was cultured at 37 °C for 12 h and then transferred to 100 ml fresh Luria-Bertani (LB) medium supplemented with kanamycin (50 μg/ml) and cultured at 37 °C. The culture was induced by the addition of 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when the OD600 reached 0.6. After induction at 20 °C for 20 h, cells were harvested at 8500×g for 10 min and resuspended in 50 mM Tris-HCl (pH 8.0). After disrupting the cells by sonication and removing cell debris/inclusion bodies by centrifugation, the soluble cell-free extract was filtered (Millipore filtration/0.22 μm) and loaded onto a nickel column pre-equilibrated with binding buffer (50 mM Tris-HCl, pH 8.0). After being washed with binding buffer, the bound recombinant enzyme was eluted by applying binding buffer with increasing concentrations of imidazole (20–200 mM). Pure TpEH1 could be obtained with 200 mM imidazole. The expression and purity of the protein were identified by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on 12 % gels.
Enzyme activity and protein assay
Epoxide hydrolase activity was determined using PGE as the substrate. One microliter of the enzyme solution (1.8 mg/ml) was added to 450 μl Tris-HCl (pH 8.0) buffer. After pre-incubating the mixture at 30 °C for 5 min, the reaction was started by adding 50 μl of 200 mM stock solution of PGE in DMSO to a total volume of 500 μl. After a 5-min incubation at 30 °C with 200 rpm shaking, the reaction was terminated by adding an equal volume of methanol. The sample was then analyzed by high-performance liquid chromatography (HPLC). One unit of the enzyme activity was defined as the amount of enzyme capable of producing 1 μmol diol/min under the standard assay conditions. The protein concentration was determined by the Bradford method, with bovine serum albumin as a standard.
Determination of kinetic parameters
Specific amounts of pure TpEH1 (0.5 μg for the (S)-enantiomer, 5 μg for the (R)-enantiomer) were added to 950 μl Tris-HCl buffer and pre-incubated for 5 min in 30 °C. The reactions were initiated with final substrate concentrations of 1, 2, 4, 8, or 16 mM, and conversion proceeded to a maximum of 10 % of the PGE. The reaction was terminated with the same volume of methanol. The initial velocities at different substrate concentrations were used for Lineweaver-Burk plots (1/v vs. 1/[S]).
Effects of pH and temperature on the TpEH1 activity
The optimal pH of TpEH1 was investigated with purified enzyme by measuring its activity in 0.1 M of the following buffers with the pH ranging from 5.2 to 9.9: citrate buffer (pH 5.2 and 6.4); phosphate buffer (pH 6.4, 7, 7.6, and 8.0); Tris-HCl buffer (pH 8.0 and 9.1); and carbonic buffer (pH 9.9). The effects of temperature on TpEH1 activity were examined over a temperature range from 20 to 40 °C in 0.1 M Tris-HCl buffer (pH 7.6).
Determination of substrate enantioselectivity
The enantioselectivity of the purified TpEH1 was assayed in the kinetic resolution of PGE and derivatives. The reaction mixture contained 5 μl of enzyme solution (7.9 mg/ml) in 445 μl of Tris-HCl buffer (pH 8.0). After pre-incubating the mixture at 30 °C for 5 min, the reaction was started by adding 50 μl of 200 mM substrate stock solution to a total volume of 500 μl. The sample was equally separated into two parts: one was terminated by adding an equal volume of methanol followed by HPLC analysis to determine the conversion ratio. The other portion was extracted with ethyl acetate. The residual epoxide and corresponding diol were analyzed by HPLC on a system equipped with a chiral column to determine enantiomeric excess. The enantiomeric excess was derived from the remaining epoxides of the two enantiomers as follows: \( ee\ \left(\%\right)=\left(S-R\right)/\left(S+R\right)\times 100 \). The enantiomeric ratio (E) was derived from the extent of conversion (c) and the enantiomeric excess of the remaining enantiomer of the substrate as follows: \( E=\mathit{\ln}\left[\left(1\hbox{--} \mathrm{c}\right)\ \left(1\hbox{--} ees\right)\right]/\mathit{\ln}\left[\left(1\hbox{--} \mathrm{c}\right)\ \left(1+ees\right)\right] \).
Analytical methods
The reaction conversion ratio was determined by HPLC (Agilent 1100) with a SB-AQ column and a UV-VIS detector at 256 or 210 nm. The enantiomeric excess of the epoxides and diols was determined with chiralcel chiral columns or supelco fused silica capillary column (Table S1). The absolute configuration was determined by comparing the retention time with previous reports (Bala et al. 2010).
