Abstract
In order to provide more alternative epoxide hydrolases for industrial production, a novel cDNA gene Rpeh-encoding epoxide hydrolase (RpEH) of Rhodotorula paludigena JNU001 identified by 26S rDNA sequence analysis was amplified by RT-PCR. The open-reading frame (ORF) of Rpeh was 1236 bp encoding RpEH of 411 amino acids and was heterologously expressed in Escherichia coli BL21(DE3). The substrate spectrum of expressed RpEH showed that the transformant E. coli/Rpeh had excellent enantioselectivity to 2a, 3a, and 5a–10a, among which E. coli/Rpeh had the highest activity (2473 U/g wet cells) and wonderful enantioselectivity (E = 101) for 8a, and its regioselectivity coefficients, αR and βS, toward (R)- and (S)-8a were 99.7 and 83.2%, respectively. Using only 10 mg wet cells/mL of E. coli/Rpeh, the near-perfect kinetic resolution of rac-8a at a high concentration (1000 mM) was achieved within 2.5 h, giving (R)-8a with more than 99% enantiomeric excess (ees) and 46.7% yield and producing (S)-8b with 93.2% eep and 51.4% yield with high space-time yield (STY) for (R)-8a and (S)-8b were 30.6 and 37.3 g/L/h.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Enantiomeric isomers of chiral compounds, such as (R)- and (S)-enantiomers of a racemic drug, usually possess different and even antagonistic biological activities and pharmacological functions (Kotik et al. 2012). Since the early 1990s, there has been an ever-increasing demand for the optically pure epoxides and vicinal diols, as they are versatile building blocks in synthesis of various pharmaceuticals, fine chemicals, and agrochemicals (Archelas et al. 2016). For example, (R)-o-methylphenyl glycidyl ether (8a) and the corresponding vicinal diol (S)-3-(2-methylphenoxy)propane-1,2-diol (8b) were used as synthetic precursors of cardiovascular hybrid drugs (Bisi et al. 2003), neuroprotective molecules (Pieper et al. 2010), β-secretase-cleaving enzyme (BACE) inhibitors (Pieper et al. 2010), and modulators of toll-like receptor 7 (Basith et al. 2011).
Epoxide hydrolases (EHs, EC 3.3.2.-), a kind of cofactor-independent biocatalysts and existing ubiquitously in nature, can catalyze the enantioselective or enantioconvergent hydrolysis of rac-epoxides or meso-epoxides and prepare chiral epoxides and vicinal diols (Lin et al. 2011). Reportedly, the EH-catalyzed hydrolysis of epoxides proceeded mainly in two steps. The nucleophilic Asp residue first attacks the carbon atom in an oxirane ring of epoxide, forming a hydroxyalkyl-EH intermediate. Then, a water molecule that is activated by His in the catalytic triad interacts with the intermediate, releasing a vicinal diol product (Lind and Himo 2016). Recently, many EHs from bacteria, fungal, and plant were discovered, cloned, and engineered for organic chemical synthesis (Kotik et al. 2012), such as bacteria EHs from Agrobacterium radiobacter AD1 (ArEH) (Zou et al. 2018), Rhodococcus erythropoli (ReLEH) (Sun et al. 2016), and Bacillus megaterium EH (BmEH) (Serrano-Hervas et al. 2017), fungal EH from Aspergillus niger (AnEH) (Reetz et al. 2009), and plant EHs from Solanum tuberosum (StEH) (Lind and Himo 2016) and Vigna radiata (VrEH1 and VrEH2) (Wu et al. 2015; Li et al. 2018). In addition, four red yeast EH genes have been cloned and heterologously expressed in Escherichia coli, Pichia pastoris, and Yarrowia lipolytica (Table S1).
With the wave of green chemistry, whole-cell catalysis at high substrate concentrations has been a hot spot in the studies of epoxide hydrolases. For example Dongzhi Wei’s team tried to use recombinant E. coli TpEH1 (10 mg/mL lyophilized cells) to hydrolyze phenyl glycidyl ether at concentrations of up to 400 mM (Wu et al. 2015). Zhi Li's team wanted to achieve kinetic resolution of multiple epoxides at high concentrations by using E. coli (SpEH) (Wu et al. 2013). Compared with pure enzymes, immobilized enzymes, and transition metal catalysts, using easily available and low-cost whole cells as biocatalysts is a much more economical alternative (De Carvalho 2011; Hwang et al. 2010). Nevertheless, the application of whole-cell biocatalyst on large-scale preparation for chiral mono-substituted epoxides is still hampered by high biocatalyst loading, low substrate concentrations, and low stereoselectivity (Wu et al. 2013). Hence, the possibility of discovering novel EHs which can product desired epoxides and vicinal diols in high enantiomeric excess (ee), high yield, and high concentration at low catalyst loading is of interest.
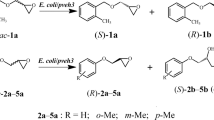
In this work, a yeast Rhodotorula paludigena JNU001 was identified by 26S rDNA D1/D2 domain sequencing. A unique EH encoding gene Rpeh was amplified by RT-PCR from R. paludigena JNU001 total RNA and expressed in E. coli BL21(DE3). Substrate scope of the engineered recombinant E. coli expressing RpEH was explored toward rac-epoxides 1a-12a (Scheme 1). The enzymatic property of purified recombinant RpEH was investigated using rac-8a as substrate. Furthermore, considering the practical application, a gram-scale kinetic resolution of rac-8a at high concentration (1000 mM) was carried out to prepare (R)-8a and (S)-8b in high ee values and yields using low loading of E. coli/Rpeh whole cells as biocatalyst.
Materials and methods
Strains, plasmids, and chemicals
The red yeast strain Rhodotorula sp. was isolated from marine sludge around Dongji Island, China, using potato dextrose agar plates, and preserved in our lab. All enzymes and kits used for gene manipulations in this work were purchased from TaKaRa (Dalian, China) and Sangon (Shanghai, China). E. coli JM109 and linearized plasmid pUCm-T (Sangon) were applied to gene cloning and sequencing while E. coli BL21(DE3) and pET-28a(+) (Novagen, Madison, WI, USA) to EH gene expression. Rac-1a, 7a, and 11a-12a were products of TCI (Tokyo, Japan), and rac-2a-6a and 8a-10a were chemically synthesized by our lab according to the methods reported previously (Cambell and Duron 2013; Stephenson et al. 2008). All other chemicals were of analytical grade and commercially available from the local companies (Wuxi, China).
