Abstract
Methanogenesis from wastewater-borne organics and organic solid wastes (e.g., food residues) can be severely suppressed by the presence of toxic phenols. In this work, ambient (20 °C) and mesophilic (37 °C) methane-producing and phenol-degrading consortia were enriched and characterized using high-throughput sequencing (HTS). 454 Pyrosequencing indicated novel W22 (25.0 % of bacterial sequences) in the WWE1 and Sulfurovum-resembled species (32.0 %) in the family Campylobacterales were the most abundant in mesophilic and ambient reactors, respectively, which challenges previous knowledge that Syntrophorhabdus was the most predominant. Previous findings may underestimate bacterial diversity and low-abundance bacteria, but overestimate abundance of Syntrophorhabdus. Illumina HTS revealed that archaeal populations were doubled in ambient reactor and tripled in mesophilic reactor, respectively, compared to the ∼4.9 % (of the bacteria and archaea sequences) in the seed sludge. Moreover, unlike the dominance of Methanosarcina in seed sludge, acetotrophic Methanosaeta predominated both (71.4–76.5 % of archaeal sequences) ambient and mesophilic enrichments. Noteworthy, this study, for the first time, discovered the co-occurrence of green sulfur bacteria Chlorobia, sulfur-reducing Desulfovibrio, and Sulfurovum-resembling species under ambient condition, which could presumably establish mutualistic relationships to compete with syntrophic bacteria and methanogens, leading to the deterioration of methanogenic activity. Taken together, this HTS-based study unravels the high microbial diversity and complicated bacterial interactions within the biogas-producing and phenol-degrading bioreactors, and the identification of novel bacterial species and dominant methanogens involved in the phenol degradation provides novel insights into the operation of full-scale bioreactors for maximizing biogas generation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
With the excessive exploitation of fossil fuels for economic development, energy crisis, global warming, and environmental pollution have become severe problems that hinder the sustainable growth of world economy. The development of renewable and clean energy has therefore become a critical and urgent mission for the scientists and engineers worldwide. Recently, anaerobic technologies that retrieve bioenergy (e.g., H2 and CH4) from organic substrates have been receiving more and more attention globally in the developed and developing countries [1, 2]. Among them, methanogenic process has been widely applied for generating biogas from various wastewater-borne organics or organic solid wastes originated from various municipal and industrial sources due to its advantages including methane generation, less sludge yield, and energy conservation. However, the presence of toxic phenol and its derivatives in the wastewater of coal gasification, petroleum refining, and manufacturing of resins, synthetic chemicals and pesticides industries [3, 4], as well as in the municipal solid wastes such as organic household residues, slaughter house wastes and animal manure, kitchen wastes can largely inhibit the methanogenesis from these organic substrates by suppressing the activity of the responsible microorganisms in the methanogenic bioreactors [5] [6]. Further, methane-generating and phenol-degrading bioreactors [3, 4, 7–10] usually suffer operational difficulties such as long start up time, limited loading, and instability. All these problems indicate the still-limited knowledge on the methanogenic phenol-degrading community.
Early studies of the biogas-producing and phenol-degrading consortia are mainly based on chemical monitoring [11] and microscopic observation [3]. Later, molecular techniques, such as cloning, fluorescence in situ hybridization (FISH), denaturing gradient gel electrophoresis, and terminal restriction fragment length polymorphism, were widely used, which greatly improved our understanding of the methanogenic consortia [4, 10, 12]. Among them, FISH is a fast method for visualizing and counting cells, but may be limited by low hybridization efficiency and coverage of probes; cloning has been the most commonly used method [4, 12], but the less abundant fermentative syntrophic bacteria and methanogens could easily fall under the detection limit due to the usually very limited number of clones (low-throughput). For example, a recent study on mesophilic phenol-degrading enrichment failed to detect methanogens with acetotrophic activity or any syntrophic acetate-oxidizing bacteria by the clone method while methanogenic degradation of phenol into acetate was chemically confirmed [13].
High-throughput sequencing makes huge number of sequences available at acceptable cost for the exploring microbial diversity in various habitats at much deeper coverage and higher resolution [14–18], and this will help explore the microbial structure and complicated interactions among different microbial populations, which cannot be clearly revealed using the low-throughput molecular techniques. In this study, two methane-yielding and phenol-degrading consortia were acclimated under ambient and mesophilic conditions and characterized using the 454 pyrosequencing and Illumina HTS. This study aims to supply more detailed knowledge on the microbial components and diversity of methanogenic consortia that are capable of methane generation from toxic phenol and mainly addresses the following issues: (1) what kinds of bacteria and archaea that accomplish methane generation from phenol-containing wastewater under ambient and mesophilic conditions, respectively?; (2) How can the difference between the microbial structure of ambient and mesophilic consortia affect the methanogenic activity and phenol-degrading capacity?; and (3) What roles do different members within ambient and mesophilic phenol-degrading consortia presumably play?
