Abstract
The white-rot fungus Phanerochaete chrysosporium secretes extracellular oxidative enzymes during secondary metabolism, but lacks versatile peroxidase, an enzyme important in ligninolysis and diverse biotechnology processes. In this study, we report the genetic modification of a P. chrysosporium strain capable of co-expressing two endogenous genes constitutively, manganese peroxidase (mnp1) and lignin peroxidase (lipH8), and the codon-optimized vpl2 gene from Pleurotus eryngii. For this purpose, we employed a highly efficient transformation method based on the use of shock waves developed by our group. The expression of recombinant genes was verified by PCR, Southern blot, quantitative real-time PCR (qRT-PCR), and assays of enzymatic activity. The production yield of ligninolytic enzymes was up to four times higher in comparison to previously published reports. These results may represent significant progress toward the stable production of ligninolytic enzymes and the development of an effective fungal strain with promising biotechnological applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lignin is a complex and heterogeneous aromatic polymer, very abundant in nature and highly recalcitrant to degradation. The interest in lignin biodegradation for industrial applications has increased over the last 20 years but the mechanisms of degradation, particularly regarding the enzymatic and regulatory mechanisms of the metabolic pathways involved, are not completely understood. Microorganisms able to depolymerize lignin include wood-rotting fungi and to a lesser extent some bacteria and actinomycetes (Abdel-Hamid et al. 2012). Nevertheless, only white-rot fungi are able to completely mineralize lignin because they have a complex enzymatic machinery (Martínez et al. 2005).
In recent years, there has been an increased interest in organisms or a consortium of microorganisms that can be utilized as a pretreatment for lignin degradation, in the biotransformation of a wide range of environmental pollutants and in the biobleaching of pulp paper (Nicolaou et al. 2010; Fischer et al. 2008; Davila-Vazquez et al. 2005). In this regard, the white-rot fungus Phanerochaete chrysosporium is considered a valuable species for lignin degradation and other bioprocesses (Dashtban et al. 2010; Singh and Chen 2008). This species secretes an exceptional array of peroxidases and oxidases with high-redox potential in comparison with other organisms (Singh and Chen 2008). However, P. chrysosporium does not produce versatile peroxidases (VPs), which, as has been demonstrated in several reports, is quite relevant for lignin degradation (Salame et al. 2014; Moreira et al. 2007; Camarero et al. 1999). VP degrades and modifies both phenolic and non-phenolic aromatic compounds, which cannot be oxidized directly by peroxidases from P. chrysosporium due to enzyme inactivation or lack of the so-called mediators (Singh and Chen 2008; Martínez et al. 2005; Kamitsuji et al. 2005; Camarero et al. 1999; Ruiz-Dueñas et al. 1999; Martínez et al. 1996). Sometimes, the presence of these redox mediators is required for providing the oxidation of a broad range of complex substrates not oxidized by the enzyme alone (Dashtban et al. 2010).
For numerous metabolic engineering applications, simultaneous expression of genes encoding several enzymes is sometimes more desirable than expression of a single enzyme to improve a specific trait. Previous reports have shown that the diversity and abundance of fungal peroxidases may increase the biotransformation of diverse compounds (Knop et al. 2014; Salame et al. 2014; Knežević et al. 2013; Elisashvili and Kachlishvili 2009). Owing to these observations, it is possible that the heterologous expression of VP may enhance the activity of endogenous enzymes of P. chrysosporium. In addition, overexpression of multiple peroxidases in P. chrysosporium can be favorable since it possesses two of the most important lignin-modifying enzymes, MnP and LiP (Sollewijn et al. 1999; Mayfield et al. 1994). MnP is able to oxidize Mn2+ to Mn3+, which facilitates the degradation of phenolic lignin, simple phenols, amines, and dyes, and additionally oxidizes a second mediator for the degradation of non-phenolic compounds, while LiP catalyze oxidative cleavage of ether and C–C interunit bonds in non-phenolic aromatic substrates of high-redox potential (Dashtban et al. 2010; Martínez et al. 2005). Nevertheless, these peroxidases are secreted only during secondary metabolism in response to nutrient limitation and in very low amounts.
Availability of fungal peroxidases for industrial applications has been limited due to lack of an efficient genetic transformation system, allowing manipulation of multiple genes in a suitable host (Fischer et al. 2008; Meyer et al. 2008; Conesa et al. 2000). Although expression of single genes encoding ligninolytic peroxidases has been explored in filamentous fungi (Conesa et al. 2000; Ruiz-Dueñas et al. 1999), yeasts (Garcia-Ruiz et al. 2012; Jiang et al. 2008), bacteria (Pérez-Boada et al. 2002), and insect cells (Johnson et al. 1991), the production levels of the recombinant enzymes have been low and the enzymes obtained did not have adequate posttranslational modifications (Jiang et al. 2008; Pérez-Boada et al. 2002; Conesa et al. 2000; Ruiz-Dueñas et al. 1999). In contrast, P. chrysosporium can offer a better processing of recombinant peroxidases in comparison with other platforms, and also, the recombinant peroxidases may act in synergy with endogenous enzymes to combine the ability of fungal oxidation for potential future applications.
The aim of this work was the genetic manipulation of this white-rot fungus for increased production of peroxidases. We overexpressed the MnP1 and LiPH8 endogenous enzymes from P. chrysosporium and co-expressed the codon-optimized VPL2 from Pleurotus eryngii. For this purpose, we employed our highly efficient transformation method based on the use of underwater shock waves in a simple and one-step process (Magaña-Ortíz et al. 2013). The expression of MnP1 and LiPH8 was significantly enhanced in the P. chrysosporium transformants strains. Also, a high VP activity was detected in the recombinant strains but not in the wild-type strain. Finally, recombinant VP enzyme was successfully purified and characterized from the crude extract of P. chrysosporium transformed strains.