Kinetic resolution of PGE in biphasic and single aqueous phase system
Lyophilized cells were used as the catalyst in these systems. Benzene, methylbenzene, cyclohexane, hexane, heptane, and isooctane were treated as the second phase, forming an organic/aqueous system. The volume ratio of this two-phase system was fixed at 1:1. The enzyme activity was measured by the extent of PGE decrease in the organic phase. Next, 4 ml of PGE in isooctane was catalyzed by 4 ml lyophilized cells (10 mg/ml) at 30 °C and 200 rpm. During this process, samples (50 μl) in the organic phase were taken periodically to measure the enantiomeric excess and conversion ratio. Same amount of catalyst was used in a 4-ml single aqueous buffer for comparison.
Kinetic resolution of PGE with different concentrations in a single aqueous phase
Lyophilized cells were used as the catalyst in this process. To this end, 15 mg/ml catalyst was added to 4.5 ml of 100 mM Tris-HCl buffer (pH 8.0). After pre-incubating the mixture at 30 °C for 5 min, the reaction was started by adding 500 μl of 2.5, 4, or 5 M substrate stock solution in DMSO. The samples were withdrawn periodically during incubation, and the reaction mixtures (200 μl) were extracted with diethyl ether (500 μl) twice. The ee of the residual epoxide and the corresponding diol were analyzed as mentioned above.
Measurement of the inactivation effect of product and substrate
To measure the inactivation effect of the substrate, 60 μl of lyophilized cells (10 mg/ml) was added to different volumes of Tris-HCl buffer (pH 8.0). After pre-incubating the mixture at 30 °C for 5 min, the reaction was started by adding various volumes of PGE to bring the reaction to a total volume of 1 ml. The reaction was terminated by cooling on ice, followed by removal of the cells by centrifugation, and the sample was immediately analyzed by HPLC to measure vicinal diol production.
Different amounts of vicinal diol were added to 470 μl Tris-HCl (pH 8.0) as reaction buffer, and 30 μl lyophilized cells (10 mg/ml) was added. After pre-incubating the mixture at 30 °C for 20 min, the reaction was started by adding 50 μl PGE (200 mM). After incubation at 30 °C with shaking at 200 rpm for 3 min, the reaction was terminated by cooling on ice, followed by removal of the cells by centrifugation. The sample was immediately analyzed by HPLC to measure the extent of epoxide decrease.
Gram-scale bioresolution of PGE and 3-methylPGE
For bioresolution of PGE and 3-methylPGE, 45 ml Tris-HCl (pH 8.0) buffer with 10 mg/ml lyophilized cells was pre-incubated at 30 °C with shaking at 200 rpm. Three grams of PGE or 3.3 g of 3-methylPGE in DMSO was then added into the mixture. The reaction was terminated after 50 min by extracting PGE with 20 ml hexane three times, and vicinal diol was then extracted by 20 ml ethyl acetate three times. After drying over anhydrous sodium sulfate, solvents were removed under vacuum.
Results
Cloning a novel EH from T. paurometabola DSM20162 with excellent enantioselectivity to PGE
The genome sequence of the T. paurometabola type strain has been reported and includes two predicted α/β fold EH proteins, TpEH1(YP_003645475) (Munk et al. 2011). Alignment of TpEH1 with the selected database EHs is displayed in Fig. 1. TpEH1 had typical characteristics of conserved EH motifs, such as HGXP, Tyr proton donor residues, the nucleophile Asp, and the GXSmXS/T sequence (where Sm = small amino acid, X = any amino acid). Analysis of these sequences showed that the TpEH1 consisted of 1038 bp encoding 345 amino acids, encoding a protein with a predicted molecular weight of 37,619 Da (Fig. 1). A phylogenetic relationship among the amino acid sequences of TpEH1 and eight other EHs was constructed (Fig. 2). A BLASTP search against the NCBI protein database revealed that the most related protein was a hypothetical EH from Candidatus entotheonella sp. TSY2, which shared only 50 % amino acid identity. The phylogenetic relationship and low identity suggested that TpEH1 was unique.