Materials and Methods
Enrichment of Phenol-Degrading Consortia
The enrichment of phenol-degrading consortia was started in two 200/288-ml sealed serum bottles incubated at 20 and 37 °C, respectively, with mild shaking at 100 rpm. The seed sludge was collected from the No. 2 Digester of Shek Wu Hui WWTP, Hong Kong. The mesophilic (37 °C) digester was operated to digest primary sludge and wasted activated sludge (sludge retention time (SRT), 25 days; total suspended solid (TSS), 17.7 g/L; volatile suspended solid (VSS), 13.0 g/L). Phenol as the sole substrate, plus balanced nutrients and alkalinity [9], was added batch by batch using concentrated phenol stock solution after complete depletion in the previous batch. For each gram of phenol, the solution was supplemented with 2.38 g NaHCO3, 618.8 mg NH4Cl, 119.0 mg MgSO4·7H2O, 71.4 mg K2HPO4, 28.6 mg KH2PO4, 35.7 mg CaCl2, 16.7 mg, NiSO4·7H2O, 11.9 mg FeCl3·6H2O, 2.6 mg MnCl2·4H2O, 1.4 mg ZnCl2, 1.4 mg CoCl2·2H2O, 1.0 mg (NH4)2MoO4·4H2O, 0.7 mg CuCl2·2H2O, and 0.5 mg Na2B4O7·10H2O. After six batches of phenol addition which lasted for 109 days, the sludge in 20 and 37 °C serum bottles was anaerobically transferred by syringe into two 0.8/1.1-L five-neck anaerobic batch reactors (ABRs) operated at ambient and mesophilic temperatures, with mild magnetic stirring, little sludge discharge and pH controlled at 7.0–7.5 (by adding 1 N sodium bicarbonate using peristaltic pumps). At the beginning of each batch, concentrated phenol solution (10 g/L) was injected anaerobically into the reactors (from the top) after drawing out equal volume of supernatant (after ∼10 min settling). On day 193 (the 24th batch), sludge samples were taken from both reactors for specific methanogenic activity (SMA) test (Supporting Information S1) and DNA extraction.
Chemical Analysis
The component and concentration of biogas, including methane, carbon dioxide, and hydrogen, were determined by a gas chromatograph (Hewlette-Packard 5890II, USA) equipped with a thermal conductivity detector following the method descripted previously [19]. The generated biogas in each reactor was collected by a 1.5-L Tedlar bag, and its volume was measured using a glass syringe at the end of each batch. The concentrations of phenol and benzoate in liquid were measured by HPLC (SHIMADZU, Japan) with a C18 column using a flow rate of 1 ml/min, an eluent consisting of MeOH–H2O (60:40) and 0.1 % H3PO4, and a UV detector set to 280 nm [20]. The composition and concentrations of other products including volatile fatty acids (VFAs) and alcohols were measured by another GC (6890N, Agilent Technologies, USA) equipped with a flame ionization detector [9]. Total suspended solids and volatile suspended solids were determined according to the standard methods [21].
Molecular Method
For each sample, genomic DNA was extracted in duplicate from 2.0 mL slurry by FastDNA SPIN Kit for Soil (MPBiomedicals, LLC, Illkirch, France). The duplicate DNA extracts were then mixed together for 454 pro-sequencing and Illumina HTS. The corresponding DNA concentration determined (NanoDrop-1000, Thermo Scientific, Wilmington, DE, USA) for the seed sludge, AT, and MT enrichment was 150.1, 148.3, and 141.2 ng/μl, with a 260/280 ratio of 1.68, 1.85, and 1.88, respectively.
For the 454 pro-sequencing, extracted DNA was amplified with forward primer 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and a cocktail of four equally mixed reverse primers, that is, R1 (5′-TACCRGGGTHTCTAATCC-3′), R2 (5′-TACCAGAGTATCTAATTC-3′), R3 (5′-CTACDSRGGTMTCTAATC-3′), and R4 (5′-TACNVGGGTATCTAATCC-3′), which targets the hyper-variable V3-V4 regions of the 16S ribosomal RNA (rRNA) genes [22]. Barcodes for sample multiplexing during sequencing were modified in the 5′ terminus of the forward primer. The polymerase chain reaction (PCR) conditions were initial 5 min denaturation at 94 °C; 30 cycles of 30 s at 94 °C, 30 s at 50 °C, and 60 s at 72 °C; and final 10 min extension at 72 °C. PCR amplicons from different sludge samples were firstly purified with a quick-spin Kit (iNtRON, Seoul, Korea), then mixed at equal mass concentrations and finally sent out for pyrosequencing on the Roche 454 FLX Titanium platform (Roche) at the Genome Research Center of Beijing. In our study, attempts to amplify archaeal amplicons using the forward primer A344F (5-AYGGGGYGCASCAGGSG-3) and reverse primer A519R (5-GGTDTTACCGCGGCKGCTG-3) targeting V3 region of 16S rRNA genes failed, although these primers have been successfully in amplifying marine water and sediment samples [23, 24].
For metagenomic sequencing, DNA samples from AT and MT reactors (on Day 193) were used to construct libraries of 180 bp for sequencing using Illumina Hiseq2000 platform by applying the 101-bp paired-end (PE) strategy (Beijing Genomics Institute, Shenzhen, China). For the seed sludge and MT enrichment, DNA replicates were sequenced to evaluate the reproducibility of the experimental results, and the average values from the two replicate data sets were reported. The raw 454 reads and shotgun metagenomic reads have been deposited into the NCBI short-reads archive database (Accession Number: SRR764554 and SRR764555).
Bioinformatics Analysis
Procedures for (1) processing of high-throughput sequencing data sets (Figure S1), (2) taxonomic analysis, and (3) construction of rarefaction curves and phylogenic trees were available at Supporting Information S2.