Materials and methods
Strains and culture conditions
P. chrysosporium wild-type strain ATCC 24725 was used throughout this study. The strain was maintained in the medium described by Tien and Kirk (1988). Conidia were obtained as has been reported previously (Alic et al. 1987).
High-carbon, high-nitrogen (HCHN) medium contained basal medium (Tien and Kirk 1988), 2 % glucose, 24 mM ammonium tartrate, and 20 mM 2,2-dimethyl succinate buffer (pH 4.5) (Ma et al. 2003). For screening of ligninolytic activity, HCHN medium was supplemented with 1.7 % agar and 0.05 % Remazol Brilliant Blue R dye (RBBR) (Sigma Aldrich, Saint Louis, MO, USA) (Moreira et al. 2001). Minimal medium contained 1 % glucose, 0.1 % thiamine, 0.1 % trace elements, and 1 % nitrate salts (Kaminskyj 2001). Liquid cultures were maintained in stationary 125 ml Erlenmeyer flasks and incubated at 37 °C. The inoculum for all growth conditions was one disk (5 mm diameter) of mycelium obtained from a freshly grown colony in solid maintenance culture.
Escherichia coli strain DH5α (Life Technologies Inc., Waltham, MA, USA) was used for cloning of constructs and cultured on LB broth supplemented with ampicillin at 37 °C.
Construction of expression cassettes
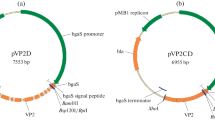
The genes and regulatory regions were amplified by PCR with the primers described in Table 1 using P. chrysosporium genomic DNA as template. PCR was performed in a MultiGene gradient thermal cycler (Labnet, Edison, NJ, USA) with Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA, USA). Expression cassettes were constructed by inserting the different genes under the control of the glyceraldehyde-3-phosphate dehydrogenase (gpd) promoter, followed by the trpC terminator from P. chrysosporium. The fragments mentioned above were cloned in the plasmid pBlueScript SK + (Stratagene, Santa Clara, CA, USA). The mnp1 (GenBank: M77513) and lipH8 (GenBank: M27401) genes obtained for PCR were subcloned into the SpeI-NcoI sites to generate pPCMNP and pPCLIP plasmids, respectively. In both cases, the complete sequence of genes was used, including the introns. PCR cycling profiles were 98 °C for 30 s, 30 cycles of 98 °C for 10 s, 60 °C for 1 min, and 72 °C for 1 min followed by a final extension at 72 °C for 10 min and then hold at 4 °C. For the vector pPCHPH, we included the hygromycin resistance gene (hph) that was commercially synthesized (GenScript, Piscataway, NJ, USA) and cloned into the cassette expression. Finally, for optimal expression in P. chrysosporium, a codon-optimized vpl2 gene of P. eryngii was synthesized by GenScript using the cDNA of a published sequence (GenBank: AF007222) with the sequence for the signal peptide of the mnp1 gene from P. chrysosporium. Codon-optimized vpl2 sequence was deposited in GenBank with the accession number KM206768. This gene was cloned into the plasmid pBlueScript SK + containing gpd promoter and trpC terminator of P. chrysosporium using restriction sites SpeI-NotI to generate the vector pPCVP.
Shock wave-mediated transformation of P. chrysosporium
Transformation of P. chrysosporium was carried out as described previously (Magaña-Ortíz et al. 2013). For co-transformation, 15 μg of each linearized construct (pPCMNP, pPCLIP, and pPCVP) were mixed with 5 μg of selection plasmid pPCHPH. In each protocol of transformation 1 × 104 conidia were employed. The P. chrysosporium conidia were exposed to 150 shock waves at a rate of 0.5 Hz. Underwater shock waves were generated using an experimental shock wave generator, based on a piezoelectric shock wave source, model Piezolith 2300 (Richard Wolf GmbH, Knittlingen, Germany), operated at a discharge voltage of 5.7 kV. The mean positive and negative pressure values, recorded with a polyvinylidene difluoride needle hydrophone (Imotec GmbH, Würselen, Germany), were 37.8 ± 4.2 and −18.2 ± 2.4 MPa, respectively (mean ± standard deviation). Details on the shock wave generator are given elsewhere (Magaña-Ortíz et al. 2013). Transformants were selected by their ability to grow in the presence of 100 μg/ml of hygromycyn on minimal medium. The resistant colonies recovered were subjected to three rounds of selection. The stability of the transformation was confirmed by subculturing colonies onto fresh selective liquid medium (up to 20 times) to verify that the phenotype was maintained. The transformants were grown in liquid minimal media without hygromycin in ten subsequent cultures, and the DNA extracted was used to verify the stability of the gene insertion by PCR.
PCR and Southern blot analysis of the P. chrysosporium transformants
Transformants were cultivated in stationary-flasks in maintenance medium. Mycelium grown for 5 days was filtered and frozen in liquid nitrogen for extraction of genomic DNA. The extraction was performed as described elsewhere (Punekar et al. 2003). For vpl2 gene detection by PCR, the primers vpF and vpR described in the Table 1 were used. PCR was performed in a MultiGene gradient thermocycler (Labnet) using Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA). The amplification program comprised an initial denaturation (94 °C for 5 min) followed by 30 cycles of denaturation at 94 °C for 50 s, annealing (60 °C for 1 min), and extension at 72 °C for 1 min. After amplification, each PCR product was analyzed by agarose gel electrophoresis.