Amino acid sequence alignment of EHs. TpEH1 Tsukamurella paurometabola epoxide hydrolase (YP_003645475), ArEH Agrobacterium radiobacter epoxide hydrolase (Y12804), CcEH Caulobacter crescentus epoxide hydrolase (WP_010919111), BmEH Bacillus megaterium epoxide hydrolase (HQ436037), RbEH Rhodobacterales bacterium epoxide hydrolase (WP_008328870), kau2 uncultured microorganism epoxide hydrolase (ACO95125), ElEH Erythrobacter litoralis epoxide hydrolase (WP_011414387), MtEH Mycobacterium tuberculosis epoxide hydrolase (WP_003899827). The conserved motifs included HGXP, two proton donor residues (Tyr158 and Tyr248), the nucleophile Asp102, and GXSmXS/T
Phylogenetic tree of EH amino acid sequences. Sequence alignment was performed using MEGA6 software. Tsukamurella paurometabola: this paper; Candidatus entotheonella sp.TSY2: ETX05676 (hypothetical EH); Bradyrhizobium japonicum: WP_018648134 (predicted EH); Caulobacter crescentus: WP_010919111 (Hwang et al. 2008); Agrobacterium tumefaciens: CAA73331 (Spelberg et al. 1998); Bacillus megaterium: ADV36302 (Zhao et al. 2011); Homo sapiens: NP_000111 (Team 2002); Aspergillus niger: CAB59812 (Arand et al. 1999); Mugil cephalus: ACQ91144 (Choi et al. 2009); Rhodobacterales bacterium HTCC2654: ZP_01014743 (Woo et al. 2010); Novosphingobium aromaticivorans: YP_497537 (Woo et al. 2009); Rhodotorula glutinis: AAF64646 (Yoo et al. 2008); Erythrobacter litoralis HTCC2594 rEEH1: YP_457985 (Woo et al. 2007)
This putative EHs were heterologously expressed in E. coli. TpEH1 was found to be highly soluble and active for the hydrolysis of racemic PGE. The TpEH1 fusion protein was purified to apparent homogeneity by Histag-affinity chromatography. SDS-PAGE analysis of the purified TpEH1 showed a single band with an apparent mass of 37 kDa (Fig. 3). The specific activity of TpEH1 was determined to be 39 U/mg using PGE as a substrate. The kinetic parameters of the two enantiomers of PGE were obtained from the plot and are summarized in Table 1: The enantioselectivity factor E was calculated as 61 from \( \left( KcatS\ /\ KmS\right)\ /\ \left( KcatR\ /\ KmR\right) \).
Temperature and pH optima of TpEH1
The activity of purified TpEH1 was determined by measuring the hydrolysis of PGE. The optimum pH and temperature of TpEH1 in the reaction with PGE were pH 8.0 and 30 °C, respectively. TpEH1 activity was maintained relatively high at temperatures ranging from 20 to 30 °C; however, activity of the protein decreased sharply when the temperature was increased to 35 °C (Fig. 4), which is consistent with the observation that the T. paurometabola type strain grows in the range from 10 to 35 °C (Munk et al. 2011). TpEH1 maintained relatively high activity (more than 80 %) between pH 6.5 and 8.0 (Fig. 5).
Kinetic resolution of racemic PGE, derivatives of PGE, styrene oxide, and cyclohexene oxide with purified enzyme
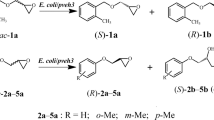
In order to explore the substrate scope of the TpEH1, a range of racemic phenyl glycidyl ethers, substituted with methyl group, nitro group, and chlorine, styrene oxide, and cyclohexene oxide were employed for biohydrolysis using TpEH1 (Table 2). The resolution of epoxides 1–10 proceeded with varying degrees of enantioselectivity (E = 2 to >100). TpEH1 showed excellent enantioselectivity for substrates 1, 3, and 6 (chiral HPLC chromatograms are provided as Supplemental Figs. S1, S2, and S3 online) and moderate enantioselectivity for substrates 4 and 9, while poor enantioselectivity was observed for substrates 7, 10, 11. TpEH1 showed very low conversion to substrates 2, 5, and 8, which was hard to detect. No conversion was detected to substrate 12.
Kinetic resolution of PGE in biphasic and single aqueous phase system
Biphasic system has been proved to be an efficient strategy in bioresolution of high concentration epoxide. The biphasic system was also investigated in our study. Six organic solvents were selected as the second phase. The results are illustrated in Fig. 6. Solvents with Log P values of more than 3 (cyclohexane Log P = 3.2) were less toxic to TpEH1 activity (Fig. 6a). The E-value was slightly increased from 65 (single aqueous buffer) to 83 (isooctane/aqueous buffer), and the substrate concentration was increased to 400 mM. The biphasic system, consisting of 50 % isooctane, was compared with the single buffer system in the resolution of 400 mM substrate (Fig. 6b). Surprisingly, we discovered that the single aqueous buffer could tolerate the same substrate concentration (400 mM) while only requiring 1 h to reach an ee of more than 99 %. No obvious nonenzymatic hydrolysis was observed due to short reaction time, whereas the biphasic system required more time (2 h) to reach an ee of more than 99 % in the resolution. Based on these data, we used the single aqueous buffer in further reactions.