Molecular biological identification of Rhodotorula sp.
A single colony of Rhodotorula sp. was inoculated into YPD medium (2.0% tryptone, 1.0% yeast extract, and 2.0% dextrose, pH 6.0) and grown at 30 °C for 24 h on a rotary incubator (220 rpm). The cultured red yeast cells were collected by centrifugation at 8000 rpm for 15 min at 4 °C and washed with 20 mM Na2HPO4–NaH2PO4 buffer (pH 7.0). This Rhodotorula sp. was identified by sequence analysis of the D1/D2 domain of 26S rDNA as in previously reported method (Fell et al. 2000). First, the DNA sequence of D1/D2 domain was amplified by PCR using Ex Taq DNA polymerase (TaKaRa, Dalian, China) from the extracted genomic DNA of Rhodotorula sp. using a pair of universal primers, FL1 and FL4 (Table S2). The PCR conditions are as follows: an initial denaturation at 98 °C for 5 min, 30 cycles at 98 °C for 10 s, 55 °C for 30 s, 72 °C for 30 s, and an extra elongation at 72 °C for 10 min. Second, the target PCR product was purified using the SanPrep column DNA gel extraction kit, ligated with pUCm-T, and transformed into E. coli JM109, followed by DNA sequencing. Finally, the correct nucleotide sequence of the D1/D2 domain was submitted to the GenBank database, and then the DNA sequence matching analysis was carried out by BLAST server in the NCBI website (https://www.ncbi.nlm.nih.gov/).
Design and synthesis of a pair of PCR primers
The three amino acid sequences of the characterized EHs from known red yeasts, Rhodotorula araucariae CBS 6031, R. toruloides CBS 14, and R. glutinis CIMW 147 (Choi et al. 2000; Visser et al. 2002), were searched in the NCBI website. After the multiple sequence alignment among three known EHs was carried out by using the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/), the three most conserved oligopeptide segments, CHGWPG, AQGGDWGSI, and DGGHFAALEKP, were located. Thereafter, using three oligonucleotide sequences corresponding to the most conserved segments as templates, a DNA sequence coding for a hypothetical EH was located in fragment 1 (GenBank accession no. SWEA01000001) of Rhodotorula paludigena CM33 genomic DNA. Furthermore, the localization of exon/intron boundaries in the DNA sequence of a hypothetical EH-encoding gene of R. paludigena CM33 was predicted using the Eukaryotic GeneMark.hmm (http://topaz.gatech.edu/GeneMark/gmhmme.cgi), while a cDNA sequence, that is, an open-reading frame (ORF), of the EH gene was located by NCBI ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). Based on the above computer-aided analysis, a pair of PCR primers, Rp-F and Rp-R (Table S2), was designed according to the 5′- and 3′-end sequences of the ORF, and synthesized by Sangon (Shanghai, China).
Cloning of both cDNA and DNA sequences of a hypothetical EH gene Rpeh
The total RNA of R. paludigena JNU001 identified by our lab was extracted using the Spin Column Yeast Total RNA purification kit, from which the ORF (i.e., cDNA sequence) of Rpeh was amplified by RT-PCR using the BcaBEST™ RNA PCR kit and a pair of specific PCR primers. In detail, the first-strand cDNA was reversely transcribed from the total RNA of JNU001 using an Oligo dT-Adaptor primer provided by RNA PCR kit. Then, the ORF coding for RpEH, flanked by Nde I and EcoR I sites, was amplified from the first-strand cDNA using specific primers Rp-F and Rp-R. The recombinant plasmid, designated pUCm-T-Rpeh, was constructed by ligating ORF with pUCm-T, and transformed into E. coli JM109 competent cells, followed by DNA sequencing. The correct cDNA sequence of Rpeh was excised from pUCm-T-Rpeh with Nde I and EcoR I and inserted into expression plasmid pET-28a(+) digested by same enzymes, followed by transforming it into E. coli BL21(DE3), thereby constructing an E. coli transformant, designated E. coli/Rpeh. Additionally, the DNA sequence of Rpeh was amplified from the genomic DNA with the same primers. The target PCR product was ligated with pUCm-T and transformed into E. coli JM109, followed by DNA sequencing.
Expression and purification of RpEH
A single colony of E. coli/Rpeh strain was inoculated into LB medium (1.0% tryptone, 0.5% yeast extract, and 1.0% NaCl, pH 7.2) supplemented with 100 μg/mL kanamycin sulfate and grown at 37 °C overnight as the seed culture. Then, the same fresh LB medium was inoculated with 1.0% (v/v) seed culture and grown at 37 °C until the OD600 reached 0.6–0.8. After induced by 0.2 mM IPTG at 20 °C for 8 h, the E. coli/Rpeh cells were collected and resuspended in 100 mM Na2HPO4–NaH2PO4 buffer (pH 7.0) to 10 mg wet cells/mL as the whole-cell biocatalyst unless stated otherwise. In this work, E. coli BL21(DE3) transformed with pET-28a(+), designated E. coli/pET-28a, was used as a negative control.
The collected E. coli/Rpeh whole cells, heterologously expressing RpEH with a 6× His tag at its N-terminus, were resuspended in buffer A (20 mM Tris–HCl, 500 mM NaCl and 20 mM imidazole, pH 7.5) to 100 mg wet cells/mL and disrupted by ultrasonication in the ice-water bath. Then, the resultant supernatant was loaded onto a nickel–nitrilotriacetic acid (Ni–NTA) column (Tiandz, Beijing, China) pre-equilibrated with buffer A, followed by elution at 0.4 mL/min with buffer B as the same as buffer A except for 200 mM imidazole. Aliquots of 1 mL eluent only containing the target protein RpEH were pooled, dialyzed against 20 mM Na2HPO4–NaH2PO4 buffer (pH 7.0), and concentrated using a Amicon® Ultra-15 10K filter device (Millipore, Billerica, MA).