The integration and copy numbers of VP in the transformants were examined by Southern blot using standard procedures (Sambrook et al. 1989). Genomic DNA was digested with BstXI overnight and transferred onto Hybond N + membranes (GE Healthcare, Amersham, Buckinghamshire, UK). The probe was prepared by PCR amplification of the VP gene fragment using primer pair vpF and vpR labeled with a α-32P-dCTP detection.
Gene expression analysis by quantitative real-time PCR
In order to determine mnp1, lipH8, and vpl2 transcript abundance and to confirm that no endogenous enzymes were detected in the transformants, mutant strains were incubated in HCHN medium in parallel for 12 days and sampled every 2 days. The wild-type strain was included in the same set of experiments for each of the abovementioned variables. The mycelium was removed by filtration, ground with liquid nitrogen, and then the total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The first-strand cDNA synthesis was performed with SuperScript III reverse transcriptase (Invitrogen) using oligo dT20 primers as recommended by the supplier. The synthesized cDNA was used as the template for quantitative real-time PCR (qRT-PCR). All measurements were independently repeated three times on two separate biological samples using the primers listed in Table 1. Primers were designed for mnp1, lipH8, vpl2, gpd, and actin (act) genes. The specificity of primer sets used for qRT-PCR amplification was evaluated by melting curve analysis. All quantitative PCRs were performed in CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Reactions were prepared in a total volume of 20 μl containing 1× iQ SYBR Green Supermix (Bio-Rad), 0.1 μM forward primer, 0.1 μM reverse primer, and as template cDNA (100-fold diluted). The following PCR protocols were followed: 15 min initial denaturation at 95 °C, followed by 40 cycles of 15 s at 95 °C and annealing at 60 °C for 1 min. The CFX Manager v2.1 software (Bio-Rad) was used to compile PCR protocols and to define plate set-ups. Transcript abundance data were normalized against the average transcript abundance of act and gpd reference genes. Results of transcription analyses were determined as relative expression ratios.
Protein extracellular preparation
Extracellular proteins were extracted from 50 ml of minimal medium or HCHN liquid cultures collected on days 2, 4, 6, 8, 10, and 12 post-inoculation. Analysis was independently conducted on two separate biological samples. Culture fluid was removed by filtering through Miracloth filters (Calbiochem, San Diego, CA, USA), and the mycelium of each strain was dried at 110 °C until a constant weight was obtained. Subsequently, the supernatant was filtered through 0.22-μm filters (Millipore Corp., Billerica, MA, USA) and concentrated 16-fold by an Amicon 10-kDa cutoff membrane (Millipore) at 4 °C. After this step, the protein concentration was determined based on the Bradford protocol (Bradford 1976) using the Bio-Rad protein assay according to the manufacturer’s instructions with crystalline bovine serum albumin (BSA) as standard. The concentrated samples were analyzed by 12 % sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) according to Laemmli (1970) and utilized for enzymatic assays.
Enzyme activity assays
MnP activity was determined for the formation of Mn3+-malonate complex in 50 mM sodium malonate buffer (pH 4.5) with 0.5 mM MnSO4 (Wariishi et al. 1992). The reaction was initiated by adding H2O2 to the final concentration of 0.1 mM at 270 nm (ε = 11,590 M−1 cm−1). LiP activity was estimated for the oxidation of veratryl alcohol to veratraldehyde at 310 nm (ε = 9300 M−1 cm−1). Reaction mixture contained 2 mM veratryl alcohol, 50 mM tartaric acid pH 3, and 0.4 mM H2O2 in a total volume of 1 ml (Tien and Kirk 1988). The enzyme activity of VP (Mn2+-independent activity) was analyzed by conversion of 2,6-dimethoxyphenol (2,6-DMP) to coerulignone at an absorbance of 469 nm (ε = 27,500 M−1 cm−1). VP activity was measured for the oxidation 1 mM of 2,6-DMP in 100 mM sodium tartrate buffer (pH 3) in the presence of 0.1 mM H2O2 (Pérez-Boada et al. 2002). Enzyme absorption spectra were determined with Pharmacia Biotech Ultrospec 1000 UV/visible spectrophotometer at room temperature by using a cuvette with a 1-cm light path. One unit of enzymatic activity (U) was defined as the amount of enzyme that transforms 1 μmol of substrate per minute.
Versatile peroxidase purification
The transformant with the highest VP activity was selected for enzyme purification and characterization. The extracellular culture was harvested on the optimal VP production day and dialyzed with cellulose dialysis tubing membrane (Sigma) in 10 mM sodium tartrate buffer (pH 4.5) at 4 °C overnight. The sample was loaded onto DEAE-Sepharose Fast Flow column (GE Healthcare, Bio-Sciences, Uppsala, Sweden) pre-equilibrated with 10 mM sodium tartrate buffer, pH 4.5. Elution was performed by a linear NaCl gradient from 0 to 1 M at 4 °C. All fractions with VP activity were pooled and concentrated on an Amicon 10 kDa cutoff membrane (Millipore) and loaded onto Sephadex-G75 gel filtration column (Sigma) equilibrated with 10 mM sodium tartrate buffer (pH 4.5). Protein concentrations were determined by Bradford protein assay (Bio-Rad) to establish specific VP activities. VP-positive fractions were concentrated and analyzed by 12 % SDS-PAGE, and protein bands were visualized by staining with Coomassie brilliant blue R250 (Sigma).
