Abstract
Previously, we reported that ten strains belonging to Erythrobacter showed epoxide hydrolase (EHase) activities toward various epoxide substrates. Three genes encoding putative EHases were identified by analyzing open reading frames of Erythrobacter litoralis HTCC2594. Despite low similarities to reported EHases, the phylogenetic analysis of the three genes showed that eeh1 was similar to microsomal EHase, while eeh2 and eeh3 could be grouped with soluble EHases. The three EHase genes were cloned, and the recombinant proteins (rEEH1, rEEH2, and rEEH3) were purified. The functionality of purified proteins was proved by hydrolytic activities toward styrene oxide. EEH1 preferentially hydrolyzed (R)-styrene oxide, whereas EEH3 preferred to hydrolyze (S)-styrene oxide, representing enantioselective hydrolysis of styrene oxide. On the other hand, EEH2 could hydrolyze (R)- and (S)-styrene oxide at an equal rate. The optimal pH and temperature for the EHases occurred largely at neutral pHs and 40–55 °C. The substrate selectivity of rEEH1, rEEH2, and rEEH3 toward various epoxide substrates were also investigated. This is the first representation that a strict marine microorganism possessed three EHases with different enantioselectivity toward styrene oxide.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epoxide hydrolases (EHases; EC 3.3.2.3) are ubiquitous enzymes that have been isolated from a wide variety of sources such bacteria, yeast, fungi, insect, plant, and mammals (Archelas and Furstoss 2001; Weijers and de Bont 1999) and hydrolyze an epoxide to its corresponding vicinal diol with the addition of a water molecule to the oxirane ring (Weijers and de Bont 1999). Enantiopure epoxides and vicinal diols are versatile synthetic intermediates for the preparation of enantiopure bioactive compounds (Archelas and Furstoss 1997). Because of the potential application in the production of enantiopure epoxides by kinetic resolution of enantioselective EHase, several EHases from microbial sources have been developed (Tokunaga et al. 1997). However, the limited number of enantioselective EHase demands studies to explore new enantioselective EHase for the production of enantiopure epoxides in pharmaceutical industries.
Most EHase are members of the α/β-hydrolase fold family (Nardini and Dijkstra 1999; Ollis et al. 1992; Rink et al. 1999), which includes lipases, esterases, and haloalkane dehalogenases (Nardini and Dijkstra 1999; van Loo et al. 2006). α/β-Domains consist of a central, parallel, or mixed β-sheet surrounded by α-helices with a variable cap domain sitting on top (Ollis et al. 1992). These enzymes characteristically employ a two-step mechanism in which a catalytic nucleophile of the enzyme attacks a polarized electrophile substrate of the covalent intermediate, followed by hydrolysis (Yamada et al. 2000). The conserved catalytic triad of α/β-hydrolase fold enzymes consists of a nucleophilic residue (Asp or Ser or Cys), an acidic residue (Asp or Glu), and a conserved histidine residue (Arand et al. 1996). The nucleophile fits the conserved amino-acid-sequence motif, Sm-X-Nu-X-Sm-Sm (Sm = small residue, X = any residue, and Nu = nucleophile). Another conserved amino acid sequence is the human genome expression profile HGXP motif containing the oxyanion hole of the enzyme (Ollis et al. 1992). The active site of EHase further contains two tyrosine located in the cap domain, which are involved in substrate binding and assist in the ring opening of the epoxide by acting as a proton donor to the epoxide oxygen (Rink et al. 1999, 2000).
Oceans cover more than three quarters of the earth’s surface and so offer abundant resources for biotechnological research and development. Marine organisms represent a dramatically different environment for biosynthesis than do terrestrial organisms and therefore represent a vast untapped resource with potential benefits in many different areas such as medicine, aquaculture and fisheries, industry, research tools, and environmental applications. Marine organisms, in particular, represent great phylogenetic diversity, making them reservoirs of unique genetic information and important natural resources for possible development (Venter et al. 2004). Furthermore, the genomic sequencing of marine microorganisms mostly made by Moore foundation (www.moore.org) can facilitate a rapid cloning and overexpression for the characterization of a putative or possible EHase originated from the marine environment as a recent report on the screening of various genomic databases for EHases of the α/β-hydrolase fold family (van Loo et al. 2006). Previously, we reported that the Erythrobacter clan could be resources for screening enantioselective EHases (unpublished data). In this paper, we searched for probable EHase genes from E. litoralis HTCC2594. As a result, three EHases from E. litoralis HTCC2594 were cloned and characterized. The activity toward epoxide substrates and the mechanism for enantioselectivity were also considered.