Enzyme activity and protein assays
The activity of RpEH for rac-1a was measured as reported previously (Hu et al. 2017), with slight modification. In detail, 900 μL cell suspension or purified RpEH solution, suitably diluted with 100 mM Na2HPO4–NaH2PO4 buffer (pH 7.0), was well mixed with 100 μL 200 mM rac-1a (dissolved in methanol) at a final concentration of 20 mM, incubated at 25 °C for 15 min, and terminated by addition of 4 mL methanol. The reaction sample was analyzed by high-performance liquid chromatography (HPLC), using a Waters e2695 apparatus (Waters, Milford, MA, USA) equipped with an XBridge BEH C18 column. The mobile phase, methanol/H2O (7:3, v/v), was used at 0.8 mL/min, and monitored using a Waters 2489 UV–Vis detector at 220 nm. One unit (U) of EH activity was defined as the amount of wet cells or purified RpEH hydrolyzing 1 μmol rac-1a per minute under the given assay conditions. Analogously, the activities of RpEH for rac-2a-12a were measured by substituting rac-1a with them, respectively.
SDS-PAGE was performed on a 12% agarose gel, and the isolated proteins were visualized by staining with Coomassie Brilliant Blue R-250 (Sigma-Aldrich, St. Louis, MO, USA). The apparent molecular weight of the expressed RpEH was estimated by comparison with those of standard proteins using a Quantity One software. The protein concentration was determined using the BCA-200 Protein Assay Kit (Pierce, Rockford, IL, USA), using bovine serum albumin as the standard.
Substrate spectrum investigation of RpEH
The asymmetric hydrolytic reactions of rac-epoxides were performed in 1.0 mL 100 mM phosphate buffer (pH 7.0) system consisting of 10 or 20 mM rac-1a–12a (200 mM, dissolved in methanol) and a certain amount of whole cells of E. coli/Rpeh (Table 1) at 25 °C for 15 min, respectively. During the hydrolytic process, aliquots of 100 μL reaction sample were periodically drawn out and then extracted with 900 μL ethyl acetate or that containing 1 mM n-hexanol (as the internal standard). The treated sample was analyzed by HPLC equipped with a chiral column or by chiral gas chromatography (GC) using a GC-2010 apparatus (Shimadzu, Tokyo, Japan) (Table S3, Fig. S1). The absolute configurations of all the single enantiomers of rac-1a–12a and their corresponding vicinal diols rac-1b–12b were confirmed, respectively, by comparing their retention times with those reported previously (Hu et al. 2017; Li et al. 2017). The conversion ratio (c) of rac-substrate was defined as the percentage of its consumed amount to initial one. The ees of retained single epoxide and the eep of produced chiral diol were calculated using the following equations: ees = |[(Rs − Ss) / (Rs + Ss)]| × 100% and eep = |[(Rp − Sp) / (Rp + Sp)]| × 100%. Rs and Ss represent the concentrations of (R)- and (S)-1a–12a, respectively, while Rp and Sp are the concentrations of (R)- and (S)-1b–12b.
The enantioselectivity of EH for a given rac-epoxide, which is quantitatively described by its enantiomeric ratio (i.e., E value), was used to evaluate the degree of enantio-preferential hydrolysis of one epoxide enantiomer over its antipode (Kotik and Kyslík 2006). Based on the hydrolytic parameters of rac-substrates (c and ees) calculated previously, E values of RpEH for rac-1a–12a were as follows calculated: E = ln [(1 − c) × (1 − ees)] / ln [(1 − c) × (1 + ees)]. The regioselectivity coefficients of EH, αR (or βR = 1 − αR) and βS (or αS = 1 − βS), were used to estimate the probabilities of attacks on Cα (a more hindered carbon in the oxirane ring) of (S)-epoxide and on Cβ (a less hindered terminal carbon) of (R)-epoxide, respectively (Zhu et al. 2014). Herein, the αS and βR values of RpEH for (S)- and (R)-8a were derived by linear regression: eep = (αR + βS − 1) + [(αR − βS) × ees × (1 − c)] / c (Kotik et al. 2010).
Effects of pH and temperature on the RpEH activity and stability
The pH optimum of purified RpEH for rac-o-methylphenyl glycidyl ether (8a) was examined under the standard EH activity assay conditions, except for using buffers (100 mM Na2HPO4–citric acid: pH 5.5–7.0 and 100 mM Tris–HCl: pH 7.5–9.0). To estimate the pH stability, aliquots of RpEH solution were preincubated, in the absence of substrate, at a pH range of 5.5–9.0 at 20 °C for 1 h. The residual EH activity was measured under the standard assay conditions. In this work, the pH stability was defined as a pH range, over which the residual RpEH activity retained over 85% of its original activity.
The temperature optimum of RpEH for rac-8a was determined, at pH optimum, at temperatures ranging from 20 to 50 °C. To evaluate the thermostability, aliquots of RpEH solution were incubated at 20–50 °C, respectively, for 1 h. Herein, the thermostability was defined as the temperature, at or below which the residual RpEH activity was more than 85% of its original activity.
Kinetic parameter assay of purified RpEH
The initial hydrolytic reaction rate of rac-8a (μmol/min/mg protein) catalyzed by purified RpEH was measured under the standard EH activity assay conditions, except for the concentrations of rac-8a ranging from 2.0 to 20 mM. Both the Michaelis constant (Km) and maximum velocity (Vmax) of RpEH were calculated, respectively, by non-linear regression analysis using an OriginPro 2016 software (http://www.orignlab.com/). The turnover rate or number (kcat) of RpEH was deduced from its Vmax and apparent molecular weight, while its catalytic efficiency was defined as the ratio of kcat to Km.
Kinetic resolution of rac-8a at elevated concentrations by E. coli/Rpeh
The asymmetric hydrolytic reactions of rac-8a at the elevated concentrations of 400, 600, 800, and 1000 mM were carried out at 30 °C, respectively, by using 10 mg wet cells/mL of E. coli/Rpeh in the 2 mL 100 mM phosphate buffer (pH 7.0) system. During the hydrolytic course, aliquots of 50 μL reaction sample were drawn out at given time points, extracted with 950 μL ethyl acetate, then diluted ten times, and analyzed by chiral HPLC equipped with a Chiralcel OD-H column (Daicel, Osaka, Japan) for calculating the c of rac-8a, ees and yield of retained (R)-8a, as well as eep and yield of produced (R)-8b. Herein, the ees and yield of (R)-8a were used as main criteria to confirm the maximum allowable concentration of rac-8a. Based on the above-mentioned experimental results, the gram-scale kinetic resolution, in the 25 mL phosphate buffer (pH 7.0) system containing 10 mg wet cells/mL and rac-8a at maximum allowable concentration, was performed at 30 °C. When the ees of (R)-8a reached over 99%, the total reaction solution was extracted with 10 mL n-hexane three times. Then, three n-hexane fractions containing (R)-8a were pooled, dried over anhydrous magnesium sulfate, and purified by silica gel column chromatography, followed by concentration under reduced pressure.