Results and Discussion
Enrichment of Methane-Generating and Phenol-Degrading Consortia
Methane-generating and phenol-degrading consortia were obtained after the 193-day acclimation in ambient temperature (AT, 20 °C) and mesophilic temperature (MT, 37 °C) reactors, respectively. The results showed that initial phenol degradation and methane production were observed in AT and MT reactors after a lag phase of ∼68 and ∼37 days, respectively, after incubation with the seed sludge (Fig. 1a). The longer acclimation period required for the AT sludge than MT sludge was likely attributed to the fact that mesophilic fermentative populations present in the seeding sludge had been previously accustomed to MT of the digester in which it was collected and, therefore, needed more time to adapt to AT under the stress of phenol. Although the phenol loadings and degrading rate in the MT reactor were always higher than that of AT reactor (Fig. 1b), the sludge in both reactors was able to completely mineralize phenol to final products (e.g., methane and carbon dioxide) without much accumulation of intermediates throughout the whole enrichment period. For instance, during the 18th batch, complete phenol degradation was observed in both AT and MT reactors within 5 days, with little benzoate and no volatile fatty acids (VFAs, e.g., butyrate and proportionate) detected, indicating that initial acidogenesis was likely to be the rate-limiting step instead of subsequent acetogenesis and methanogenesis for complete phenol degradation. However, considerable concentration of ethanol (18.1∼164.5 mg/L in AT reactor; 58.8∼156.5 mg/L in the MT reactor) was continuously detected in both reactors (Figure S2), possibly reflecting partially inhibited methanogenic activity [25]. Likewise, acetic acid was also successively detected in both reactors but at much lower levels (14.5∼37.5 mg/L). Notably, concentrations of benzoate monitored in the AT reactor (27.6 mg/L in average) were much higher than in the MT reactor (2.4 mg/L), regardless of the relatively lower initial phenol concentration in the AT reactor. This validates the activity of phenol-degrading consortia and suggests difference of phenol-degrading dynamics driven by phenol-degrading consortia under ambient and mesophilic conditions.
SMA Tests
The phenol-degrading profiles and SMA of the AT and MT enrichments were investigated at different initial phenol concentrations (100∼1,000 mg L−1). As has been expected, the time required for the complete degradation increased with initial phenol concentrations (Figure S3). As shown in Fig. 2, the highest SMA for AT and MT enrichments was observed at an initial phenol concentration of 200 mg L−1, corresponding to phenol loadings of 274.0 and 363.6 g phenol/g VSS/day, respectively. At the starting concentrations of 400 and 600 mg L−1, the SMA of the AT and MT enrichments dropped down to 52.8∼80 % and 14.8∼19.9 % of the highest SMA values, respectively. Almost 92.3∼100 % of methane-producing activity of the AT and MT enrichments were suppressed at the initial phenol concentration of 1,000 mg L−1.
The highest SMA for AT and MT enrichments was 200 and 283 mg CH4 COD g−1 VSS day−1 (Fig. 2), respectively, which were comparable to the levels that were reported for previous methanogenic phenol-degrading consortia cultivated in upflow anaerobic sludge bed (UASB) reactors and full-scale granular activated carbon-anaerobic fluidized bed (GAC-AFB) reactors [3, 10, 26]. The higher SMA of the MT over AT enrichment was in accordance with previous studies [3, 8], which was likely associated with both the differences in microbial community structure and enzyme activity of phenol-degrading consortia at 20 and 37 °C.
16S and Shotgun Metagenomic Data Sets
Table S1 shows the basic information of the 454 data sets. The rarefaction curves show that the OTU numbers in seed sludge (at the dissimilarity cutoffs of both 3 and 6 %) were the highest, followed by AT and MT enrichments (Figure S4), indicating that microbial diversity decreased with the enrichment time. Also, it can be seen that the curves of AT and MT were almost flat (with the slope of the end points at the dissimilarity cutoffs of 3 and 6 % between 0.22∼0.44 % and 0.11∼0.22 %, respectively), indicating that the sequencing depth adopted in this study was deep enough to reflect the operational taxonomic unit (OTU) diversities in the two reactors. Additionally, 454 pyrosequencing applied in this study identified about 5∼10 times more OTUs than the cloning method used previously [4, 6, 9, 10, 12, 13] (Table S1), implicating that many novel OTUs previously undetected in the ambient and mesophilic phenol-degrading enrichments can now be well identified by 454 pyrosequencing.
Table S2 shows the basic information of the Illumina metagenomic data sets. After de-replication and reads overlapping, tags number in each data set was normalized to 13,106,796, of which 11,291∼11,672 were identified as tags containing 16S rRNA genes by BLASTN at an e-value cutoff of 1e-20. Those 16S rRNA gene tags with length between 150 and 190 bp ranged from 8,516 to 9,614 and were applied for taxonomic analysis using LCA algorithm in MEGAN.
Shared and Distinct Bacterial Classes and Genera
Figure 3 shows the number of bacterial classes and genera shared between the seed sludge, AT, and MT enrichments. Considering the well-known phenol toxicity on microbial growth, it is assumed that those classes and genera well-detected in the AT and MT enrichments but absent from the seed sludge are the best candidates associated with phenol degradation, while those merely detected in the seed sludge may play no roles or at least much less roles in phenol degradation.