The selection of organic solvents and comparison between biphasic and single aqueous buffer. a Effects of different organic solvents used in biphasic buffer. b Comparison of the biphase (isooctane/Tris-HCl) and single-phase systems (Tris-HCl). The time curve of the PGE kinetic resolution when the single-phase (filled square) and biphasic (open circle) systems were used
High substrate tolerance of TpEH1 and the inactivating effect of product
We found that TpEH1 could complete resolution below a substrate concentration of 400 mM in a single aqueous buffer, obtaining enantiopure (R)-PGE in 60 min. However, at higher substrate concentrations (500 mM), TpEH1 failed to reach 99 % ee (Fig. 7). To explore the mechanisms preventing TpEH1 from completing the resolution, the inhibitory effect of PGE and the corresponding diol were investigated (Fig. 8). During this process, continuous production of diol caused loss of TpEH1 activity and the higher concentration led to more dramatic inactivation (Fig. 8a). However, the high concentration of PGE did not repress TpEH1 (Fig. 8b). These results demonstrated that TpEH1 was a unique biocatalyst with high substrate tolerance.
Gram-scale bioresolution of PGE and 3-methylPGE
In order to facilitate the development of more practical applications, we performed a scale-up production assay of 50 ml to produce enantiopure (R)-PGE and (R)-3-methylPGE. We obtained (R)-PGE in nearly enantiopure form (98.0 % ee) and (S)-diol (84.3 % ee). The isolated yields of (R)-PGE and (S)-diol were 46.3 % (1.38 g) and 44.1 % (1.48 g), respectively. (R)-3-methylPGE was obtained with 99.3 % ee, and the (S)-diol was obtained with 79.8 % ee. The isolated yields of (R)-3-methylPGE and the corresponding (S)-diol were 40.5 % (1.33 g) and 43.9 % (1.60 g), respectively. These results indicated that TpEH1 was a potential biocatalyst for further large-scale applications.
Discussion
In this study, we identified and characterized a novel EH (TpEH1) from T. paurometabola. TpEH1 showed excellent enantioselectivity to the substrates PGE, 3-methylPGE, and 3-nitroPGE. The easily available and highly active E. coli TpEH1 reported herein is the best biocatalyst described to date for the practical preparation of optically pure (R)-PGE.
Chiral epoxides and vicinal diols are extensively employed in the synthesis of chiral high-value intermediates due to their ability to react with a broad variety of nucleophiles (Faber et al. 1996). Many chemical asymmetric syntheses of enantiopure epoxides have been described in recent years, such as the Sharpless method and salen ligands catalysts (Hwang et al. 2010). However, all these elegant chemical methods suffer from the fact that they use potentially toxic heavy metal-based catalysts and/or exhibit only low to moderate turnover frequencies (Kotik et al. 2012). Chemical and biological methods for preparing enantiopure epoxides are complementary approaches, as their stereoselectivities depend heavily on the nature of the substrate and the method/catalyst applied (Genzel et al. 2000). EHs are ubiquitous in nature and can be produced easily from various microorganisms as recombinant proteins. Moreover, EHs are relatively stable proteins and require neither cofactors nor metal ions for their activities. These characteristics make EHs promising alternatives for organic chemists.
On one hand, due to the recent increase in the amount of available genomic sequence information, we could easily obtain novel biocatalysts with excellent potential. EHs are present in various species. The results of the database search showed that more than 20 % of sequenced organisms contain one or more putative EHs (van Loo et al. 2006). One practical approach, dubbed genome mining, is to search for enzymes within a specified microorganism. For example, open reading frames were searched in the genome of a certain microorganism selected from either soil samples or culture collections (Luo et al. 2012). Then, sequences that are annotated as putative enzymes are subjected to multiple alignments, confirmed by manual or computational methods. In our lab, using the above genome mining methods, we had overexpressed 47 putative EHs from different bacteria (data not shown). The obtained EHs were employed to catalyze racemic PGE, screening for high enantioselectivity. Among these EHs, TpEH1 was found with high enantioselectivity to PGE, thus chosen for further study.
PGE and its derivatives have been used as substrates for various EHs, including those in B. megaterium ECU1001(Zhao et al. 2011), Trichosporon loubierii ECU1040 (Xu et al. 2004), and Rhodobacterales bacterium HTCC2654 (Woo et al. 2010). Among all the known native EHs, the novel TpEH1 from T. paurometabola identified in this study had the best E and the best substrate tolerance to PGE (Table 3). Moreover, the reported EHs from Bacillus alcalophilus MTCC10234 and B. megaterium as whole cell catalysts are able to prepare enantiopure (S)-PGE and derivatives. The EH from B. megaterium is highly selective toward (R)-PGE, complementary to TpEH1. This unusual EH with (R)-enantioselectivity exhibited excellent activity (80 U) and enantioselectivity (E = 58) for PGE. Additionally, this EH has been overexpressed in E. coli successfully. Thus, both enantiomers of PGE could be obtained by EH efficiently. Some studies have examined the application of directed evolution for improvement of the enantioselectivity of EH from A. niger, using PGE as substrate. Reetz introduced an active-site combinatorial saturation test (CAST) as an efficient means to improve EH enantioselectivity (Reetz et al. 2006). A dramatic increase in E (E = 4.6 versus E = 115) was found in the LW202 mutant. Thus, this directed evolution revealed that the enantioselectivity of EH may result from the difference in the distance to the catalytic residue (Asp192) for the two enantiomers (Reetz et al. 2009).