Results
Molecular identification of Rhodotorula sp.
The pink elliptical shape red yeast was isolated from marine sludge in China, which showed the EH activity of 0.98 U/g wet cells toward rac-1a (Fig. 1a). The PCR-amplified D1/D2 domain sequence of 26 s rDNA from the red yeast Rhodotorula sp. was 585 bp in length (Fig. 1b, c), which shared the highest identity (> 99%) with a known R. paludigena HB77-2 by BLAST search in GenBank database. As a result, the isolated strain was identified as R. paludigena and named as R. paludigena JNU001, and then submitted to the Culture and Information Center of Industrial Microorganisms of China University (accession no. CICIM Y7069).
Analysis of the primary structure of deduced RpEH
A 1236-bp coding sequence (CDS) and a 1600-bp DNA sequences of Rpeh (GenBank: MK748445) were amplified from the total RNA and the genomic DNA of R. paludigena JNU001, respectively, using primers Rp-F and Rp-R. The result showed that the DNA-coding region of Rpeh DNA contained six introns ranging from 58 to 66 bp. The ORF of Rpeh encodes 411 amino acid protein (RpEH). The primary structure of RpEH shares less than 75% identity with the reported four red yeast EHs from R. araucariae (74.1%, AAN32663) (Visser et al. 2002), R. glutinis (AAF64646, 63.1%) (Yoo et al. 2008), Rhodosporidium toruloides (63.6%, AAN32662) (Visser et al. 2002), and R. mucilaginosa (AAV64029, 45.2%) (Labuschagne and Albertyn 2007), respectively (Table S1). Multiple alignments of RpEH and other four known EHs indicated that RpEH had typical characteristics of EH, including the conserved motifs GXSmXS/T and HGXP (where Sm is small amino acid and X is any amino acid) (Barth et al. 2004), the catalytic triad Asp190-His385-Glu359, and two conserved Tyr262 and Tyr331 residues (Fig. 2).
The multiple sequence alignment of four EHs. RpEH (QDD56409, in this work); RaEH (AAN32663, 74.1% identity with RpEH); RtEH (AAN32662, 63.6%); RgEH (AAF64646, 63.1%); RmEH (AAV64029, 45.2%). The conserved motifs of HGXP, GXSmXS/T, and SmXNuXSmSm are boxed. A catalytic triad (Asp190-His385-Glu359) and both Tyr262 and Tyr331 in RpEH are marked with stars
Expression and purification of RpEH
After induction by 0.2 mM IPTG at 20 °C for 8 h, the activity of recombinant E. coli/Rpeh toward rac-1a was 2132 U/g wet cells, which was 2173-fold higher than that of the wild-type strain R. paludigena JNU001 (0.98 U/g wet cells). A total of 15 mL 100 mg wet cells/mL of E. coli/Rpeh (the total number of units of EH activity, 3710 U) were purified to obtain 63.8 mg (2244 U) pure enzyme. The specific activity of purified RpEH toward rac-8a was determined to be 35.2 U/mg protein.
The SDS-PAGE analysis results indicated that RpEH with an apparent molecular weight of 48.4 kDa was expressed as a soluble form in E. coli cells (Fig. 3, lanes 1 and 2) and was purified to apparent homogeneity with 3.8-fold purification and 60.5% yield (Fig. 3, lanes 3 and 4).
Substrate scope analysis of RpEH toward rac-1a–12a
In order to explore the practical applications, a range of racemic styrene oxide derivatives 1a–6a, phenyl glycidyl ether derivatives 7a–10a, and straight-chain aliphatic epoxides 11a–12a was investigated using the easily available E. coli/Rpeh wet cells as biocatalyst (Scheme 1). As shown in Table 1, E. coli/Rpeh displayed high activities of 743–9081 U/g wet cells toward 1a and 5a–12a, while relative low activity toward nitro-substituted styrene oxide 2a–4a (24–276 U/g wet cells). The determination of enantiomeric ratio (E value) toward given rac-epoxides displayed that E. coli/Rpeh possessed the low enantioselectivity toward 1a (E = 3.5), 11a (E = 1.6), and 12a (E = 2.3), moderate enantioselectivity toward 4a (E = 19), and high enantioselectivity toward 2a (E > 200), 3a (E = 64), and 5a–10a (E = 49–160).
Enzymatic properties of the purified RpEH
The enzymatic properties of purified RpEH were investigated using rac-8a as substrate. As shown in Fig. 4, RpEH exhibited high catalytic activity at a pH range of 5.5–9.0, over which the pH optimum was 7.0. It was highly stable at pH values ranging from 6.0 to 7.5, retaining more than 85% of its original activity (Fig. 4a). Additionally, the temperature optimum of RpEH, at pH optimum of 7.0, was 30 °C. After being incubated at 20–50 °C for 1 h, RpEH displayed high stability at 45 °C or below and still retained 61% activity at 50 °C (Fig. 4b). In addition, the kinetic parameters of purified RpEH toward rac-8a were determined. The resultant Km, Vmax, kcat, and kcat/Km of purified RpEH toward rac-8a were 8.35 mM, 57.2 U/mg, 46.1 s−1, and 5.5 mM−1 s−1, respectively.