Weighed Venn diagrams of the shared and distinct bacterial classes and genera among the samples. AT, 20 °C enrichment; MT, 37 °C enrichment; SEED, the seed sludge. The statistics was based on the taxonomic results of 454 pyrosequencing data sets. The number of total bacterial sequences in each sample was 8,150. The numbers outside and inside the brackets indicated the number and total abundances (calculated as reads count) of shared taxa among different samples
As shown in Fig. 3a and Table S3, eight bacterial classes appeared merely in the AT and/or MT enrichment after accustomed to phenol. Among them, four bacterial classes (e.g., Epsilonproteobacteria and WWE1) were shared by the AT and MT enrichments, three (e.g., green sulfur bacteria Chlorobia) were only represented in the AT enrichment and one (Thermotogae) was merely detected in the MT enrichment. Figure 3b and Table S4 show that 28 out of the total 70 bacterial genera were merely detected in the AT and/or MT enrichment. Among them, seven genera such as W22, Pelotomaculum, Syntrophus, Desulfovibrio, and Moorella, were shared by the AT and MT enrichments, corresponding to 3,765 and 3,151 of the total 8,150 bacterial sequences, respectively. These genera were most likely the core bacterial consortia closely related to the methanogenic conversion of phenol or its degradation products (like VFAs and alcohols). Moreover, ten genera (e.g., Rhodopseudomonas, Chlorobaculum, Rhodococcus, and Geobacter), which accounted for 361 bacterial sequences, were merely represented in the AT enrichment, whereas eleven genera (e.g., Brachymonas, Moorella, Thermonema, and Turicibacter), which corresponded to 1,301 bacterial sequences, were only detected in the MT enrichment. These genera uniquely occurred either under ambient condition or mesophilic condition may reflect the influence of temperature on the microbial components of methanogenic phenol-degrading consortia, which, in turn, revealed different metabolism pathways that could exist to transform phenol to methane under the ambient and mesophilic conditions.
Shift in Bacterial Populations After Enrichment
Methanogenic degradation of phenol has been demonstrated to be strongly influenced by temperature, with much higher degradation capacity and methanogenic activity observed under mesophilic condition compared with ambient and thermophilic conditions [3, 4, 8, 9, 13]. Similar results were obtained in this study with higher degradation capacity (Fig. 1) and SMA value (Fig. 2) detected under the mesophilic condition compared with ambient condition. Possible reasons for such phenomenon may lie in difference in both abundance and diversity of the key phenol-degrading bacteria that functioned under these two temperatures. Previous studies demonstrated that Deltaproteobacteria (e.g., Syntrophorhabdaceae) and Clostridia (e.g., Pelotomaculum) were the key syntrophic populations of methanogenic phenol-degrading consortia in different types of anaerobic reactors treating phenol at various temperatures [4, 9, 13].
Apparent shift in relative sequences percentage of Proteobacteria was observed (Fig. 4a; Figure S5), with its percentage increased dramatically from 19.1 % of bacterial sequences in seed sludge to 53.5 % in AT enrichment and 43.9 % in MT enrichment, respectively (Figure S5), confirming that members of Proteobacteria are largely involved in the phenol degradation under both conditions. Moreover, Fig. 4a demonstrates that in the seed sludge, alpha subdivision was the most abundant (53 % of total proteobacterial sequences, Figure S5), followed by gamma and beta subdivisions, while delta subdivision merely occupied a very small proportion (12 %, Figure S5) and epsilon subdivision was not even detected. However, epsilon and delta subdivisions predominated proteobacterial sequences of the AT and MT enrichments (Fig. 4a, Figure S5). This phenomenon illustrated the selectivity of high concentrations of phenol (875∼1,000 mg L−1) on Proteobacteria subdivisions, implicating significant difference in their adaptability to phenol toxicity as well as their capacity to utilize phenol or its intermediates. Intriguingly, the abundance of Betaproteobacteria in the seed sludge (3.1 %) was decreased to 0.64 % in the AT enrichment, but increased to 12.9 % in the MT enrichment (Fig. 4a), revealing that beta subdivision played much more significant roles in phenol metabolism under 37 than 20 °C.
Bacterial classes (a) and major genera (b) in the seed sludge ambient and mesophilic enrichments. AT, 20 °C enrichment; MT, 37 °C enrichment; SEED, the seed sludge. The result was based on the taxonomic analysis of 454 pyrosequencing data sets and major genera referred to those with relative abundance >0.5 % in at least one sample. The thickness of each ribbon represents the abundance of each taxon. The absolute tick above the inner segment and the relative tick above the outer segment stand for the reads abundances and relative abundance of each taxon (in total 8,150 bacterial sequences of each sample), respectively. The taxa names in green, red, and blue refer to those taxa shared by 3 (AT, MT, and SEED), 2 (AT, MT, AT, and SEED; MT and SEED), and 1 (AT, MT, and SEED) samples, respectively. Others refer to those unassigned reads. The data were visualized using Circos (v 0.63, [27])
Moreover, candidate division WWE1 in the phylum Spirochaetes dominated (as illustrated by the thickest ribbon for MT segment in Fig. 4a) and accounted for 25.0 % of bacterial sequences in the MT enrichment. All those WWE1-affiliated sequences were closely related to the genus W22 in the family SHA-4 (Fig. 4b), which is a genus with frequent occurrence in various anaerobic digesters but whose roles still remaining unknown [28, 29]. The sequence similarity-based phylogenetic analysis (Figure S5b) showed that all W22-affiliated sequences were closely (>99.0 % similarity) related to Candidatus Cloacamonas acidaminovorans, which is the only sequenced fermentative bacterium within the candidate class WWE1 with the genetic potentials to derive most of its carbon and energy from the fermentation of amino acids and to syntrophically oxidize butyrate to CO2, hydrogen, and acetate [29, 36]. Thus, the putative role of W22-related species in the MT enrichment was to metabolize amino acids and fatty acids (e.g., butyrate) produced during phenol metabolism. In addition, the much higher abundance of W22 in 37 °C, compared with 20 °C enrichment and the seed sludge, implicated a much more significant role played by W22 under mesophilic than ambient conditions during phenol degradation under methanogenic condition.