Effect of pH and temperature on VP activity
The pH optimum of recombinant VP was determined in the universal McIlvaine buffer within a pH range of 2.0 to 9.0 at room temperature. Reaction mixtures containing 100 mM McIlvaine buffer (previously pH adjusted), 1 mM 2,6-DMP and 0.1 mM H2O2. The effect of temperature on the VP activity was studied by incubating the enzyme solution at different temperatures between 20 and 80 °C. In each case, the substrate was pre-incubated at the required temperature for 10 min in the presence of 1 mM 2,6-DMP in 100 mM McIlvaine buffer (pH 3.0) and 0.1 mM H2O2. VP activity was normalized to the optimum activity value, and results were calculated as relative activity.
Kinetic analysis of recombinant VP
The Michaelis-Menten and catalytic constants (K m, k cat, and k cat/K m) of recombinant VP toward three typical VP substrates, 2,6-DMP, Mn2+, and 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) were determined. The reactions were conducted in 100 mM sodium tartrate buffer containing 0.1 mM H2O2 at pH 3 for ABTS and 2,6-DMP, and at pH 5 for Mn2+ in 50 mM sodium malonate buffer. Steady-state enzyme kinetics were determined using the following molar absorption coefficients: DMP, ε469 = 27,500 M−1 cm−1; Mn2+, ε270 = 11,590 M−1 cm−1; and ABTS, ε420 = 36,000 M−1 cm−1. The kinetic constants for the purified enzyme were determined from Lineweaver-Burk transformation of Michaelis-Menten model using the statistical program GraphPad Prism (Graph Pad Software, San Diego, USA).
Results
Cloning of peroxidases genes and selection marker in the expression cassettes
The mnp1, lipH8, vpl2, and hph genes were cloned in independent expression cassettes under the control of the gpd promoter and the trpC terminator from P. chrysosporium for constitutive expression (Fig. 1a), yielding vectors pPCMNP, pPCLIP, pPCVP, and pPCHPH. In all cases, restriction analysis and sequencing confirmed the presence of the inserts in the open reading frame.
Molecular analysis and protein profile of the recombinant peroxidases expressed in Phanerochaete chrysosporium. a Schematic diagram of the pPCVP expression vector used in this study to transform P. chrysosporium. b PCR detection of the vpl2 gene in P. chrysosporium transformed with shock waves. The size of the amplicon was 650 bp (arrow). c Southern blot analysis of the four independent transformants. Genomic DNA was digested with BstXI. A specific probe, a fragment vpl2 from pPCVP vector, was labeled with α-32P-dCTP. d SDS-PAGE analysis of extracellular proteins of the transformants producing recombinant peroxidases grown on minimal media. Proteins were visualized by staining the gel with Coomassie brilliant blue R250. The arrows indicate bands detectable only in the recombinant strains. The putative identity of each band has been included
Production of transformants with the four recombinant genes using shock wave-mediated transformation
To obtain P. chrysosporium strains co-expressing the peroxidases and the hph resistance gene, fungal genetic transformation was performed with shock waves. Forty hygromycin-resistant transformants were obtained using shock wave-mediated transformation, but only 20 colonies were analyzed. A positive PCR of the hph gene was observed in all of them (data not shown). For a rapid screening of the ligninolytic ability in the transformants, selected positive strains were inoculated in HCHN medium with RBBR dye. The appearance of a decolorization zone around the colonies indicated positive peroxidase activity. This dye has a complex molecular structure that makes it more difficult to degrade; nevertheless, only fungal peroxidases can catalyze complete degradation of this aromatic dye (Moreira et al. 2001). For further studies, four mutants (PcT1, PcT2, PcT3, and PcT4) were selected for their ability to decolorize RBBR dye in HCHN medium in a short period of time (data not shown). The presence of the VP gene in the recombinant strains was confirmed in all cases by PCR (Fig. 1b).
In addition, the presence of the vpl2 gene in the fungal genome was determined by Southern blot hybridization. Southern blot analysis of the four mutants showed that the vpl2 gene was integrated in the genome of P. chrysosporium. The PcT1 and PcT2 mutants showed two copies of the vpl2 gene, and only one copy was evident in PcT3 and PcT4 (Fig. 1c).
qRT-PCR analysis of recombinant strains
In order to verify the expression of recombinant genes and to confirm that the detectable enzyme activity was produced by recombinant peroxidases, qRT-PCR was performed using total RNA isolated from HCHN-cultures. A significant increase in the levels of transcripts of the three recombinant genes in all four transformed strains was detected by qRT-PCR. MnP1 transcripts of PcT1 were 20 % more abundant than PcT2 and PcT3 from 6 days onwards. PcT4 showed the lowest levels of transcripts. In contrast, the presence of transcripts was not detected during growth on HCHN and stationary culture conditions in the wild-type strain (Fig. 2).
Quantitative RT-PCR analysis of the mnp1, lipH8, and vpl2 transcripts in the transformants. Total RNAs were extracted from transformant mycelia grown in HCHN-supplemented medium from 2 to 12 days. cDNAs were subjected to qRT-PCR analysis using specific primers as described in Table 1. The relative expression level for each gene was normalized to gpd and act genes. Data are presented as the mean of three independent experiments and error bars indicate standard deviation
Furthermore, the levels of LiPH8-specific mRNA in the transformant PcT3 were consistently more abundant than in PcT1 throughout the experiment (Fig. 2). After the sixth day, the levels of LiPH8 transcripts increased approximately 40-fold (over basal levels), with no detectable transcripts in the wild-type strain. PcT2 showed transcript levels lower than those of PcT3 and PcT1, and PcT4 presented the lowest transcript levels.