Effects of pH values (a) and temperatures (b) on the catalytic activity and stability of RpEH. (a) pH ranging from 5.5 to 9.0: Na2HPO4–citric acid buffer (pH 5.5–7.0) and Tris–HCl buffer (pH 7.5–9.0). The value at pH 7.0 was set as 100%. (b) The value at 30 °C was set as 100%. All experiments were performed in triplicate
Enantiopreference and regioselectivity of RpEH toward 8a
The kinetic resolution of rac-8a at low substrate concentration (20 mM) showed that (S)-8a was preferentially hydrolyzed, while (R)-8a was retained (Fig. 6a), indicating that RpEH displayed (S)-enantiopreference toward 8a. The regioselectivity coefficients, βS and αR, toward the two enantiomers of 8a were calculated to be 99.7 and 83.2%, indicating that the Cβ of favored (S)-8a was specifically attacked to form (S)-8b in reversion configuration, while the Cα of disfavored (R)-8a was mainly to attacked to form (S)-8b in retention configuration (Fig. 6b).
Kinetic resolution of rac-8a at high concentration
In order to facilitate the development of practical applications, with only 10 g/L E. coli/Rpeh wet cells (about 1.6 g/L dry cells) as the catalyst, the kinetic resolution of rac-8a was performed in 100 mM phosphate buffer (pH 7.0) at a higher substrate loading of 400, 600, 800, and 1000 mM, respectively (Fig. 5). The (S)-8a was quickly hydrolyzed to corresponding (S)-8b and retaind (R)-8a with both high ee and yeild values within a short reaction time (Table 2). Especially, the kinetic resolution of rac-8a was smoothly achieved at high substrate concentration (1000 mM,164.2 g/L) and a very low catalyst loading (in terms of substrate/catalyst ratio, 16.4 g/g wet cells) with 2.5 h, obtaining (R)-8a with > 99% ees and 46.7% yields, producing (S)-8b with 93.2% eep and 51.4% yieldp. The space-time yield (STY) values of (R)-8a and (S)-8b reached 30.6 and 37.3 g/L/h, respectively.
Preparation of (R)-8a by E. coli/Rpeh
Furthermore, a gram-scale kinetic resolution of rac-8a (4.1 g, 1000 mM) was performed in 25 mL phosphate buffer (pH 7.0) using 0.25 g E. coli/Rpeh wet cells as biocatalyst at 30 °C for 2.5 h. (R)-o-Methylphenyl glycidyl ether (R)-8a: colorless oil; 1.89 g; isolated yield 46.0%; ees > 99.0% (HPLC); 1H NMR (400 MHz, CDCl3, TMS): δ 7.13–7.16 (m, 2H), 6.9 (t, J = 7.6 Hz, 1H), 6.8 (d, J = 8.0 Hz, 1H), 4.25 (dd, J1 = 3.2 Hz, J2 = 11.2 Hz, 1H), 4.00 (q, J = 5.2 Hz, 1H), 3.36–3.40 (m, 1H), 2.92 (t, J = 4.4 Hz, 1H), 2.8 (dd, J1 = 2.8 Hz, J2 = 4.8 Hz, 1H), 2.25 (s, 3H). (S)-3-(2-methylphenoxy)propane-1,2-diol (S)-8b: white solid;2.2 g; isolated yield 48.2%; eep 92.8% (HPLC); 1H NMR (400 MHz, CDCl3, TMS): δ 7.13–7.17 (m, 2H), 6.88 (t, J = 12.0 Hz, 1H), 6.81 (d, J = 8.0 Hz, 1H), 4.11–4.14 (m, 1H), 4.04–4.02 (m, 2H), 3.76–3.87 (m, 2H), 2.22 (s, 1H). The HPLC spectra and the NMR spectra for (R)-8a and (S)-8b are in the supplementary information (Figs. S2, S3, and S4).
As compared to other reported EHs (Table 3), obviously, the easily available and highly active E. coli/Rpeh with high enantioselectivity is a technically competitive and economically viable biocatalyst for preparing chiral (R)-8a.
Discussion
Since Weijers and co-workers discovered that one red yeast R. glutinis CIMW 147 displayed enantioselective toward aryl, alicyclic, and aliphatic epoxides, yeast EHs have been attracted much attention (Weijers 1997). By screening of 187 yeast strains from 25 different genera, Botes and co-workers found that 54 yeast strain displayed EH activity toward 1,2-epoxyoctane (12a), while only 8 yeasts belonging to Trichosporon, Rhodotorula, and Rhodosporidium genera displayed enantioselectivity (Botes 1998). In the current study, we identified a new red yeast R. paludigena JNU001 by analyzing its D1/D2 domain of 26S rDNA, which showed the EH activity of 0.98 U/g wet cells toward typical substrate rac-1a.
Furthermore, an EH-encoding sequence Rpeh from R. paludigena JNU001 was successfully cloned and expresed in E. coli BL21. The whole cell activity of E. coli/Rpeh toward rac-1a was improved by 2173-fold compared to the R. paludigena JNU001. The primary structure of RpEH shares less than 75% identity with the reported four red yeast EHs from R. araucariae, R. glutinis, Rhodosporidium toruloides, and R. mucilaginosa, which demostrated that RpEH as a novel EH may display different catalyst characteristics.