By contrast, the most abundant genus in the AT enrichment was affiliated with order Campylobacterales (32.0 % of bacterial sequences in the AT enrichment) in the class Epsilonproteobacteria. The occurrence of Epsilonproteobacteria had been reported previously in phenol-degrading UASB granule sludge, but its role remained unclear [8]. The phylogenetic tree (Figure S6a) shows that all Campylobacterales-related sequences discovered in the AT and MT enrichments were most closely related to two environmental clones affiliated to an unassigned family in the order Campylobacterales in GreenGenes database. However, these sequences only showed <96.0 % similarity to two well-known hydrogen- and sulfur-oxidizing chemolithoautotrophs, namely Sulfurovum lithotrophicum and Sulfurovum sp. NBC37-1, implicating that the Campylobacterales-related sequences highly represented in the AT enrichment were quite likely to be affiliated with a novel species in the genus Sulfurovum. Stable-isotope probing and substrate enrichment experiments indicated that Sulfurovum could cooperate with species related to Desulfovibrio and Pelotomaculum to syntrophically degrade benzene under sulfate-reducing conditions [37, 38]. Given the co-occurrence of these three types of bacteria in the AT reactor at considerable abundance, as well as the similar chemical structure between benzene and phenol, it is assumed that Sulfurovum-like species could establish syntrophic associations with Desulfovibrio and Pelotomaculum, leading to anaerobically syntrophic degradation of phenol at ambient temperature.
Moreover, multiple fermentative syntrophic bacteria emerged in the AT and MT bacterial consortia at high abundances after enrichment (Fig. 4b). Among them, Syntrophorhabdus in a novel family Syntrophorhabdaceae (previously known as clone cluster Deltaproteobacteria group TA [30]) was highly represented in both AT (8.2 %) and MT (17.6 %) enrichments, but almost undetected in seed sludge. All sequences within this genus showed >99.87 % similarity to Syntrophorhabdus aromaticivorans UI (GenBank Accession No.: AB212873), which has been recently identified as the first cultured anaerobe capable of degrading phenol via benzoate in syntrophic association with a hydrogenotrophic methanogen or a sulfate-reducing bacterium [30]. Therefore, S. aromaticivorans was likely to be the key anaerobe which degrades phenol to benzoate in the ambient and mesophilic reactors of this study. The other two syntrophs in the class Clostridia that is Pelotomaculum in the family Peptococcaceae and Moorella in the family Thermoanaerobacteraceae were also detected, but at quite different abundances in the AT and MT enrichments (Fig. 5). Exactly, Pelotomaculum-related species had much higher abundance in the AT (6.3 %) than MT enrichments (1.1 %). This genus, also termed Desulfotomaculum subcluster Ih, could degrade benzoate and propionate in syntrophy, but had lost their ancestral ability of reducing sulfate, sulfite, and thiosulfate as they adopted a syntrophic lifestyle in methanogenic environments [31]. Moorella-related species occupied 6.0 % of bacterial populations in the MT enrichment, but was undetectable in the AT enrichment. This genus has been well-known as obligate anaerobes capable of syntrophic degradation of methanol and formate [32, 33]. Overall, the apparent difference in the abundances of Pelotomaculum and Moorella under AT and MT conditions may reflect significant effect temperature that plays in shaping the microbial components of syntrophic consortia that engaged in the metabolism of intermediate VFAs and alcohols during methanogenic degradation of phenol. However, unlike Pelotomaculum and Moorella, Syntrophus-related species showed no significant difference on abundances in AT (2.0 %) and MT enrichments (1.2 %). This genus, regardless of its relatively lower abundance, has a higher affinity for benzoate and plays critical roles in degrading this substrate into acetate and hydrogen through syntrophic associations with methanogens [4, 9].
Comparison of consistency between the taxonomic results of key bacteria identified by 16S rRNA sequences in 454 data and metagenomic data. Key bacteria referred to those genera with relative percentage >0.5 % in at least one sample. The numbers in the parentheses after each genus tag represent the ratio of abundance of this genus in AT enrichment to MT enrichment
Notably, besides the dominance of multiple fermentative syntrophic bacteria, considerable amount of green sulfur bacteria (GSB) Chlorobia (Fig. 4a) and sulfate-reducing bacteria (SRB) Desulfovibrio (Fig. 4b) in the class Deltaproteobacteria were detected mainly in the AT enrichment (Fig. 5). The Desulfovibrio-related sequences were closely related to a sulfate-reducing species, i.e., Desulfovibrio mexicanus (4.0 % in AT enrichment), which could syntrophically metabolize a large variety of substrates, such as ethanol, lactate, pyruvate, and formate [34, 35]. The co-occurrence of GSB Chlorobia, SRB D. mexicanus, and Sulfurovum-like bacteria in the AT enrichment could establish tightly coupled mutualistic interactions that exchange sulfur or sulfate compound between the partners [33], which could compete with fermentative syntrophic bacteria for organic substrates and with methanogenic archaea for acetate and hydrogen, respectively, leading to the deterioration of methanogenic activity.
Overall, given that syntrophic aromatic acid (e.g., phenol) and fatty acid (e.g., benzoate, propionate, butyrate, and formate) metabolisms are often the rate-limiting steps in methanogenic degradation of organic compounds [33, 39], the much higher temperature, as well as higher proportions of S. aromaticivorans and other syntrophic bacteria (e.g., W22 and Moorella) under mesophilic condition, compared with ambient condition, could probably explain the higher phenol-degrading rate observed in MT than AT reactor.