Analysis of the expression of VP showed that VPL2-specific mRNA is produced by 4 days of growth, whereas in the wild-type strain it is not detectable at any time (Fig. 2). The number of transcripts of PcT1 was about 20 % lower than that of PcT3 by the eighth day. In contrast, PcT1 transcripts were 35 % higher than those of PcT3 by 10 days of cultivation. PcT4 again presented the lowest transcript levels except at day sixth, when the levels were similar than those of PcT3.
Analysis of extracellular recombinant proteins in P. chrysosporium
We identified some of the recombinant proteins preliminary by the molecular size on SDS-PAGE. Analysis of extracellular proteins of the recombinant strains showed several bands, not detectable in the wild-type strain, with apparent molecular weights comparable to those of the recombinant proteins expected. Three bands of about 42, 43, and 46 kDa were similar in molecular weights to that of the native LiP, VP, and MnP, respectively (Fig. 1d). One of them (VP, see below) has been purified to determinate its identity; nevertheless, protein sequencing is in progress to confirm the identity of the rest of bands.
Determination of the enzyme activity of transformed strains
To determine the specific activity of secreted recombinant proteins, the transformants were grown in minimal and HCHN liquid media and enzymatic activity was measured. In all recombinant strains, enzyme activity apparently increased in a more or less regular pattern, but after 12 days of growth we observed a decline in activity in a more irregular pattern (data not shown).
MnP activities corresponding to the formation of Mn3+-malonate complex in the culture supernatants of PcT1 reached the highest level with 9.084 U/ml in HNCN medium. In contrast, no MnP activity was detected in the wild-type strain in HCHN medium. Meanwhile, the enzymatic activity of PcT1 in minimal medium reached its maximum peak with 43.08 U/ml in comparison with 7.11 U/ml of the wild-type strain (Fig. 3a). In the case of PcT3 and PcT4 transformants, after the tenth day, the activity gradually decreased when the activity of PcT1 reached its maximum.
Comparison of extracellular MnP, LiP, and VP activities produced by the P. chrysosporium transformants and wild type during 12 days of cultivation in HCHN (discontinuous lines) and minimal media (continuous lines). a MnP, b LiP, and c VP activity detected after 2 to 12 days of incubation. Data are shown as means ± standard deviations of three independent experiments
On the other hand, strong LiP activity in PcT3 was observed on the tenth day with the highest peak by the twelfth day in HCHN medium. While the opposite result, the wild-type strain did not show any activity under the same conditions. The LiP activity was threefold higher in comparison with the wild-type control at this point in minimal medium (Fig. 3b). Moreover, the highest VP activity on 2,6-DMP was displayed for PcT1 (20 U/ml) in HCHN media, while the enzyme activity in the wild-type strain was not detected at any time (Fig. 3c).
Fungal biomass and the enzymatic activity of recombinant peroxidases
Co-expression of peroxidases did not affect the growth of the transformants nor the active form of the recombinant proteins. The transformants and the wild-type strain showed a similar pattern of growth according to nutrient availability and no significant differences among them were observed, particularly in the early stage of the culture (Fig. 4). In most cases, production of biomass was consistent with elevated peroxidase activities throughout the cultivation period. However, the vigorous growth and increase in fungal biomass did not correlate with the enhanced peroxidase activities in the mutants.
Versatile peroxidase purification
The PcT1 transformant with the highest VP activity was chosen for purification and characterization studies. We knew that the enzyme was secreted into the medium and although the purification was not optimized to obtain large amounts of enzyme, the rVPL2 was purified in only two steps in large amounts. A single band of approximately 43 kDa, as shown by SDS-PAGE analysis in Fig. 5, was obtained. The molecular weight determination indicated that the recombinant enzyme was similar to native VPL2 from P. eryngii.
Evaluation of the pH and temperature on the VP activity profile
In order to characterize rVP, the effect of the temperature and the pH on the activity of the peroxidase was studied using 2,6-DMP as substrate. Despite that only the recombinant protein from PcT1 was purified, all the enzymes had an optimal pH of 3.0, but the activities of all recombinant enzymes decreased dramatically at pH levels greater than 4.0 (Fig. 6). On the other hand, the recombinant enzymes maintained 80–100 % activity at 30 and 40 °C, but decreased to 30 % with incubation at 50 °C (Fig. 6). Interestingly, there were no obvious differences among mutants containing diverse enzymatic activities.
Effect of pH and temperature on the enzymatic activity of VPL2. Activities were measured at different pHs (2–9) and temperatures (20–80 °C) with 1 mM 2,6-DMP and 0.1 mM H2O2. Relative VP activity was obtained of VP normalized to the optimum activity value. Discontinuous lines show the relative activity of purified rVPL2
Kinetic properties of purified enzyme
The steady-state kinetic parameters with three different substrates were determined for the purified rVPL2, as summarized in Table 2. The rVLP2 showed some differences in catalytic properties when it was compared with the native VPL2 from P. eryngii. K m values of DMP (K m = 198 μM) and ABTS (K m = 4 μM) were very similar in comparison with the native enzyme (Table 2), whereas the K m for Mn2+ (K m = 54 μM) was slightly higher with respect to the wild-type VPL2. The highest activity was found with Mn2+(k cat = 29 s−1) followed by similar value for ABTS (k cat = 28 s−1). However, the catalytic efficiency (k cat/K m) was greater for ABTS (7000 s−1 M−1) than with Mn2+. This indicates that the substrates were oxidized by the rVPL2 in a similar manner to those by the native enzyme.