For further research, we studied the properties of E. coli/Rpeh catalysis 1a to 12a. As is well known, styrene oxide (1a) is a typical substrate for EHs. The highest substrate concentration was reported with the EH from R. glutinis, with a concentration of 1.8 M and a yield of 41% with 98% ee (Yoo et al. 2008). Unfortunately, although RpEH had high catalytic activity for 1a, the enantioselectivity was low (E = 3.5). But interestingly, RpEH had superior enantioselectivity and catalytic activity for both nitrostyrene oxides (2a, 3a, 4a) and chlorosytrene oxides (5a, 6a). Among them, the most representative catalytic properties were for 6a (E = 58, 1463 U/g wet cells), which was higher than previously reported for Sphingomonas sp. HXN-200 (E = 14, 920 U/g lyophilized cell) (Wu et al. 2013). RpEH had high activity and enantioselectivity for both PGE (7a) and methyl substituted PGE (8a, 9a, 10a). In these substrates, the highest enantioselectivity (E = 101) was achieved when 8a was used as the substrate. 8a was used as substrate for various EHs, including TpEH (Wu et al. 2015), VrEH3 (Hu et al. 2017), and REH (Woo et al. 2010). Among all the known native EHs, the novel RpEH from R. paludigena JNU001 identified in this study had the best E and the highest whole cell catalytic activity to 8a (Table 3). RpEH had not only a high (S)-enantiopreference (E = 101) for 8a but also a high and complementary regioselectivity (αR = 99.7%, βS = 83.2%). The high (S)-enantiopreference means during the kinetic resolution of rac-8a, (S)-8a will be preferentially hydrolyzed but (R)-8a will be retained (Fig. 6a). The high and complementary regioselectivity indicated that the Cβ of favored (S)-8a was specifically attacked to form (S)-8b in reversion configuration, while the Cα of disfavored (R)-8a was mainly to attacked to form (S)-8b in retention configuration (Fig. 6b). Therefore, under the synergistic action of those two characteristics, a near perfect kinetic resolution of rac-8a was achieved, affording both (R)-8a in > 99% ees and (S)-8b in 91.4% eep and near-theoretical yield of 50% (Table 2). Moreover, the reported EHs from Bacillus megaterium ECU1001 (Zhao et al. 2011) as whole cell catalysts are able to prepare enantiopure (S)-8a and derivatives. The EH from B. megaterium is highly selective toward (R)-8a, complementary to RpEH. This unusual EH with (R)-enantioselectivity exhibited excellent activity and enantioselectivity for 8a. Additionally, this EH has been overexpressed in E. coli successfully. Thus, both enantiomers of 8a could be obtained by EH efficiently. Some studies reported that red yeast-derived EHs like RaEH, RtEH, and RgEH had excellent catalytic activity and enantioselectivity for aliphatic oxides (Botes et al. 1998; Matsumoto et al. 2014). For example, RgEH from R. glutinis strain CIMW 147 was able to hydrolyze rac-11a resulted in (S)-11a (ee > 98%, yield = 48%) and (R)-11b (ee = 83%, yield = 47%) (Weijers et al. 1998). Unlike other red yeast-derived EHs, RpEH had high catalytic activity for 11a, 12a but very poor enantioselectivity for such substrates. In future research, we may be able to improve the enantioselectivity of RpEH for 11a and 12a by directed evolution and broaden its application range.
By comparing the pure enzyme activity (35.2 U/mg) with whole cell catalytic activity (2473 U/g wet cells) using 8a as a substrate, the total number of units of EH activity for the expression (3710 U ) with for the purification (2244 U), observing the result of protein purification, we found that the expression of RpEH in E. coli was very successful and the expression quantity was high. That makes it possible to prepare chiral epoxides and diols at high substrate concentrations using E. coli/Rpeh wet cells and avoids cumbersome purification process. Furthermore, RpEH maintained high activity over more wide pH and temperature ranges (Figs. 3 and 4) and had good stability in those range. These properties are important in industrial applications (Zheng and Xu 2011), revealing the enormous potential of RpEH.
In this study, we achieved near-perfect kinetic resolution in a simple phosphate buffer system. A small amount of E. coli/Rpeh (10 mg/ml wet cells) could catalyze the asymmetric hydrolysis of rac-8a at high concentrations (1000 mM), giving (R)-8a with more than 99% enantiomeric excess (ees) and producing (S)-8b with 93.2% eep. By comparing with reported native EHs (Table 3), we found that the STY for producing (R)-8a was the highest in our knowledge. We achieved preparation of optically pure (R)-8a (1.89 g) and (S)-8b (2.2 g) in a amplification system, which is composed of 25 mL pH 7.0 phosphate buffer, 25 mmol rac-8a, and 0.25 g E. coli/Rpeh wet cells. Under normal circumstances, when the kinetic resolution is carried out under high concentration conditions, many problems will seriously affect the progress of the reaction, like substrate inhibition and product inhibition. In order to overcome these problems and enable EH to be used in industrial production, several approaches were developed to overcome stringent bottlenecks, like using water-miscible cosolvent, water-immiscible biphasic system (Deregnaucourt et al. 2007), and hollow fiber membrane bioreactor–based aqueous/organic biphasic system (Gao et al. 2017). However, these methods will bring about a rising cost and inevitably introduce a large amount of organic solvents that are difficult to be processed, which is contrary to the concept of green chemistry. For example, in the water-immiscible biphasic system, n-hexane is often used as the organic phase to reduce product inhibition. But, it is unrealistic to use large amounts of this solvent in industrial production, because n-hexane is expensive, toxic, and will be miscible with the products making it difficult to separate. E. coli/Rpeh made it possible to perform high-concentration kinetic resolution without complex systems.
In conclusion, we successfully cloned and expressed a novel EH (RpEH) from R. paludigena JNU001, which was identified by 26S rDNA sequence analysis. The substrate spectrum of expressed RpEH showed that the transformant E. coli/Rpeh had excellent enantioselectivity to 2a (E > 200), 3a (E = 64), and 5a–10a (E = 49–160), among which E. coli/Rpeh had the highest activity (2473 U/g wet cells) for 8a, and its regioselectivity coefficients, αR and βS, toward (R)- and (S)-8a were 99.7 and 83.2%, respectively. The resultant Km, Vmax, kcat, and kcat/Km of purified RpEH toward rac-8a were 8.35 mM, 57.2 U/mg, 46.1 s−1, and 5.5 mM−1 s−1, respectively. Furthermore, we used 8a as a model substrate to study the application potential of E. coli/Rpeh in industrial production. Using only 10 mg wet cells/mL of E. coli/Rpeh, the near-perfect kinetic resolution of rac-8a at a high concentration (1000 mM) was achieved within 2.5 h, giving (R)-8a with more than 99% enantiomeric excess (ees) and 46.7% yield and producing (S)-8b with 93.2% eep and 51.4% yield with high STY for (R)-8a and (S)-8b were 30.6 and 37.3 g/L/h. We also achieved efficient scale preparation of optically pure (R)-8a (1.89 g, isolated yield 46.0%; ees > 99.0%) and (S)-8b (2.2 g, isolated yield 48.2%; eep 92.8%) in 25 mL phosphate buffer (pH 7.0) at high substrate concentration (1000 mM) by using a small amount of E. coli/Rpeh wet cells (0.25 g). Therefore, the E. coli/Rpeh, which can produce desired epoxides and vicinal diols in high enantiomeric excess (ee), high yield, and high concentration at low catalyst loading, is an attractive biocatalyst for potential utilization in chiral synthesis.