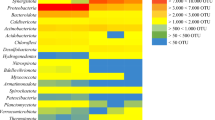
Comparison of 16S-Based Bacterial Abundance in 454 and Metagenomic Data Sets
Figure 5 shows the comparison of consistency between the taxonomic results of key bacteria involved in phenol degradation using 16S rRNA gene sequences from two different sources, that is, 454 data sets and metagenomic data sets. In general, relative abundance of bacterial genera/species indicated by 454 data sets is somewhat different from that by metagenomic data sets (Fig. 5, heatmap). However, ratios of their abundance in AT enrichment to MT enrichment indicated by 454 data were comparable to those indicated by metagenomic data sets (blue bars, Fig. 5) with few exceptions (e.g., Aminobacterium), indicating consistency of these two sequencing technologies in 16S rRNA-based comparison of bacterial components among different samples even at the genus level. In addition, comparison between the last two columns of the heatmap (Fig. 5) revealed that genera abundances derived from the two replicate metagenomic data sets of MT (that is MT1 and MT2) highly resembles each other, revealing good reproducibility of the experimental results from metagenomic data sets.
The Archaeal Populations Revealed by Metagenomic Data Sets
The relative abundance of bacterial and archaeal populations in methanogenic phenol-degrading consortia were evaluated based on the number of 16S rRNA gene tags identified from the metagenomic data sets. The 16S rRNA gene tags were identified by BLASTN against two 16S rRNA gene databases, i.e., GreenGenes and Silva SSU, at an e-value cutoff of 1e-20. Table 1 shows that 4.9 % of the 16S rRNA gene tags in the seed sludge were identified to be affiliated with archaea, and this percentage was increased to 8.8∼9.2 % and 12.7∼13.5 % in the AT and MT enrichments, respectively. Such increase in the relative abundance of overall archaeal populations after enrichment went hand in hand with variations in terms of both the species diversity and abundance, implicating underlying adjustment in the syntrophic associations between syntrophic bacteria and methanogenic archaea after the introduction of phenol to the reactors. Further, the higher relative abundance of archaea in MT than AT enrichment could partially explain the higher SMA value of MT than the AT enrichment detected in SMA tests.
Figure 6 shows the relative abundances of archaeal populations before and after enrichment. All the classified archaeal tags from the three samples were affiliated with three methanogenic orders, i.e., Methanobacteriales, Methanomicrobiales, and Methanosarcinales. At the genus level, two acetoclastic (Methanosaeta and Methanosarcina) and five hydrogenotrophic (Methanobacterium, Methanobrevibacter, Methanoculleus, Methanolinea, and Methanospirillum) methanogens were identified. Although these hydrogenotrophic methanogens merely occupied 11.2∼15.7 % of all archaeal populations, they play very significant roles in hydrogen consumption and in maintaining hydrogen partial pressure at a level low enough for phenol degradation. Moreover, unlike the dominance of Methanosarcina (57.1 % of total archaeal populations) in the seed sludge, Methanosaeta (71.4∼76.5 %) predominated the AT and MT enrichments, implicating potentially stronger adaptability of Methanosaeta to phenol toxicity as well as the low acetate concentrations detected in the AT and MT reactors.
Comparison of diversity and abundances of archaeal populations in the seed, ambient, and mesophilic enrichments. The result was based on the taxonomic analysis of 16S rRNA gene tags from metagenomic data sets. Tags that were assigned to Archaea (phylum Euryachaeota) in the GreenGenes database for the seed sludge, AT, and MT1 enrichments were 472 (100 %), 884 (100 %), and 1165 (100 %), respectively
The Putative Roles of Different Microbial Groups During Phenol Degradation
The putative roles of different microbial groups during ambient and mesophilic degradation of phenol under methanogenic conditions were shown in Fig. 7. Phenol was first converted mainly by S. aromaticivorans to benzoate, which was further converted by acidogens affiliated with Syntrophus and Pelotomaculum to intermediates like VFAs and alcohols. These intermediates could then further be utilized by syntrophic acetogens (e.g., W22, Pelotomaculum, and Moorella) to acetate and hydrogen, both were finally converted by respective methanogens into methane. Notably, syntrophic bacteria like S. aromaticivorans, W22, and Moorella and Aminobacterium were detected with much higher abundances in the MT than AT reactor, indicating that mesophilic temperature, compared with ambient temperature, was more favorable for their prevalence in methanogenic reactors to outcompete other competitive bacterial groups (e.g., SRB).
However, instead of being outcompeted by those typical syntrophic bacteria (as observed in the MT reactor), SRB affiliated with genus Desulfovibrio was quite abundant in the methanogenic AT reactor. This SRB could utilized sulfate and reduce it to sulfite using a wide variety of substrates, such as propionate, butyrate, ethanol, and acetate, for supplies of carbon and electrons. Presumably, Desulfovibrio could establish syntrophic associations with GSB Chlorobia and Campylobacterales-related (Sulfurovum-like) species highly represented in the AT reactor by exchanging sulfur or sulfate compounds between the partners. In this way, they could compete with syntrophic acidogens (e.g., Syntrophorhabdus and Syntrophus) and acetogens (e.g., W22, Pelotomaculum, and Moorella) for organic substrates and with methanogens for acetate and hydrogen, respectively, which is to the disadvantage of methanogenesis. Therefore, from a perspective of maximizing methanogenesis, mesophilic temperature, instead of the ambient temperature, was recommended for anaerobic bioreactors treating phenol-containing wastewater.