Materials and methods
Materials
The epoxides used in this study are indicated in Fig. 1. Racemic styrene oxide was purchased from Fluka. Pure (R)-styrene oxide, pure (S)-styrene oxide, and all other racemic epoxides were purchased from Aldrich. All materials were of analytical or of reagent grade. The chiraldex γ-cyclodextrin trifluoroacetyl (G-TA) capillary gas chromatography (GC) column was purchased from Astec (Whippany, NJ). Other medium components were purchased Merck and Difco.
Strains and growth conditions
E. litoralis HTCC2594 was cultured at 30°C in ZoBell 2216E broth (Oppenheimer and ZoBell 1952) consisting of 0.5% peptone, 0.1% yeast extract, and 75% seawater (pH 7.5) for 1 day. For the storage, the bacterial cells were suspended in ZoBell 2216E broth with 20% glycerol and stored at −80 °C until used.
Escherichia coli DH5α and E. coli BL21-CodonPlus(DE3)-RIL cells (Stratagene, LaJolla, CA) were used for plasmid propagation and gene expression, respectively. E. coli strains were cultured in Luria–Bertani medium at 37 °C, and an appropriate antibiotic was added.
DNA manipulation and DNA sequencing
DNA manipulations were performed using standard procedures (Sambrook and Russell 2001). Restriction enzymes and other modifying enzymes were purchased from Promega (Madison, WI). Small-scale preparation of plasmid DNA from E. coli cells was performed with a plasmid mini-kit (Qiagen, Hilden, Germany). DNA sequencing was performed with an automated sequencer (ABI3100) using a BigDye terminator kit (PE Applied Biosystems, Foster City, CA).
BLAST search and multiple sequence alignments
To clone EHases from E. litoralis HTCC2594, sequence searches (Sm-X-Nu-X-Sm-Sm motif and H-G-X-P) against open reading frames (ORFs) of E. litoralis HTCC2594 whose genome sequence was determined by Moore foundation (www.moore.org) were performed using the ProteinFinder program of Ensoltek (www.ensoltek.com) and the basic local alignment search tool (BLAST) program. The pairwise comparison of the candidate EHase and reported EHases were performed with the CLUSTAL W program (Thompson et al. 1994). The resulting candidates were manually confirmed for the presence of the putative EHase active-site residues. Sequences that contained ring-opening tyrosine, HGXP motif, and Sm-X-Nu-X-Sm-Sm motif were selected and aligned together with the known EHase sequences.
Phylogenetic analysis
For phylogenetic analysis, reported EHase sequences were retrieved from the SwissProt or European Molecular Biology Laboratory protein database and analyzed in comparison with eeh1, eeh2, and eeh3. Phylogenetic distances were calculated by using the CLUSTAL W program, and phylogenetic trees were drawn by the Molecular Evolutionary Genetics Analysis 3.1 software (The Biodesign Institute, Tempe, AZ; Kumar et al. 2004).
Cloning of eeh genes from E. litoralis HTCC2594
Genomic DNA of E. litoralis HTCC2594 was isolated using the Genomic DNA extraction kit (Promega) following the manufacturer’s instructions. The full-length of eeh1, eeh2, and eeh3 genes flanked by NdeI and XhoI/NotI sites was amplified by a polymerase chain reaction (PCR) with the forward primers (eeh1F: 5′-CGACCCGGCATATGAGCGAGATCAGGCCCTTCGTTCT-3′; eeh2F: 5′-CGACC-CGGCATATGGCCGGACCAAGCCTGGGCGAATGG-3′; eeh3F: 5′-CGACCCGG-CATATGCCCGATCCTGCGAGCGGGATT-3′) and reverse primers (eeh1R: 5′-CTCCACATCTCGAGTCGCATGAGTGAAAAACAGGCGCG-3′; eeh2R: 5′-CTCCACATCTCGAGGCGTGCGAGCCAATCCAGCGTCACGC-3′; eeh3R: 5′-CT-CCACATGCGGCCGCGGATGCCGGAGCGGGCTTAGG-3′). The sequences in italics indicate the NdeI site in the forward primer and the XhoI/NotI site in the reverse primer. For the expression of eeh1, eeh2, and eeh3 without the His-tag, the reverse primers (eeh1RX: 5′-CTCCACATCTCGAGCTATCGCATGAGTGAAAAACAGGC-3′; eeh2RX: 5′-CTCCACATCTCGAGTTAGCGTGCG-AGCCAATCCAGCGTCACGC-3′; eeh3RX: 5′-CTCCACATGCGGCCGCTCAG-GATGCCGGAGCGGGCTTAG-3′) were also designed. The amplified DNA fragment was digested with NdeI and XhoI/NotI, the fragment was ligated to NdeI/XhoI- or NdeI/NotI-digested plasmid pET-24a (+), and then the recombinant plasmid was used to transform E. coli DH5α. The recombinant plasmid was introduced into BL21-CodonPlus (DE3)-RP (Novagen) for expression after sequence confirmation.