References
Archelas A, Iacazio G, Kotik M (2016) Epoxide hydrolases and their application in organic synthesis. In R N Patel (ed) Green biocatalysis. Wiley, New York, pp 210–216
Bala N, Chimni SS (2010) Recent developments in the asymmetric hydrolytic ring opening of epoxides catalysed by microbial epoxide hydrolase. Tetrahedron-Asymmetry 21(24):2879–2898. https://doi.org/10.1016/j.tetasy.2010.11.013
Barth S, Fischer M, Schmid RD, Pleiss J (2004) Sequence and structure of epoxide hydrolases: a systematic analysis. Proteins 55(4):846–855. https://doi.org/10.1002/prot.20013
Basith S, Manavalan B, Lee G, Kim SG, Choi S (2011) Toll-like receptor modulators: a patent review (2006–2010). Expert Opin Ther Patents 21(6):927–944. https://doi.org/10.1517/13543776.2011.569494
Bisi A, Rampa A, Budriesi R, Gobbi S, Belluti F, Ioan P, Valoti E, Chiarini A, Valenti P (2003) Cardiovascular hybrid drugs: new benzazepinone derivatives as bradycardic agents endowed with selective β1-non-competitive antagonism. Bioorg Med Chem 11(7):1353–1361. https://doi.org/10.1016/s0968-0896(02)00621-1
Botes AL, Weijers CAGM, Van Dyk MS (1998) Biocatalytic resolution of 1,2-epoxyoctane using resting cells of different yeast strains with novel epoxide hydrolase activities. Biotechnol Lett 20(4):421–426
Cambell D, Duron SG (2013) Preparation of 8-ethyl-6-(aryl)-pyrido[2,3-d]pyrimidin-7 (8H)-one compds. as PAK inhibitors useful in treatment of nervous system disorders and cancer. U.S. Patent Appl 2013043232
Choi WJ, Choi CY, De Bont JAM, Weijers CAGM (2000) Continuous production of enantiopure 1,2-epoxyhexane by yeast epoxide hydrolase in a two-phase membrane bioreactor. Appl Microbiol Biotechnol 54(5):641–646. https://doi.org/10.1007/s002530000451
De Carvalho CC (2011) Enzymatic and whole cell catalysis: finding new strategies for old processes. Biotechnol Adv 29(1):75–83. https://doi.org/10.1016/j.biotechadv.2010.09.001
Deregnaucourt J, Archelas A, Barbirato F, Paris J-M, Furstoss R (2007) Enzymatic transformations 63. High-concentration two liquid-liquid phase Aspergillus niger epoxide hydrolase-catalysed resolution: application to trifluoromethyl-substituted aromatic epoxides. Adv Synth Catal 349(8–9):1405–1417. https://doi.org/10.1002/adsc.200700085
Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A (2000) Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol 50(3):1351–1371. https://doi.org/10.1099/00207713-50-3-1351
Gao P, Wu S, Praveen P, Loh KC, Li Z (2017) Enhancing productivity for cascade biotransformation of styrene to (S)-vicinal diol with biphasic system in hollow fiber membrane bioreactor. Appl Microbiol Biotechnol 101(5):1857–1868. https://doi.org/10.1007/s00253-016-7954-1
Hu D, Tang C, Li C, Kan T, Shi X, Feng L, Wu M (2017) Stereoselective hydrolysis of epoxides by reVrEH3, a Novel Vigna radiata epoxide hydrolase with high enantioselectivity or high and complementary regioselectivity. J Agric Food Chem 65(45):9861–9870. https://doi.org/10.1021/acs.jafc.7b03804
Hwang S, Choi CY, Lee EY (2010) Bio- and chemo-catalytic preparations of chiral epoxides. J Ind Eng Chem 16(1):1–6. https://doi.org/10.1016/j.jiec.2010.01.001
Kotik M, Kyslík P (2006) Purification and characterisation of a novel enantioselective epoxide hydrolase from Aspergillus niger M200. Biochim Biophys Acta-Gen Subj 1760(2):245–252. https://doi.org/10.1016/j.bbagen.2005.11.002
Kotik M, Štěpánek V, Grulich M, Kyslík P, Archelas A (2010) Access to enantiopure aromatic epoxides and diols using epoxide hydrolases derived from total biofilter DNA. J Mol Catal B-Enzym 65(1–4):41–48. https://doi.org/10.1016/j.molcatb.2010.01.016
Kotik M, Archelas A, Wohlgemuth R (2012) Epoxide hydrolases and their application in organic synthesis. Curr Org Chem 16(4):451–482. https://doi.org/10.2174/138527212799499840
Li C, Hu D, Zong X-C, Deng C, Feng L, Wu M-C, Li J-F (2017) Asymmetric hydrolysis of styrene oxide by Pv EH2, a novel Phaseolus vulgaris epoxide hydrolase with extremely high enantioselectivity and regioselectivity. Catal Commun 102:57–61. https://doi.org/10.1016/j.catcom.2017.08.026
Li F-L, Kong X-D, Chen Q, Zheng Y-C, Xu Q, Chen F-F, Fan L-Q, Lin G-Q, Zhou J, Yu H-L, Xu J-H (2018) Regioselectivity engineering of epoxide hydrolase: near-perfect enantioconvergence through a single site mutation. ACS Catal 8(9):8314–8317. https://doi.org/10.1021/acscatal.8b02622
Lin H, Liu J-Y, Wang H-B, Ahmed AAQ, Wu Z-L (2011) Biocatalysis as an alternative for the production of chiral epoxides: a comparative review. J Mol Catal B-Enzym 72(3–4):77–89. https://doi.org/10.1016/j.molcatb.2011.07.012
Lind MES, Himo F (2016) Quantum chemical modeling of enantioconvergency in soluble epoxide hydrolase. ACS Catal 6(12):8145–8155. https://doi.org/10.1021/acscatal.6b01562
Matsumoto M, Sugimoto T, Ishiguro Y, Yamaguchi H, Kondo K (2014) Effect of organic solvents and ionic liquids on resolution of 2-epoxyhexane by whole cells of Rhodotorula glutinis in a two-liquid phase system. J Chem Technol Biotechnol 89(4):522–527. https://doi.org/10.1002/jctb.4148