Compared with Previous Studies
This is the first systematic and comprehensive profiling of methanogenic phenol-degrading consortia by HTS approach. Similar to previous studies, Syntrophorhabdus is identified as the key phenol degrader and Methanosaeta as the most predominant methanogen. However, previous findings based on cloning method demonstrate that Syntrophorhabdus-affiliated sequences dominate total bacterial 16S rRNA clones [4, 10, 13], whereas high-throughput 454 pyrosequencing adopted in this study implicated that Syntrophorhabdus, despite of its key role in initial phenol degradation, was not necessarily the most abundant (based on 16S rRNA sequences). Moreover, more OTUs (at similarity threshold of 97 %) are identified from 454 data sets (106–150 OTUs) than from clone sequences (<21 OTUs). These results implicated that compared with 454 pyrosequencing, clone method may underestimate the OTUs diversity but overestimate the proportion of Syntrophorhabdus in total bacterial 16S rRNA gene sequences of methanogenic community. Moreover, this study indicates that Campylobacterales-related genus and W22 were the most abundant genera in the AT and MT phenol-degrading consortia, respectively, which challenges previous findings that Syntrophorhabdus was the most predominant [4, 6]. In addition, this study identifies several more syntrophic genera (e.g., W22, Moorella, and Aminobacterium) that were not identified previously. Noteworthy, the results of this study, for the first time, discovered the co-occurrence of GSB Chlorobia, SRB Desulfovibrio, and Campylobacterales-related genus in the ambient phenol-degrading methanogenic consortia, which could presumably establish tightly coupled mutualistic interactions to compete with fermentative syntrophic bacteria for organic substrates and with methanogens for acetate and hydrogen, respectively, possibly leading to the decrease of methanogenic activity.
Several limitations may affect our results and the corresponding conclusions. First, the length of 16S rRNA gene tags used for identification of bacteria and archaea in this study was still short, about 150∼190 bp in length, although taxonomic classification of this length using LCA algorithm in MEGAN indicates that reads length of 100∼200 bp could be long enough to identify a species [40]. Second, biases are also likely to be introduced during the DNA extraction and PCR amplification of V3∼V4 amplicons, although sequencing of 16S rRNA amplicons is the general practice for bacterial identification. Third, the lack of replicated setups of reactors renders the reproducibility or statistical significance of observed differences in community structure less reliable. In addition, sequence-based comparison of microbial abundance in this study is different from traditional cell number-based estimation (i.e., using in situ detection methods, FISH, and CARD-FISH); thus, difference in length and copy number of 16S rRNA gene of different bacteria should be taken into consideration during comparison. Finally, this study relies on sequence similarity-based method for taxonomic analysis and role prediction, which may be affected by the completeness of 16S rRNA gene databases whose data size keeps on increasing, as well as the difference between the genotype and phenotype of some species.
References
Sreekrishnan T, Kohli S, Rana V (2004) Enhancement of biogas production from solid substrates using different techniques––a review. Bioresour Technol 95:1–10
Khanal S (2009) Anaerobic biotechnology for bioenergy production: principles and applications. Wiley, North America, USA
Fang HHP, Chen T, Li YY, Chui HK (1996) Degradation of phenol in wastewater in an upflow anaerobic sludge blanket reactor. Water Res 30:1353–1360
Chen CL, Wu JH, Liu WT (2008) Identification of important microbial populations in the mesophilic and thermophilic phenol-degrading methanogenic consortia. Water Res 42:1963–1976
Fang HHP, Chan OC (1997) Toxicity of phenol towards anaerobic biogranules. Water Res 31:2229–2242
Levén L, Nyberg K, Schnürer A (2011) Conversion of phenols during anaerobic digestion of organic solid waste—a review of important microorganisms and impact of temperature. J Environ Manage. doi:10.1016/j.jenvman.2010.10.021
Veeresh GS, Kumar P, Mehrotra I (2005) Treatment of phenol and cresols in upflow anaerobic sludge blanket (UASB) process: a review. Water Res 39:154
Fang H, Liu Y, Ke S, Zhang T (2004) Anaerobic degradation of phenol in wastewater at ambient temperature. Water Sci Technol: J Int Assoc Water Pollut Res 49:95
Fang HHP, Liang DW, Zhang T, Liu Y (2006) Anaerobic treatment of phenol in wastewater under thermophilic condition. Water Res 40:427–434. doi:10.1016/j.watres.2005.11.025
Chen CL, Wu JH, Tseng IC, Liang TM, Liu WT (2009) Characterization of active microbes in a full-scale anaerobic fluidized bed reactor treating phenolic wastewater. Microbes and environments: 904220077
Boyd SA, Shelton DR, Berry D, Tiedje JM (1983) Anaerobic biodegradation of phenolic compounds in digested sludge. Appl Environ Microbiol 46:50
Zhang T, Ke S, Liu Y, Fang H (2005) Microbial characteristics of a methanogenic phenol-degrading sludge. Water Sci Technol: J Int Assoc Water Pollut Res 52:73
Levén L, Schnürer A (2010) Molecular characterisation of two anaerobic phenol-degrading enrichment cultures. Int Biodeterior Biodegrad 64:427–433. doi:10.1016/j.ibiod.2010.04.009
Shendure J, Ji H (2008) Next-generation DNA sequencing. Nat Biotechnol 26:1135–1145