TpEH1 also shows high enantioselectivity to substituted PGE, especially high E to m-substituted PGE, but low E to p-substituted PGE. TpEH1 had excellent enantioselectivity (E = 52) for 3-methylPGE, which was higher than previously reported for T. loubierii (E = 21), B. megaterium ECU1001 (E = 19), and the LW202 mutant of the epoxide hydrolase from A. niger (E = 31). The best E value was from B. alcalophilus MTCC10234 (E = 67), producing (S)-3-methylPGE, contrary to TpEH1. The highest enantioselectivity (E > 100) was achieved when 3-nitroPGE was used as the substrate. The E-value is the best representative of (R)-3-nitroPGE production, although nitro-substituted PGE can cause severe inactivation, leading to a low substrate concentration (2 mM). Almost no activity was detected when o-substituted PGE was used as the substrate, and this may be attributed to the steric hindrance caused by the ortho-substitution (ESTELL et al. 1986). Desymmetrization of meso-epoxides can yield the corresponding enantiopure diol in 100 % theoretical yield. The highest substrate tolerance of cyclohexene oxide was observed with SpEH from Sphingomonas sp. HXN-200, which completed enantioselective hydrolysis of cyclohexene oxide at 500 mM with 86 % ee. However, TpEH1 showed no activity for cyclohexene oxide. No activity was detected with o-substituted PGE and cyclohexene, suggesting that the catalytic pocket of TpEH1 may be quite narrow. Based on these findings, we will investigate the enantioselectivity for epichlorohydrin and aliphatic oxide in future studies. Styrene oxide is also a typical substrate for EHs. The highest substrate concentration was reported with the EH from Rhodotorula glutinis, with a concentration of 1.8 M and a yield of 41 % with 98 % ee (Yoo et al. 2008). However, TpEH1 showed relatively low activity and very poor (R)-enantioselectivity for styrene oxide, which was contrary to the (S)-enantioselectivity of PGE.
Thus far, there have been few reported examples of using EH to prepare enantiopure epoxides at high concentrations. Several approaches have been developed in order to overcome this stringent bottleneck, which severely hampers large-scale industrial development. Different solutions, including addition of a water-miscible cosolvent and the use of a water-immiscible biphasic system, have been proposed to solve this problem. Biphasic buffer has been widely applied in biocatalytic processes. Deregnaucourt et al. used isooctane as a cosolvent, allowing kinetic resolution of trifluoromethyl-substituted aromatic epoxide by operating at room temperature within a few hours at a very high global volume substrate concentration (Deregnaucourt et al. 2007). Additionally, Gong and Xu developed an isooctane/aqueous system to overcome low solubility and instability of PGE in the aqueous solution. The E was dramatically increased from 39.5 to 94, with 1:5 (v/v) isooctane and potassium phosphate buffer, but only 15 g/L substrate (Gong and Xu 2005). Under this system, the loss of epoxide by spontaneous, nonenantioselective chemical hydrolysis may be reduced by partitioning of a large proportion of the epoxide to the organic phase, leading to the increase in E. Additionally, the biphasic buffer maintains the substrate in the organic phase while the product is separated automatically into the aqueous phase, making downstream purification much easier. In this study, we also investigated an isooctane/aqueous (1:1) system, which obtained as high as 400 mM PGE with 99.1 % ee and an analytical yield of 46.3 %. The E was slightly improved from 65 to 83. However, this system required a relatively long reaction time, which may have been due to the interfacial inactivation and unfavorable energetics of the reaction resulting from the hydrophobic substrate (Baldascini and Janssen 2005; Klibanov 1997).
During the investigation of two-phase system, we observed that with the increase in aqueous volume, resolution of PGE at more than 1 M could be completed (data not shown), suggesting that enzyme inactivation was caused by vicinal diol maintained in the aqueous phase. According to the data revealed above, we concluded that while the product exhibited considerable inhibition, high concentrations of substrate did not suppress the reaction. Thus, we inferred that the limited concentration of substrate in the EH catalyzing process maybe due to inhibition from diol. It may be possible to use a membrane reactor to avoid product inhibition in further studies. For example, Choi et al. used an aqueous/organic cascade, hydrophilic, hollow-fiber membrane bioreactor, which could separate inhibitory diol (Choi et al. 2000). With this approach, enantiopure (S)-1, 2-epoxyhexane could be obtained with a volumetric productivity of 3.8 g/L/h. Thus, for application of TpEH in industrial-scale processes, the stability of this EH, particularly inactivation from diol, remains a challenge.