Pieper AA, Xie S, Capota E, Estill SJ, Zhong J, Long JM, Becker GL, Huntington P, Goldman SE, Shen CH, Capota M, Britt JK, Kotti T, Ure K, Brat DJ, Williams NS, MacMillan KS, Naidoo J, Melito L, Hsieh J, De Brabander J, Ready JM, McKnight SL (2010) Discovery of a proneurogenic, neuroprotective chemical. Cell 142(1):39–51. https://doi.org/10.1016/j.cell.2010.06.018
Reetz MT, Bocola M, Wang L-W, Sanchis J, Cronin A, Arand M, Zou J, Archelas A, Bottalla A-L, Naworyta A, Mowbray SL (2009) Directed evolution of an enantioselective epoxide hydrolase: uncovering the source of enantioselectivity at each evolutionary stage. J Am Chem Soc 131(21):7334–7343. https://doi.org/10.1021/ja809673d
Serrano-Hervas E, Garcia-Borras M, Osuna S (2017) Exploring the origins of selectivity in soluble epoxide hydrolase from Bacillus megaterium. Org Biomol Chem 15(41):8827–8835. https://doi.org/10.1039/c7ob01847a
Stephenson KA, Wilson AA, Meyer JH, Houle S, Vasdev N (2008) Facile radiosynthesis of fluorine-18 labeled β-blockers. Synthesis, radiolabeling, and ex vivo biodistribution of [18F]-(2S and 2R)-1-(1-fluoropropan-2-ylamino)-3-(m-tolyloxy)propan-2-ol. J Med Chem 51(16):5093–5100. https://doi.org/10.1021/jm800227h
Sun Z, Lonsdale R, Wu L, Li G, Li A, Wang J, Zhou J, Reetz MT (2016) Structure-guided triple-code saturation mutagenesis: efficient tuning of the stereoselectivity of an epoxide hydrolase. ACS Catal 6(3):1590–1597. https://doi.org/10.1021/acscatal.5b02751
Visser H, Weijers CAGM, van Ooyen AJJ, Verdoes JC (2002) Cloning, characterization and heterologous expression of epoxide hydrolase-encoding cDNA sequences from yeasts belonging to the genera Rhodotorula and Rhodosporidium. Biotechnol Lett 24(20):1687–1694. https://doi.org/10.1023/a:1020613803342
Weijers CAGM (1997) Enantioselective hydrolysis of aryl, alicyclic and aliphatic epoxides by Rhodotorula glutinis. Tetrahedron-Asymmetry 8(4):639–647. https://doi.org/10.1016/S0957-4166(97)00012-8
Weijers CAGM, Botes AL, van Dyk MS, de Bont JAM (1998) Enantioselective hydrolysis of unbranched aliphatic 1,2-epoxides by Rhodotorula glutinis. Tetrahedron-Asymmetry 9(3):467–473. https://doi.org/10.1016/S0957-4166(97)00639-3
Woo JH, Kang JH, Hwang YO, Cho JC, Kim SJ, Kang SG (2010) Biocatalytic resolution of glycidyl phenyl ether using a novel epoxide hydrolase from a marine bacterium, Maritimibacter alkaliphilus KCCM 42376. J Biosci Bioeng 109(6):539–544. https://doi.org/10.1016/j.jbiosc.2009.11.019
Wu S, Li A, Chin YS, Li Z (2013) Enantioselective hydrolysis of racemic and meso-epoxides with recombinant Escherichia coli expressing epoxide hydrolase from Sphingomonas sp. HXN-200: preparation of epoxides and vicinal diols in high ee and high concentration. ACS Catal 3(4):752–759. https://doi.org/10.1021/cs300804v
Xu Y, Xu J-H, Pan J, Tang Y-F (2004) Biocatalytic resolution of glycidyl aryl ethers by Trichosporon loubierii: cell/substrate ratio influences the optical purity of (R)-epoxides. Biotechnol Lett 26(15):1217–1221. https://doi.org/10.1023/B:BILE.0000036598.35494.de
Yoo SS, Park S, Lee EY (2008) Enantioselective resolution of racemic styrene oxide at high concentration using recombinant Pichia pastoris expressing epoxide hydrolase of Rhodotorula glutinis in the presence of surfactant and glycerol. Biotechnol Lett 30(10):1807–1810. https://doi.org/10.1007/s10529-008-9762-x
Zhao J, Chu Y-Y, Li A-T, Ju X, Kong X-D, Pan J, Tang Y, Xu J-H (2011) An unusual (R)-selective epoxide hydrolase with high activity for facile preparation of enantiopure glycidyl ethers. Adv Synth Catal 353(9):1510–1518. https://doi.org/10.1002/adsc.201100031
Zheng GW, Xu JH (2011) New opportunities for biocatalysis: driving the synthesis of chiral chemicals. Curr Opin Biotechnol 22(6):784–792. https://doi.org/10.1016/j.copbio.2011.07.002
Zhu QQ, He WH, Kong XD, Fan LQ, Zhao J, Li SX, Xu JH (2014) Heterologous overexpression of Vigna radiata epoxide hydrolase in Escherichia coli and its catalytic performance in enantioconvergent hydrolysis of p-nitrostyrene oxide into (R)-p-nitrophenyl glycol. Appl Microbiol Biotechnol 98(1):207–218. https://doi.org/10.1007/s00253-013-4845-6
Zou SP, Zheng YG, Wu Q, Wang ZC, Xue YP, Liu ZQ (2018) Enhanced catalytic efficiency and enantioselectivity of epoxide hydrolase from Agrobacterium radiobacter AD1 by iterative saturation mutagenesis for (R)-epichlorohydrin synthesis. Appl Microbiol Biotechnol 102(2):733–742. https://doi.org/10.1007/s00253-017-8634-5
Acknowledgments
The authors are grateful to Prof. Xianzhang Wu (School of Biotechnology, Jiangnan University, Jiangsu, China) for providing technical assistance.
Funding
This work was financially supported by the China Postdoctoral Science Foundation (No. 2018M630522) and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_1804).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 710 kb)
Rights and permissions
About this article
Cite this article
Xu, XF., Hu, D., Hu, BC. et al. Near-perfect kinetic resolution of o-methylphenyl glycidyl ether by RpEH, a novel epoxide hydrolase from Rhodotorula paludigena JNU001 with high stereoselectivity. Appl Microbiol Biotechnol 104, 6199–6210 (2020). https://doi.org/10.1007/s00253-020-10694-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10694-w