Kircher M, Kelso J (2010) High-throughput DNA sequencing—concepts and limitations. BioEssays 32:524–536
Ma J, Wang Z, Yang Y, Mei X, Wu Z (2013) Correlating microbial community structure and composition with aeration intensity in submerged membrane bioreactors by 454 high-throughput pyrosequencing. Water Res 47:859–869
Lee TK, Van Doan T, Yoo K, Choi S, Kim C, Park J (2010) Discovery of commonly existing anode biofilm microbes in two different wastewater treatment MFCs using FLX titanium pyrosequencing. Appl Microbiol Biotechnol 87:2335–2343
Ishak HD, Plowes R, Sen R, Kellner K, Meyer E, Estrada DA, Dowd SE, Mueller UG (2011) Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb Ecol 61:821–831
Xia Y, Cai L, Zhang T, Fang HH (2012) Effects of substrate loading and co-substrates on thermophilic anaerobic conversion of microcrystalline cellulose and microbial communities revealed using high-throughput sequencing. Int J Hydrogen Energy 37:13652–13659
Holmes DE, Risso C, Smith JA, Lovley DR (2012) Genome-scale analysis of anaerobic benzoate and phenol metabolism in the hyperthermophilic archaeon Ferroglobus placidus. The ISME J 6:146–157
EatonAD CLS, RiceEW GAE (2005) Standard Methods for the Examination of Water and Wastewater. Centennial Edition, 21st edn. American Public Health Association, Washington
Zhang T, Shao M-F, Ye L (2012) 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. The ISME J 6:1137–1147
Qian PY, Wang Y, Lee OO, Lau SCK, Yang J, Lafi FF, Al-Suwailem A, Wong TYH (2010) Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J 5:507–518
Teske A, Sørensen KB (2007) Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J 2:3–18
Wang Y, Zhang Y, Wang J, Meng L (2009) Effects of volatile fatty acid concentrations on methane yield and methanogenic bacteria. Biomass Bioenergy 33:848–853
Shui-zhou K, SHI Z, Tong Z, Herbert H, FANG P (2004) Degradation of phenol in an upflow anaerobic sludge blanket (UASB) reactor at ambient temperature. J Environ Sci 16
Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D et al (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Chouari R, Le Paslier D, Dauga C, Daegelen P, Weissenbach J, Sghir A (2005) Novel major bacterial candidate division within a municipal anaerobic sludge digester. Appl Environ Microbiol 71:2145–2153
Pelletier E, Kreimeyer A, Bocs S, Rouy Z, Gyapay G, Chouari R, Rivière D, Ganesan A, Daegelen P, Sghir A (2008) “Candidatus Cloacamonas acidaminovorans”: genome sequence reconstruction provides a first glimpse of a new bacterial division. J Bacteriol 190:2572–2579
Qiu YL, Hanada S, Ohashi A, Harada H, Kamagata Y, Sekiguchi Y (2008) Syntrophorhabdus aromaticivorans gen. nov., sp. nov., the first cultured anaerobe capable of degrading phenol to acetate in obligate syntrophic associations with a hydrogenotrophic methanogen. Appl Environ Microbiol 74:2051–2058
Imachi H, Sakai S, Ohashi A, Harada H, Hanada S, Kamagata Y, Sekiguchi Y (2007) Pelotomaculum propionicicum sp. nov., an anaerobic, mesophilic, obligately syntrophic, propionate-oxidizing bacterium. Int J Syst Evol Microbiol 57:1487–1492
Balk M, Weijma J, Friedrich MW, Stams AJM (2003) Methanol utilization by a novel thermophilic homoacetogenic bacterium, Moorella mulderi sp. nov., isolated from a bioreactor. Arch Microbiol 179:315–320
Sieber JR, McInerney MJ, Gunsalus RP (2012) Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol 66
Hernandez-Eugenio G, Fardeau ML, Patel BKC, Macarie H, Garcia JL, Ollivier B (2000) Desulfovibrio mexicanus sp. nov., a sulfate-reducing bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor treating cheese wastewaters. Anaerobe 6:305–312
Walker CB, Stolyar S, Chivian D, Pinel N, Gabster JA, Dehal PS, He Z, Yang ZK, Yen HCB, Zhou J (2009) Contribution of mobile genetic elements to Desulfovibrio vulgaris genome plasticity. Environ Microbiol 11:2244–2252
Lykidis A, Chen CL, Tringe SG, McHardy AC, Copeland A, Kyrpides NC, Hugenholtz P, Macarie H, Olmos A, Monroy O (2010) Multiple syntrophic interactions in a terephthalate-degrading methanogenic consortium. ISME J 5:122–130
Kleinsteuber S, Schleinitz KM, Breitfeld J, Harms H, Richnow HH, Vogt C (2008) Molecular characterization of bacterial communities mineralizing benzene under sulfate‐reducing conditions. FEMS Microbiol Ecol 66:143–157
Herrmann S, Kleinsteuber S, Chatzinotas A, Kuppardt S, Lueders T, Richnow HH, Vogt C (2010) Functional characterization of an anaerobic benzene‐degrading enrichment culture by DNA stable isotope probing. Environ Microbiol 12:401–411
Wüst PK, Horn MA, Drake HL (2009) Trophic links between fermenters and methanogens in a moderately acidic fen soil. Environ Microbiol 11:1395–1409
Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17:377–386
Acknowledgments
The authors would like to thank GRF HKU 7190/12E for the financial support of this research. Feng Ju would like to thank the University of Hong Kong (HKU) for the postgraduate scholarship. Moreover, the authors would like to thank Dr. Feng Guo for the helpful discussion. Finally, the authors would like to thank Ms. Danping Huang for the assistance in reactor operation and Ms. Vicky Fung for the technical support.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 1225 kb)
Rights and permissions
About this article
Cite this article
Ju, F., Zhang, T. Novel Microbial Populations in Ambient and Mesophilic Biogas-Producing and Phenol-Degrading Consortia Unraveled by High-Throughput Sequencing. Microb Ecol 68, 235–246 (2014). https://doi.org/10.1007/s00248-014-0405-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0405-6