Expression of eeh genes from E. litoralis HTCC2594
A transformant was cultivated at 37 °C, and overexpression was induced at 37 °C by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside when the optical density at 600 nm reached 0.4–0.6. After induction for 3 h, the cells were harvested by centrifugation at 5,000 × g for 20 min, resuspended in a buffer (50 mM phosphate [pH 7.0], 0.5 M KCl and 10% glycerol), and disrupted by sonication. Cell debris was removed by centrifugation at 15,000 × g for 30 min, with a His∙Bind Purification Kit (Novagen). The soluble fraction was applied to a Ni–nitrilotriacetic column equilibrated with a binding buffer (500 mM NaCl, 20 mM phosphate [pH 7.0], and 5 mM imidazole). After washing with a washing buffer (500 mM NaCl, 20 mM phosphate [pH 7.0], and 60 mM imidazole), the bound enzyme was eluted with an elution buffer (500 mM NaCl, 20 mM phosphate [pH 7.0], and 1 M imidazole) and then dialyzed against 50 mM phosphate buffer (pH 7.0). The purity of the protein was examined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) under denaturing conditions as described by Laemmli 1970. The protein concentration was measured by the method of Bradford (1976) using the Bio-Rad protein assay kit with bovine serum albumin as a standard.
EHase assay
EHase activity was determined by a spectrophotometric assay based on the extraction of the epoxide from the reaction mixture, followed by spectrometric quantification of the nonextracted diol (Bhatnagar et al. 2001). Four millimolars of styrene oxide (100 mM stock in dimethyl formamide [DMF]) was mixed with 100 μl of purified EHase in a buffer (10 mM sodium phosphate [pH 6.8]), and the mixture was incubated for 15 min at 30 °C. Then, 40 μl of the NaIO4 stock solution (200 mM stock in DMF) was added and immediately vortexed for 2 min. After centrifugation at 16,500 × g for 90 s, the supernatant was quantified at 290 nm.
The measurement of enantioselective EHase activity was performed by a GC analysis as follows. One hundred microliters of purified EHase showing EHase activity in the spectrophotometric measurement was mixed with 2 mM styrene oxide in a 10-ml vial containing 1 ml of 100 mM Tris–HCl (pH 8.0) and incubated at 30°C. Then, the samples were withdrawn periodically during incubation, the reaction mixtures were extracted with hexane (2 ml), and the extracts were analyzed on a chiraldex G-TA capillary GC column (0.25 mm inside diameter, 30 m length; van Loo et al. 2004) using a GC system equipped with a flame ionization detection detector (Hewlett-Packard, Avondale, PA). The temperatures of the oven, injector, and detector in GC analysis for racemic styrene oxide were 90, 220, and 230 °C, respectively, and the carrier gas was He.
Effects of pH and temperature on EHase activity
The pH dependence of EHase activity was investigated with the following buffers: 50 mM sodium acetate–acetic acid buffer (pH 4.0 and 6.0), 50 mM 2-(N-morpholino)ethanesulfonic acid buffer (pH 6.0 to 7.0), 50 mM phosphate buffer (pH 7.0 to 9.0), and 50 mM glycine buffer (pH 9.0 and 10.0). For determination of the optimal reaction temperature, EHase activities were measured at pH 7.5 over a temperature range of 10 to 70 °C.
Determination of kinetic parameters
Kinetic parameters of rEEH1, rEEH2, and rEEH3 were determined by a GC analysis using (R) or (S)-styrene oxide as a substrate. One hundred microliters of purified EHases were mixed with various concentrations of (R) or (S)-styrene oxide in a 10-ml vial containing 1 ml of the 100 mM Tris–HCl (pH 8.0) and incubated at 30 °C with shaking at 200 rpm. The reaction mixtures were extracted with 2 ml hexane, and enantiomeric excess (ee; ee = 100 × (S − R)/(S + R)) for enantiopure styrene oxide was analyzed on a chiraldex G-TA capillary GC column. Kinetic parameters were estimated by nonlinear regression using a Sigma Plot program. Various substrates as shown Fig. 1 were also tested for the enantioselective hydrolysis by rEEH1, rEEH2, and rEEH3.