In a single aqueous buffer, TpEH1 allowed the resolution to be completed within 1 h at a high concentration of 400 mM PGE when DMSO was used as a water-miscible cosolvent to enhance PGE solubility. This high concentration was beyond our expectation, which is the same as that achieved by the biphasic system. These data indicated that the single aqueous system could achieve higher space-time yield than the biphasic system: 30 g/L/h versus 7.5 g/L/h, respectively. In a single aqueous buffer, a gram-scale preparation of (R)-1 was successfully achieved within 60 min in the presence of 400 mM substrate. This afforded (R)-1 in a nearly enantiopure form (98.0 % ee) in 46.1 % isolated yield and the antipodal (S)-diol (84.3 % ee) in 44.3 % isolated yield. TpEH1 showed excellent enantioselectivity and high substrate tolerance both in single aqueous buffer and biphasic buffer, which revealed the great potential for production of enantiopure (R)-PGE.
In conclusion, we successfully cloned and expressed a novel EH from T. paurometabola with moderate to excellent enantioselectivity for PGE and its derivatives. To the best of our knowledge, 400 mM is the highest concentration in bioresolution of racemic PGE reported to date. Moreover, PGE and 3-nitroPGE exhibit the best enantioselectivity in the activity of native EHs. Therefore, the unique EH TpEH1 is an attractive biocatalyst for potential utilization in chiral synthesis.
References
Arand M, Hemmer H, Durk H, Baratti J, Archelas A, Furstoss R, Oesch F (1999) Cloning and molecular characterization of a soluble epoxide hydrolase from Aspergillus niger that is related to mammalian microsomal epoxide hydrolase. Biochem J 344:273–280. doi:10.1042/0264-6021:3440273
Bala N, Chimni SS (2010) Recent developments in the asymmetric hydrolytic ring opening of epoxides catalysed by microbial epoxide hydrolase. Tetrahedron-Asymmetry 21(24):2879–2898. doi:10.1016/j.tetasy.2010.11.013
Bala N, Chimni SS, Saini HS, Chadha BS (2010) Bacillus alcalophilus MTCC10234 catalyzed enantioselective kinetic resolution of aryl glycidyl ethers. J Mol Catal B Enzym 63(3–4):128–134. doi:10.1016/j.molcatb.2009.12.019
Baldascini H, Janssen DB (2005) Interfacial inactivation of epoxide hydrolase in a two-liquid-phase system. Enzym Eng 36(2–3):285–293. doi:10.1016/j.enzmictec.2003.08.007
Barth S, Fischer M, Schmid RD, Pleiss J (2004) Sequence and structure of epoxide hydrolases: a systematic analysis. Proteins 55(4):846–855. doi:10.1002/prot.20013
Basith S, Manavalan B, Lee G, Kim SG, Choi S (2011) Toll-like receptor modulators: a patent review (2006–2010). Expert Opin Ther Pat 21(6):927–944. doi:10.1517/13543776.2011.569494
Bisi A, Rampa A, Budriesi R, Gobbi S, Belluti F, Ioan P, Valoti E, Chiarini A, Valenti P (2003) Cardiovascular hybrid drugs: new benzazepinone derivatives as bradycardic agents endowed with selective β1-non-competitive antagonism. Bioorg Med Chem 11(7):1353–1361. doi:10.1016/s0968-0896(02)00621-1
Choi W, Choi C, De Bont J, Weijers C (2000) Continuous production of enantiopure 1, 2-epoxyhexane by yeast epoxide hydrolase in a two-phase membrane bioreactor. Appl Microbiol Biotechnol 54(5):641–646. doi:10.1007/s002530000451
Choi SH, Kim HS, Lee EY (2009) Comparative homology modeling-inspired protein engineering for improvement of catalytic activity of Mugil cephalus epoxide hydrolase. Biotechnol Lett 31(10):1617–1624. doi:10.1007/s10529-009-0055-9
Deregnaucourt J, Archelas A, Barbirato F, Paris J-M, Furstoss R (2007) Enzymatic transformations 63. High-concentration two liquid–liquid phase Aspergillus niger epoxide hydrolase-catalysed resolution: application to trifluoromethyl-substituted aromatic epoxides. Adv Synth Catal 349(8–9):1405–1417. doi:10.1002/adsc.200700085