Discussion
In this work, we demonstrated that overexpression of three peroxidases was effectively achieved in a single P. chrysosporium strain. These studies represent an important advance to improve the production yield of ligninolytic enzymes in filamentous fungi.
Shock wave-mediated transformation is an efficient method for gene transfer into intact fungi cells (Magaña-Ortíz et al. 2013). Successful transformation was evidenced by stable expression of the introduced genes. Analysis of four co-transformants expressing the vpl2 showed that the patterns of DNA integration detected are similar to that described by Ma et al. (2003), where mixtures of single and multiple integration events are observed after transformation of P. chrysosporium.
Comparing the relative expression levels of the three recombinant genes in the transformants examined, an increase of transcripts abundance was observed. In an earlier study, Gettemy et al. (1998) reported that MnP1 transcripts were present at low basal levels in wild-type strain of P. chrysosporium grown on nitrogen-rich cultures with or without Mn2+. This observation was consistent with the qRT-PCR analysis of the parental strain reported here. Our data suggest that overexpression of MnP1 in P. chrysosporium induces an increase of MnP1 transcripts that do not correspond to the endogenous enzyme. In the same way, it has been documented that LiP genes are differentially regulated in response to nutrient starvation (Stewart et al. 1992). In contrast, under HCHN conditions, our analysis demonstrated that LiPH8 transcripts of the mutants were present at relatively high levels on days 8 to 12. Under the conditions tested, expression of recombinant lipH8 genes was not impacted by medium composition. Although the levels of expression in individual transformants varied considerably throughout the cultivation period, we detected transcripts of the mnp1, lipH8, and vpl2 genes in relative abundance in the four mutants, without exception. These findings indicate that the recombinant peroxidase genes were constitutively and highly expressed in the host.
It has been shown that recombinant proteins in heterologous host often are not efficiently processed as the native proteins. In a previous study, Jiang et al. (2008) observed that the MnP expressed in Pichia pastoris is hyperglycosylated, with a molecular weight much larger than the native MnP, from 75 to 100 kDa. On the contrary, peroxidases expressed in E. coli have shown a similar molecular weight to deglycosylated native peroxidases, probably because of lack of glycosylation (Bao et al. 2012; Pérez-Boada et al. 2002). Even though these differences in glycosylation may not interfere with enzyme activity, the kinetic properties (such as thermal stability), protein folding, and secretion efficiency could be affected (Bao et al. 2012; Palma et al. 2000). Nevertheless, our results show that the recombinant proteins expressed in P. chrysosporium share very similar molecular weight than native enzymes. These results allow us to infer that heterologous expression in related species may be a suitable option for the production of recombinant peroxidases.
Interestingly, one way to facilitate heterologous gene expression in other hosts is to codon optimize the nucleotide sequence without altering the amino acid sequence (Tokuoka et al. 2008). Likewise, several studies had suggested that changing the native signal peptide by a secretion signal of the host might improve secretion of the heterologous enzyme avoiding adverse effects during the process (Garcia-Ruiz et al. 2012). Although these strategies have been applied individually to improve the expression of VP in E. coli, Saccharomyces cerevisiae, and P. pastoris, the obtained results have showed limited success (Bao et al. 2012; Garcia-Ruiz et al. 2012; Jiang et al. 2008). In our experiments, probably the combination of these strategies contributed to a correct processing and increase of the amount of recombinant enzyme produced in P. chrysosporium transformants than those reported previously (Bao et al. 2012; Pérez-Boada et al. 2002; Ruiz-Dueñas et al. 1999).
According to extracellular activities, production of both MnP and LiP was significantly enhanced in the recombinant strains in comparison with the wild-type strain under the same conditions. The enzymatic activity in MnP was fourfold higher than that previously reported for MnP of P. chrysosporium (2 U/ml) (Mayfield et al. 1994). Similarly, Sollewijn et al. (1999) reported the homologous expression of LiP in P. chrysosporium but low levels of recombinant enzyme were detected with respect to the control. Remarkably, Mn2+-independent activity in the transformants was detected in both enriched and minimal medium and no activity was observed in the wild type in the absence of redox mediators as described previously (Kamitsuji et al. 2005; Palma et al. 2000). Because LiP is not able to oxidize phenolic compounds in the absence of veratryl alcohol and MnP only oxidize phenols in the presence of Mn2+, this ruled out the possibility of any contamination in the assays (Ruiz-Dueñas et al. 2009).
As can be seen in Figs. 1d and 3, a strong increase in recombinant protein and enzyme activity was observed in all the transformants grown in minimal medium. This suggests a synergistic action of the enzymes present in the extracts of recombinants strains and it would clearly be an advantage if we want to employ the strains for biotechnological purposes. In nature, synergy is required among several enzymes for lignin solubilization and the modification of the lignin structure (Dashtban et al. 2010; Martínez et al. 2005). However, studies are in progress to try demonstrating an enhanced ligninolytic activity of these strains on natural substrates, and if the overexpression of these recombinant peroxidases may be associated with the strain efficiency to oxidize a variety of aromatic and recalcitrant compounds.