Results
Identification and phylogenetic analysis of the EHase genes from E. litoralis HTCC2594
Previously, we reported that ten strains belonging to Erythrobacter clan showed EHase activity toward various epoxide substrates (unpublished data). To characterize EHases from Erythrobacter, E. litoralis HTCC2594 was selected because whole genomic sequencing of the strain is under progress by Moore foundation, facilitating rapid cloning and characterization. The whole cell of E. litoralis HTCC2594 displayed the hydrolyzing activity toward styrene oxide, implicating that the strain could also retain EHases (data not shown). By analyzing the ORFs of E. litoralis HTCC2594 as described in “Materials and methods,” three genes consisting of 1,122 bp (eeh1; GenBank accession number YP_457985), 870 bp (eeh2; GenBank accession number YP_458376), and 888 bp (eeh3; GenBank accession number YP_458350) were selected as EHase candidates. The sequence analysis of selected ORFs showed that Sm-X-Nu-X-Sm-Sm motif, catalytic triad, and oxyanion hole shared in most of EHases could be found (Figs. 2 and 3). Firstly, eeh1 showed similarity to human microsomal EHase (35%) and retained GGD173WGS motif, catalytic triad (Asp173, Glu324, and His351), and oxyanion hole HGXP (HGW99P). Secondly, eeh2 and eeh3 showed low similarity (below 30%) to soluble EHases from mammals, plants, or bacteria, retaining Sm-X-Nu-X-Sm-Sm motif (VHD107YGV for eeh2, AHD106WGA for eeh3), catalytic triad (Asp107, Glu250, and His269 for eeh2, Asp106, Glu251, and His270 for eeh3), and oxyanion hole HGXP (HGY42P for eeh2, HGF38P for eeh3) conserved in EHases (Arahira et al. 2000; Kaneko et al. 2002; Knehr et al. 1993; Stapleton et al. 1994; Strausberg et al. 2002; Fig. 3) could be found.
Sequence alignment of EHase. The protein accession numbers are: Rhodotorula glutinis (EPH1), AAF64646; Rattus norvegicus (Ephx1), P07687; Homo sapiens (EPHX1), AAH08291; Xanthophyllomyces dendrorhous (Eph1), AAF18956; Aspergillus niger (hyl1), CAB59813; Erythrobacter litoralis HTCC2594 (EEH1, this paper). The identical, conserved, and conserved residues are highlighted by nucleophilic residue, acidic residue, oxyanion hole, and histidine. Regions of putative motif are boxed. The amino acid sequence corresponding to the equivalent positions to the two tyrosines of active site motif is underlined, and the residues supporting two tyrosines putative motif are italicized
Sequence alignment of EHase. The protein accession numbers are: Homo sapiens (EPHX2, Human sEH), AAH11628; Rattus norvegicus (Ephx2, Rat sEH), CAA46211; Solanum tuberosum (pEHSt, potato; sEH), AAA81890; Glycine max (sEHGm, soybean sEH), CAA55293; Bradyrhizobium japonicum (ephA, BAC46379); Erythrobacter litoralis HTCC2594 (EEH2, this paper); Erythrobacter litoralis HTCC2594 (EEH3, this paper). The identical, conserved, and conserved residues are highlighted by nucleophilic residue, acidic residue, oxyanion hole, and histidine. Regions of putative motif are boxed. The amino acid sequence corresponding to the equivalent positions to the two tyrosines of active site motif is underlined
A phylogenetic analysis of the three ORFs with various EHases was conducted based on the neighbor-joining method as shown in Fig. 4. Obviously, eeh1 could be grouped together with microsomal EHases while eeh2 and eeh3 were related to soluble EHases (Fig. 4). eeh1 was lacking in the common membrane anchor found in most members of microsomal EHases (mEHases). eeh2 and eeh3 were also lacking in the common N-terminal domain of the mammalian sEHase; however, the genes displayed an overall homology to C-terminal domains of plant and mammalian sEHases (Beetham et al. 1995; Morisseau et al. 2000), suggesting these EHases are structurally and mechanistically similar. Taken together, it seemed likely that the ORFs were EHases responsible for the activity.