Estell DA, Graycar TP, Miller JV, Powers DB, Wells JA, Burnier JP, Ng PG (1986) Probing steric and hydrophobic effects on enzyme-substrate interactions by protein engineering. Science 233(4764):659–663. doi:10.1126/science.233.4764.659
Faber K, Mischitz M, Kroutil W (1996) Microbial epoxide hydrolases. Acta Chem Scand 50(3):249–258. doi:10.3891/acta.chem.scand.50-0249
Genzel Y, Archelas A, Broxterman QB, Schulze B, Furstoss R (2000) Microbiological transformations. 47. A step toward a green chemistry preparation of enantiopure (S)-2-, -3-, and -4-pyridyloxirane via an epoxide hydrolase catalyzed kinetic resolution. J Org Chem 66(2):538–543. doi:10.1021/jo001406x
Gong P-F, Xu J-H (2005) Bio-resolution of a chiral epoxide using whole cells of Bacillus megaterium ECU1001 in a biphasic system. Enzym Eng 36(2):252–257. doi:10.1016/j.enzmictec.2004.07.014
Hwang S, Choi CY, Lee EY (2008) Enantioconvergent bioconversion of p-chlorostyrene oxide to (R)-p-chlorophenyl-1,2-ethandiol by the bacterial epoxide hydrolase of Caulobacter crescentus. Biotechnol Lett 30(7):1219–1225. doi:10.1007/s10529-008-9668-7
Hwang S, Choi CY, Lee EY (2010) Bio- and chemo-catalytic preparations of chiral epoxides. J Ind Eng Chem 16(1):1–6. doi:10.1016/j.jiec.2010.01.001
John V, Beck JP, Bienkowski MJ, Sinha S, Heinrikson RL (2003) Human β-secretase (BACE) and BACE inhibitors. J Med Chem 46(22):4625–4630. doi:10.1021/jm030247h
Klibanov AM (1997) Why are enzymes less active in organic solvents than in water? Trends Biotechnol 15(3):97–101. doi:10.1016/S0167-7799(97)01013-5
Kotik M, Archelas A, Wohlgemuth R (2012) Epoxide hydrolases and their application in organic synthesis. Curr Org Chem 16(4):451–482. doi:10.2174/138527212799499840
Lin H, Liu J-Y, Wang H-B, Ahmed AAQ, Wu Z-L (2011) Biocatalysis as an alternative for the production of chiral epoxides: a comparative review. J Mol Catal B Enzym 72(3–4):77–89. doi:10.1016/j.molcatb.2011.07.012
Luo X, Yu H, Xu J (2012) Genomic data mining: an efficient way to find new and better enzymes. Enzym Eng 1:104–108. doi:10.4172/2329-6674.1000104
Monterde MI, Lombard M, Archelas A, Cronin A, Arand M, Furstoss R (2004) Enzymatic transformations. Part 58: enantioconvergent biohydrolysis of styrene oxide derivatives catalysed by the Solanum tuberosum epoxide hydrolase. Tetrahedron-Asymmetry 15(18):2801–2805. doi:10.1016/j.tetasy.2004.06.032
Munk AC, Lapidus A, Lucas S, Nolan M, Tice H, Cheng J-F, Del Rio TG, Goodwin L, Pitluck S, Liolios K (2011) Complete genome sequence of Tsukamurella paurometabola type strain (no. 33T). Stand Genomic Sci 4(3):342. doi:10.4056/sigs.1894556
Pieper AA, Xie S, Capota E, Estill SJ, Zhong J, Long JM, Becker GL, Huntington P, Goldman SE, Shen C-H, Capota M, Britt JK, Kotti T, Ure K, Brat DJ, Williams NS, MacMillan KS, Naidoo J, Melito L, Hsieh J, De Brabander J, Ready JM, McKnight SL (2010) Discovery of a proneurogenic, neuroprotective chemical. Cell 142(1):39–51. doi:10.1016/j.cell.2010.06.018
Reetz MT, Wang LW, Bocola M (2006) Directed evolution of enantioselective enzymes: iterative cycles of casting for probing protein—sequence space. Angew Chem 118(8):1258–1263. doi:10.1002/ange.200502746
Reetz MT, Bocola M, Wang L-W, Sanchis J, Cronin A, Arand M, Zou J, Archelas A, Bottalla A-L, Naworyta A, Mowbray SL (2009) Directed evolution of an enantioselective epoxide hydrolase: uncovering the source of enantioselectivity at each evolutionary stage. J Am Chem Soc 131(21):7334–7343. doi:10.1021/ja809673d
Rui L, Cao L, Chen W, Reardon KF, Wood TK (2005) Protein engineering of epoxide hydrolase from Agrobacterium radiobacter AD1 for enhanced activity and enantioselective production of (R)-1-phenylethane-1,2-diol. Appl Environ Microbiol 71(7):3995–4003. doi:10.1128/AEM.71.7.3995-4003.2005