Although only one recombinant protein was purified, all the extracellular protein extracts analyzed showed a pH optimum of 3.0 on 2,6-DMP. These results are consistent with the reported optimal pH for native VPL2 (Garcia-Ruiz et al. 2012; Martínez et al. 1996). On the other hand, PcT1 and PcT4 samples maintained an optimal temperature activity at 40 °C, similar to parental VPL2 (Garcia-Ruiz et al. 2012). In contrast, PcT2 and PcT3 showed a temperature optimum of 30 °C that decreased to less than 30 % at 50 °C. Previously, it was demonstrated that a small increase of temperature above 28 °C drastically reduced the activity of VP, while reducing the temperature from 28 to 19 °C, the activity increased 5.8-fold (Eibes et al. 2009). We hypothesized that these differences may be due to variants displayed in these recombinant peroxidases that modified its biochemical properties (Garcia-Ruiz et al. 2012). However, this reduction in temperature did not affect the enzyme activity of these transformants. Thus, further purification and characterization analysis of these recombinant proteins will be necessary for a better understanding of its biochemical properties.
The steady-state kinetic study of rVPL2 demonstrated that the oxidation of DMP and ABTS had similar K m and k cat values in comparison to the previously reported in parental VPL2 (Camarero et al. 1999; Heinfling et al. 1998). An unexpected result in the present work was the decrease in both k cat and k cat/K m of the rVPL2 activity toward Mn2+, in relation to the parental enzyme. A possible explanation is a conformational change at the Mn2+-binding site near the haem of the recombinant protein (Kamitsuji et al. 2005; Camarero et al. 1999). Although specific activities and kinetic parameters of rVPL2 were not improved against DMP, Mn2+, and ABTS, the catalytic efficiencies were not affected by heterologous expression. The results described here demonstrate that rVPL2 has the catalytic function to oxidize polymeric substrates without redox mediators. Taking into account the findings of our work as well as those previously reported, we confirmed that rVPL2 expressed in P. chrysosporium has similar characteristics to native VPL2 such as molecular weight (43 kDa), pH-temperature activity profile, Mn2+-independent peroxidase activities, and kinetic parameters.
Several groups have attempted the heterologous expression of fungal peroxidases in different hosts, as basidiomycetes secrete these enzymes in low levels and only during secondary metabolism (Singh and Chen 2008). Fungal peroxidases have been notoriously difficult to produce in large amounts as recombinant proteins, despite the use of fungal species capable of secreting large amounts of proteins (Eibes et al. 2009; Conesa et al. 2000; Ruiz-Dueñas et al. 1999). Most of these failures are probably due to inadequate posttranslational modifications, improper folding, and need for exogenous heme group, which together may result in low or null activity (Jiang et al. 2008; Pérez-Boada et al. 2002; Conesa et al. 2000; Ruiz-Dueñas et al. 1999). Our current study fully confirms that the selection of an adequate host for heterologous expression of peroxidases is a key component of success.
The similar characteristics of the VP from P. eryngii and the peroxidases from P. chrysosporium indicates that P. chrysosporium may be more suitable than any other ascomycete fungus in expressing recombinant peroxidases.
Availability of fungal peroxidases for several industrial applications is an area of interest in research. There are many recent studies on isolation and characterization of well-known or novel peroxidases, in addition to several reports on the expression of the enzymes in homologous and heterologous fungal and bacterial hosts, with the aim to overproduce peroxidases (Janusz et al. 2013). Clearly, inefficiencies of current methods for fungal genetic transformation have limited progress in this field. Ideally, peroxidases should be produced and secreted into the medium and, in order to enhance secretion, many genetic transformation approaches have tried homologous expression (Singh and Chen 2008; Tsukihara et al. 2006; Irie et al. 2001). However, homologous expression of MnP and LiP in P. chrysosporium has had very limited success (Sollewijn et al. 1999; Mayfield et al. 1994). In the present study, our results demonstrate that up to 119.7 U/mg of MnP, 29.9 U/mg of LiP, and 61.4 U/mg of VP can be produced in a single transformant of P. chrysosporium in minimal medium, without the use of expensive compounds such as heme or veratryl alcohol. In contrast, only 19.8 U/mg of MnP and 19.4 U/mg of LiP in the parental strain can be obtained under the same conditions.
We believe that several factors contributed to the improvement of the expression. Two genes, mnp1 and lipH8, were amplified from the P. chrysosporium genome and were cloned and expressed in the homologous host. The third gene vpl2 was codon-optimized for P. chrysosporium expression and the native signal peptide replaced with that of MnP. Shock wave-mediated transformation without manipulation of the cell wall in a simple and one-step that, together with the selection of a host with appropriate genetic background, probably contributed to obtain the reported results. Although further works are required to determine the capacity of the recombinant enzymes in degradation of lignocellulosic substrates, our findings confirm that the co-expression of mnp1, lipH8, and vpl2 in a single strain of P. chrysosporium is possible. Since the recombinant peroxidases from P. chrysosporium exhibit multiple enzymatic activities, these strains may have the potential for a rapid oxidation of phenolic and non-phenolic compounds.