Phylogenetic analysis of EHase. Sequence alignment on the desired amino acid sequence was performed using the CLUSTAL W software package. Rhodotorula glutinis (EPH1; Visser et al. 2000; AAF64646), Rattus norvegicus (Ephx1, Rat mEH; Falany et al. 1987; P07687), Homo sapiens (EPHX1, Human mEH; Strausberg et al. 2002; AAH08291), Xanthophyllomyces dendrorhous (Eph1; Visser et al. 1999; AAF18956), Aspergillus niger (hyl1; Arand et al. 1999; CAB59813), Homo sapiens (EPHX2, Human sEH; Strausberg et al. 2002; AAH11628), Rattus norvegicus (Ephx2, Rat sEH; Knehr et al. 1993; CAA46211), Solanum tuberosum (pEHSt, potato; sEH; Stapleton et al. 1994; AAA81890), Glycine max (sEHGm, soybean sEH; Arahira et al. 2000; CAA55293), Agrobactrium radiobacter sEH (Rink et al. 1997; O31243), Corynebacterium sp. sEH (Misawa et al. 1998; O52866), and Haloalkane dehalogenase (Janssen et al. 1989; P22643)
Cloning and expression of the EHase genes from E. litoralis HTCC2594
To confirm the functionality of eeh1, eeh2, and eeh3 genes, the full ORFs were amplified by PCR, and the recombinant enzymes (rEEH1, rEEH2, and rEEH3) were purified as described above. To facilitate the rapid purification, a His-tag was inserted at the C terminus (C-rEEH1, C-rEEH2, and C-rEEH3) of the expressed polypeptide. The His-tagged rEEH1, rEEH2, and rEEH3 could be purified to an apparent homogeneity by His-tag-affinity chromatography. SDS-PAGE analysis of the purified rEEH1, rEEH2, and rEEH3 showed a single band with an apparent mass of 41, 33.4, and 34.5 kDa, respectively (Fig. 5a and b).
Purification of the recombinant EHases (the purified rEEH1 a, rEEH2, and rEEH3 b). EHases were purified from recombinant E. coil cell by His-tag affinity column. aLane M, the protein size standard; lane 1, the soluble cell lysate; lane 2, the purified rEEH1. bLane M, the protein size standard; lane 1, the purified rEEH2; lane 2, the purified rEEH3
Effects of pH and temperature on the EHase activity
The activity of the purified rEEH1, rEEH2, and rEEH3 was determined by measuring the hydrolysis of styrene oxide, and the three enzymes could hydrolyze styrene oxide, proving the functionality. The effects of pH on the EHases (rEEH1, rEEH2, and rEEH3) activity were investigated by varying pH from 4.0 to 10.0. Optimum activity of rEEH1, rEEH2, and rEEH3 toward styrene oxide occurred at pH 6.5, 7.5, and 8.0, respectively (Fig. 6a). The EHases were stable largely at neutral pHs but unstable under pH 6.0 (data not shown).
Effects of pH and temperature on the purified rEEH1, rEEH2, and rEEH3 activity. a Enzyme activity was determined toward 2 mM styrene oxide in 50 mM sodium acetate–acetic acid buffer (pH 4.0–6.0), 50 mM MES buffer (pH 6.0–7.0), 50 mM Tris–HCl buffer (pH 7.0–9.0), and 50 mM Glycine buffer (pH 9.0 and 10.0) at 45 °C. b Enzyme activity was between 10 and 70 °C determined in 50 mM Tris–HCl buffer (pH 7.5). Filled circles, rEEH1; empty circles, rEEH2; filled inverted triangles, rEEH3
The effect of temperature on the activity of the EHases (rEEH1, rEEH2, and rEEH3) was determined in the range of 10–70 °C. The hydrolysis rate of rEEH1, rEEH2, and rEEH3 toward styrene oxide occurred at 50, 55, and 45 °C, respectively. EHase activity increased as the temperature were increased from 10 to 50 °C, then sharply decreased above the optimum temperature (Fig. 6b).
Catalytic parameters and substrate selectivity
To determine enantioselective hydrolyzing activity of the purified enzymes, hydrolysis rates of rEEH1, rEEH2, and rEEH3 toward (S) or (R)-enantiopure styrene oxide were determined (Fig. 7), and kinetic parameters (V max, K m, and k cat) were determined by nonlinear regression using a Sigma Plot program (Table 1). V max R and K m R of the purified rEEH1 toward (R)-styrene oxide were 285.7 μmol min−1 mg−1 and 16.0 mM, respectively, while V max S and K m S of rEEH1 toward (S)-styrene oxide were 147.4 μmol min−1 mg−1 and 10.9 mM, respectively, indicating that (R)-styrene oxide is hydrolyzed twofold faster than (S)-styrene oxide. The faster hydrolyzing rate toward an enantiomer might be the reason why rEEH1 is enantioselectively hydrolyzing styrene oxide. In contrast, V max S and K m S of the purified rEEH3 toward (S)-styrene oxide were 5.81 μmol min−1 mg−1 and 14.04 mM, respectively, while V max R and K m R of rEEH3 toward (R)-styrene oxide were 1.88 μmol min−1 mg−1 and 12.29 mM, respectively, favoring the hydrolysis of (S)-styrene oxide than (R)-styrene oxide. On the other hand, V max S and K m S of the purified rEEH2 toward (S)-styrene oxide were 4.03 μmol min−1 mg−1 and 9.90 mM, respectively, while V max R and K m R of rEEH2 toward (R)-styrene oxide were 3.90 μmol min−1 mg−1 and 12.88 mM, respectively, indicating that rEEH2 could hydrolyze both of (S)-styrene oxide and (R)-styrene oxide at an equal rate (Table 1 and Fig. 7). Consequently, it is intriguing that E. litoralis HTCC2594 retained three EHases showing different enantioselectivity toward styrene oxide even if the cellular localization, endogenous substrate, or physiological function of three EHases were not yet understood. Nonetheless, the calculation of catalytic efficiency (k cat/K m) of rEEH1, rEEH2, and rEEH3 indicates that the hydrolyzing activity of rEEH1 was prevalent, showing approximately 150- to 740-fold much higher than rEEH2 and rEEH3 (Table 1). It seems likely that the enantioselective activity of whole cell resulted from the dominant activity of rEEH1, although the cellular expression of the proteins or the regulation mechanism needs further investigation.