Spelberg JHL, Rink R, Kellogg RM, Janssen DB (1998) Enantioselectivity of a recombinant epoxide hydrolase from Agrobacterium radiobacter. Tetrahedron-Asymmetry 9(3):459–466. doi:10.1016/S0957-4166(98)00003-2
Team MGCP (2002) Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A 99(26):16899–16903. doi:10.1073/pnas.242603899
van Loo B, Kingma J, Arand M, Wubbolts MG, Janssen DB (2006) Diversity and biocatalytic potential of epoxide hydrolases identified by genome analysis. Appl Environ Microbiol 72(4):2905–2917. doi:10.1128/AEM.72.4.2905-2917.2006
Widersten M, Gurell A, Lindberg D (2010) Structure-function relationships of epoxide hydrolases and their potential use in biocatalysis. Biochim Biophys Acta 1800(3):316–326. doi:10.1016/j.bbagen.2009.11.014
Woo JH, Hwang YO, Kang SG, Lee HS, Cho JC, Kim SJ (2007) Cloning and characterization of three epoxide hydrolases from a marine bacterium, Erythrobacter litoralis HTCC2594. Appl Microbiol Biotechnol 76(2):365–375. doi:10.1007/s00253-007-1011-z
Woo JH, Kang JH, Kang SG, Hwang YO, Kim SJ (2009) Cloning and characterization of an epoxide hydrolase from Novosphingobium aromaticivorans. Appl Microbiol Biotechnol 82(5):873–881. doi:10.1007/s00253-008-1791-9
Woo J-H, Kang J-H, Hwang Y-O, Cho J-C, Kim S-J, Kang SG (2010) Biocatalytic resolution of glycidyl phenyl ether using a novel epoxide hydrolase from a marine bacterium, Rhodobacterales bacterium HTCC2654. J Biosci Bioeng 109(6):539–544. doi:10.1016/j.jbiosc.2009.11.019
Wu S, Li A, Chin YS, Li Z (2013) Enantioselective hydrolysis of racemic and meso-epoxides with recombinant Escherichia coli expressing epoxide hydrolase from Sphingomonas sp. HXN-200: preparation of epoxides and vicinal diols in high ee and high concentration. ACS Catal 3(4):752–759. doi:10.1021/cs300804v
Xu Y, Xu J-H, Pan J, Tang Y-F (2004) Biocatalytic resolution of glycidyl aryl ethers by Trichosporon loubierii: cell/substrate ratio influences the optical purity of (R)-epoxides. Biotechnol Lett 26(15):1217–1221. doi:10.1023/B:BILE.0000036598.35494.de
Yoo SS, Park S, Lee EY (2008) Enantioselective resolution of racemic styrene oxide at high concentration using recombinant Pichia pastoris expressing epoxide hydrolase of Rhodotorula glutinis in the presence of surfactant and glycerol. Biotechnol Lett 30(10):1807–1810. doi:10.1007/s10529-008-9762-x
Zhang Z, Sheng Y, Jiang K, Wang Z, Zheng Y, Zhu Q (2010) Bio-resolution of glycidyl (o, m, p)-methylphenyl ethers by Bacillus megaterium. Biotechnol Lett 32(4):513–516. doi:10.1007/s10529-009-0181-4
Zhao J, Chu Y-Y, Li A-T, Ju X, Kong X-D, Pan J, Tang Y, Xu J-H (2011) An unusual (R)-selective epoxide hydrolase with high activity for facile preparation of enantiopure glycidyl ethers. Adv Synth Catal 353(9):1510–1518. doi:10.1002/adsc.201100031
Zhu QQ, He WH, Kong XD, Fan LQ, Zhao J, Li SX, Xu JH (2013) Heterologous overexpression of Vigna radiata epoxide hydrolase in Escherichia coli and its catalytic performance in enantioconvergent hydrolysis of p-nitrostyrene oxide into (R)-p-nitrophenyl glycol. Appl Microbiol Biotechnol 98(1):207–218. doi:10.1007/s00253-013-4845-6
Acknowledgments
This work was supported by the National major science and technology projects of China (2012ZX09304009).
Conflict of interests
The authors declare that they have no conflict of interests.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
ESM 1
(PDF 325 kb)
Rights and permissions
About this article
Cite this article
Wu, K., Wang, H., Sun, H. et al. Efficient kinetic resolution of phenyl glycidyl ether by a novel epoxide hydrolase from Tsukamurella paurometabola . Appl Microbiol Biotechnol 99, 9511–9521 (2015). https://doi.org/10.1007/s00253-015-6716-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6716-9