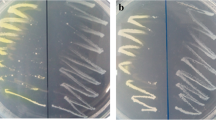
The schematic representation of enantioselective hydrolysis of three EHases (a, rEEH1; b, rEEH2; and c rEEH3) toward racemic styrene oxide (RSO) with GC analysis. Analysis method described in “Materials and methods”. Solid line, RSO a, b, and c; bold solid line, RSO incubated with rEEH1 a; long dashed line, RSO incubated with rEEH2 b; dotted line, RSO incubated with rEEH3 c
The substrate selectivity of rEEH1, rEEH2, and rEEH3 toward various epoxide substrates depicted in Fig. 1 were investigated as shown in Table 2. The purified rEEH1 showed an enantioselective hydrolysis toward monosubstituted epoxides at the C-1 position with a bulky ring such as styrene oxide and glycidyl phenyl ether, whereas both (R)- and (S)-monosubstituted epoxides with aliphatic chains were hydrolyzed equally by rEEH1. It is note to worthy that rEEH1 hydrolyzed preferentially (S)-epifluorohydrin. In contrast, the purified rEEH2 and rEEH3 were not highly enantioselective, although rEEH3 hydrolyzed preferentially (S)-styrene oxide or (R)-epoxyhexane. Several questions arise how EEH1 is differentiating (R)- and (S)-monosubstituted epoxides with a bulky ring, why the enantiomeric preference of EEH3 toward styrene oxide and 1,2-epoxyhexane was different, and so on. Further investigation is under process to address the issues.
Discussion
This study represents the characterization of three EHases from a marine bacterium, E. litoralis HTCC2594. The previous survey on EHase activities of Erythrobacter strains indicated that the Erythrobacter clan would be valuable to screen the EHase activity (unpublished data), and the possibility was proven by this study combining the genomic study and conventional genetic engineering.
We found three genes (eeh1, eeh2, and eeh3) encoding putative EHases by using the ProteinFinder program of Ensoltek (www.ensoltek.com) and the BLAST program of National Center for Biotechnology Information (NCBI) against the whole genome sequence of E. litoralis HTCC2594 (Giovannoni and Stingl 2005; Venter et al. 2004; www.moore.org). Firstly, the deduced sequence of eeh1 showed below 40% similarity to mEHases. The search against nonredundant database at NCBI showed that the homologues could be found from bacteria to a higher mammalian system, claiming that the gene is conserved throughout most of taxa (data not shown). The 38% sequence similarity to the EHase from Aspergillus niger (Arand et al. 1999; Zou et al. 2000) allowed us to rationalize the mechanism of hydrolysis. The residues of catalytic triad can be pointed with great confidence, and Glu108 involved in water activation was found. Tyr231 and Tyr300 appeared in equivalent positions to the two tyrosines of active sites, assisting in ring opening. The residues supporting two tyrosines could be aligned as Gln235 (equivalent to His255) and Trp256 (equivalent to Trp276). As suggested by Zou et al. 2000, a model of the binding of 4-nitrostyrene oxide into an active site could explain why EHases hydrolyze preferentially (R)-enantiomer of monosubstituted oxiran substrates, attacking at the least-hindered carbon of the ring while the binding of (S)-enantiomer is reduced because of the stereo-hindrance in the active site. It seemed that the mechanism might be also true in rEEH1, but the difference only in V max not K m toward (R)- or (S)-styrene oxide suggests that the binding affinity of (S)-enantiomer to rEEH1 may be similar to the (R)-enantiomer probably caused by the changes at Ala177, Ile326, and Tyr224 corresponding to Phe196, Cys350, and Phe244. Nevertheless, the high V max toward (R)-enantiomer could result from optimal arrangement of the epoxide for the catalysis and contributing to the high enantioselectivity.
Secondly, eeh2 and eeh3 genes showed low similarities to soluble EHases including bacteria, mammalian, and plant sEHases. Despite the low similarity, the alignment with the EHase from Agrobacterium radiobacter (Cao et al. 2006; Rink and Janssen 1998) allowed the rEEH2 and rEEH3 to rationalize the mechanism of hydrolysis. Two tyrosine residues assisting in the opening of the epoxide ring through hydrogen bonding could be positioned (Rink et al. 1999, 2000; Rui et al. 2005). Tyr152 and Tyr213 in rEEH3 appeared in equivalent positions to the two tyrosines. Interestingly, the Tyr211 residue in rEEH2 appeared in an equivalent position to the ring opening tyrosines, but the other tyrosine residue could be mutated to Pro156 and Leu157, which may be a reason why EEH2 showed low catalytic efficiency.
Conclusively, E. litoralis HTCC2594 possessed three EHases with different phylogenetic origins. It is intriguing that EEH3 toward (S)-styrene oxide was enantioselective while EEH1 was toward (R)-styrene oxide. The hydrolysis rate of EEH1 toward various epoxide substrates was superior to those of EEH2 or EEH3. There are still questions to be investigated: What could be the physiological roles of three EHases in Erythrobacter; is the presence of three EHases with different origins general in Erythrobacter, understanding the enantioselective hydrolysis of epoxide substrates in EEH1 or EEH3, etc.? This is the first representation that a microorganism, especially a marine microorganism, possesses three EHases. It is not certain whether the three EHases are actually expressed in the original strain. Further investigation is necessary to answer the physiological role of the proteins. The development of novel EHases from marine microorganisms presented in this study emphasize that the marine microorganisms could be valuable natural resources.
References
Arahira M, Nong VH, Udaka K, Fukazawa C (2000) Purification, molecular cloning and ethylene-inducible expression of a soluble-type epoxide hydrolase from soybean (Glycine max [L.] Merr.). Eur J Biochem 267:2649–2657
Arand M, Wagner H, Oesch F (1996) Asp333, Asp495, and His523 form the catalytic triad of rat soluble epoxide hydrolase. J Biol Chem 271:4223–4229
Arand M, Hemmer H, Durk H, Baratti J, Archelas A, Furstoss R, Oesch F (1999) Cloning and molecular characterization of a soluble epoxide hydrolase from Aspergillus niger that is related to mammalian microsomal epoxide hydrolase. Biochem J 344:273–280
Archelas A, Furstoss R (1997) Synthesis of enantiopure epoxides through biocatalytic approaches. Annu Rev Microbiol 51:491–525
Archelas A, Furstoss R (2001) Synthetic applications of epoxide hydrolases. Curr Opin Chem Biol 5:112–119
Beetham JK, Grant D, Arand M, Garbarino J, Kiyosue T, Pinot F, Oesch F, Belknap WR, Shinozaki K, Hammock BD (1995) Gene evolution of epoxide hydrolase and recommended nomenclature. DNA Cell Biol 14:61–71
Bhatnagar T, Manoj KM, Baratti JC (2001) A spectrophotometric method to assay epoxide hydrolase activity. J Biochem Biophys Methods 50:1–13
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cao L, Lee J, Chen W, Wood TK (2006) Enantioconvergent production of (R)-1-phenyl-1,2-ethanediol from styrene oxide by combining the Solanum tuberosum and an evolved Agrobacterium radiobacter AD1 epoxide hydrolases. Biotechnol Bioeng 94:522–529
Falany CN, McQuiddy P, Kasper CB (1987) Structure and organization of the microsomal xenobiotic epoxide hydrolase gene. J Biol Chem 262:5924–5930
Giovannoni SJ, Stingl U (2005) Molecular diversity and ecology of microbial plankton. Nature 437:343–348
Janssen DB, Pries F, Ploeg J, Kazemier B, Terpstra P, Witholt B (1989) Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J Bacteriol 171:6791–6799
Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, Kohara M, Matsumoto M, Shimpo S, Tsuruoka H, Wada T, Yamada M, Tabata S (2002) Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res 9:189–197
Knehr M, Thomas H, Arand M, Gebel T, Zeller HD, Oesch F (1993) Isolation and characterization of a cDNA encoding rat liver cytosolic epoxide hydrolase and its functional expression in Escherichia coli. J Biol Chem 268:17623–17627
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the heal of bacteriophage T4. Nature 227:680–685
Misawa E, Chion CK, Archer IV, Woodland MP, Zhou NY, Carter SF, Widdowson DA, Leak DJ (1998) Characterisation of a catabolic epoxide hydrolase from a Corynebacterium sp. Eur J Biochem 253:173–183
Morisseau C, Beetham JK, Pinot F, Debernard S, Newman JWB, Hammock D (2000) Cress and potato soluble epoxide hydrolases: purification, biochemical characterization, and comparison to mammalian enzymes. Arch Biochem Biophys 378:321–332
Nardini M, Dijkstra BW (1999) α/β hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol 9:732–737
Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, Sussman JL, Verschueren KHG, Goldman A (1992) The α/β hydrolase fold. Protein Eng 5:197–211
Oppenheimer CH, ZoBell CE (1952) The growth and viability of sixty-three species of marine bacteria as influenced by hydrostatic pressure. J Mar Res 11:10–18
Rink R, Janssen DB (1998) Kinetic mechanism of the enantioselective conversion of styrene oxide by epoxide hydrolase from Agrobacterium radiobacter AD1. Biochemistry 37:18119–18127
Rink R, Fennema M, Smids M, Dehmel U, Janssen DB (1997) Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J Biol Chem 272:14650–14657
Rink R, Spelberg JHL, Pieters RJ, Kingma J, Nardini M, Kellogg RM, Dijkstra BW, Janssen DB (1999) Mutation of tyrosine residues involved in the alkylation half reaction of epoxide hydrolase from Agrobacterium radiobacter AD1 results in improved enantioselectivity. J Am Chem Soc 121:7417–7418
Rink R, Kingma J, Spelberg JH, Janssen DB (2000) Tyrosine residues serve as proton donor in the catalytic mechanism of epoxide hydrolase from Agrobacterium radiobacter. Biochemistry 39:5600–5613
Rui L, Cao L, Chen W, Reardon KF, Wood TK (2005) Protein engineering of epoxide hydrolase from Agrobacterium radiobacter AD1 for enhanced activity and enantioselective production of (R)-1-phenylethane-1,2-diol. Appl Environ Microbiol 71:3995–4003
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual (vol 2, 3rd edn.). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 8.65–8.71
Stapleton A, Beetham JK, Pinot F, Garbarino JE, Rockhold DR, Friedman M, Hammock BD, Belknap WR (1994) Cloning and expression of soluble epoxide hydrolase from potato. Plant J 6:251–258
Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer A, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA (2002) Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA 99:16899–16903
Thompson JD, Higgin DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tokunaga M, Larrow JF, Kakiuchi F, Jacobsen EN (1997) Asymmetric catalysis with water: efficient kinetic resolution of terminal epoxides by means of catalytic hydrolysis. Science 277:936–938
van Loo B, Spelberg JHL, Kingma J, Sonke T, Wubbolts MG, Janssen DB (2004) Directed evolution of epoxide hydrolase from A. radiobacter toward higher enantioselectivity by error-prone PCR and DNA shuffling. Chem Biol 11:981–990
van Loo B, Kingma J, Arand M, Wubbolts MG, Janssen DB (2006) Diversity and biocatalytic potential of epoxide hydrolases identified by genome analysis. Appl Environ Microbiol 72:2905–2917
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso sea. Science 304:66–74
Visser H, Bont JAM, Verdoes JC (1999) Isolation and characterization of the epoxide hydrolase-encoding gene from Xanthophyllomyces dendrorhous. Appl Environ Microbiol 65:5459–5463
Visser H, Vreugdenhil S, Bont JAM, Verdoes JC (2000) Cloning and characterization of an epoxide hydrolase-encoding gene from Rhodotorula glutinis. Appl Microbiol Biotechnol 53:415–419
Weijers CAGM, Bont JAM (1999) Epoxide hydrolases from yeasts and other sources: versatile tools in biocatalysis. J Mol Catal, B Enzym 6:199–214
Yamada T, Morisseau C, Maxwell JE, Argiriadi MA, Christianson DW, Hammock BD (2000) Biochemical evidence for the involvement of tyrosine in epoxide activation during the catalytic cycle of epoxide hydrolase. J Biol Chem 275:23082–23088
Zou J, Hallberg BM, Bergfors T, Oesch F, Arand M, Mowbray SL, Jones TA (2000) Structure of Aspergillus niger epoxide hydrolase at 1.8 A resolution: implications for the structure and function of the mammalian microsomal class of epoxide hydrolases. Structure 15:111–122
Acknowledgement
This work was supported by KORDI in-house program (PE97803) and the Marine and Extreme Genome Research Center Program, Ministry of Marine Affairs and Fisheries, Republic of Korea. We would like to appreciate the Gordon and Betty Moore Foundation for the great effort on Microbial Genome Sequencing Project (Moore 155).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Woo, JH., Hwang, YO., Kang, S.G. et al. Cloning and characterization of three epoxide hydrolases from a marine bacterium, Erythrobacter litoralis HTCC2594. Appl Microbiol Biotechnol 76, 365–375 (2007). https://doi.org/10.1007/s00253-007-1011-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-1011-z