References
Abdel-Hamid AM, Solbiati JO, Cann IK (2012) Insights into lignin degradation and its potential industrial applications. Adv Appl Microbiol 82:1–28
Alic M, Letzring C, Gold MH (1987) Mating system and basidiospore formation in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol 53:1464–1469
Bao X, Liu A, Lu X, Li JJ (2012) Direct over-expression, characterization and H2O2 stability study of active Pleurotus eryngii versatile peroxidase in Escherichia coli. Biotechnol Lett 34:1537–1543
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Camarero S, Sarkar S, Ruiz-Dueñas FJ, Martínez MJ, Martínez ÁT (1999) Description of a versatile peroxidase involved in the natural degradation of lignin that has both manganese peroxidase and lignin peroxidase substrate interaction sites. J Biol Chem 274:10324–10330
Conesa A, van den Hondel CA, Punt PJ (2000) Studies on the production of fungal peroxidases in Aspergillus niger. Appl Environ Microbiol 66:3016–3023
Dashtban M, Schraft H, Syed TA, Qin W (2010) Fungal biodegradation and enzymatic modification of lignin. Int J Biochem Mol Biol 1:36–50
Davila-Vazquez G, Tinoco R, Pickard MA, Vazquez-Duhalt R (2005) Transformation of halogenated pesticides by versatile peroxidase from Bjerkandera adusta. Enzym Microb Technol 36:223–231
Eibes GM, Lu-Chau TA, Ruiz-Dueñas FJ, Feijoo G, Martínez MJ, Martínez AT, Lema JM (2009) Effect of culture temperature on the heterologous expression of Pleurotus eryngii versatile peroxidase in Aspergillus hosts. Bioprocess Biosyst Eng 32:129–134
Elisashvili V, Kachlishvili E (2009) Physiological regulation of laccase and manganese peroxidase production by white-rot Basidiomycetes. J Biotechnol 144:37–42
Fischer CR, Klein-Marcuschamer D, Stephanopoulos G (2008) Selection and optimization of microbial hosts for biofuels production. Metab Eng 10:295–304
Garcia-Ruiz E, Gonzalez-Perez D, Ruiz-Dueñas FJ, Martínez AT, Alcalde M (2012) Directed evolution of a temperature-, peroxide-and alkaline pH-tolerant versatile peroxidase. Biochem J 441:487–498
Gettemy JM, Ma B, Alic M, Gold MH (1998) Reverse transcription-PCR analysis of the regulation of the manganese peroxidase gene family. Appl Environ Microbiol 64:569–574
Harmsen MC, Schuren FHJ, Moukha SM, van Zuilen CM, Punt PJ, Wessels JGH (1992) Sequence analysis of the glyceraldehyde-3-phosphate dehydrogenase genes from the basidiomycetes Schizophyllum commune, Phanerochaete chrysosporium and Agaricus bisporus. Curr Genet 22:447–454
Heinfling A, Ruiz-Dueñas FJ, Martínez MJ, Bergbauer M, Szewzyk U, Martínez AT (1998) A study on reducing substrates of manganese-oxidizing peroxidases from Pleurotus eryngii and Bjerkandera adusta. FEBS Lett 428:141–146
Irie T, Honda Y, Watanabe T, Kuwahara M (2001) Homologous expression of recombinant manganese peroxidase genes in ligninolytic fungus Pleurotus ostreatus. Appl Microbiol Biotechnol 55:566–570
Janusz G, Kucharzyk KH, Pawlik A, Staszczak M, Paszczynski AJ (2013) Fungal laccase, manganese peroxidase and lignin peroxidase: gene expression and regulation. Enzym Microb Technol 52:1–12
Jiang F, Kongsaeree P, Charron R, Lajoie C, Xu H, Scott G, Kelly C (2008) Production and separation of manganese peroxidase from heme amended yeast cultures. Biotechnol Bioeng 99:540–549
Johnson TM, Li JKK (1991) Heterologous expression and characterization of an active lignin peroxidase from Phanerochaete chrysosporium using recombinant baculovirus. Arch Biochem Biophys 291:371–378
Kaminskyj SGW (2001) Fundamentals of growth, storage, genetics and microscopy of Aspergillus nidulans. Fungal Genet Newsl 48:25–31
Kamitsuji H, Watanabe T, Honda Y, Kuwahara M (2005) Direct oxidation of polymeric substrates by multifunctional manganese peroxidase isoenzyme from Pleurotus ostreatus without redox mediators. Biochem J 386:387–93
Knežević A, Milovanović I, Stajić M, Lončar N, Brčeski I, Vukojević J, Cilerdžić J (2013) Lignin degradation by selected fungal species. Bioresour Technol 138:117–123
Knop D, Ben-Ari J, Salame TM, Levinson D, Yarden O, Hadar Y (2014) Mn2+-deficiency reveals a key role for the Pleurotus ostreatus versatile peroxidase (VP4) in oxidation of aromatic compounds. Appl Microbiol Biotechnol 98:6795–6804
Laemmli UK (1970) Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature 227:680–685
Ma B, Mayfield MB, Gold MH (2003) Homologous expression of Phanerochaete chrysosporium manganese peroxidase, using bialaphos resistance as a dominant selectable marker. Curr Genet 43:407–414
Magaña-Ortíz D, Coconi-Linares N, Ortiz-Vazquez E, Fernández F, Loske AM, Gómez-Lim MA (2013) A novel and highly efficient method for genetic transformation of fungi employing shock waves. Fungal Genet Biol 56:9–16
Martínez MJ, Ruiz-Dueñas FJ, Guillén F, Martínez ÁT (1996) Purification and catalytic properties of two manganese peroxidase isoenzymes from Pleurotus eryngii. Eur J Biochem 237:424–432
Martínez AT, Speranza M, Ruiz-Dueñas FJ, Ferreira P, Camarero S, Guillén F, Martínez MJ, Gutiérrez A, del Río JC (2005) Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int Microbiol 8:195–204
Mayfield MB, Kishi K, Alic M, Gold MH (1994) Homologous expression of recombinant manganese peroxidase in Phanerochaete chrysosporium. Appl Environ Microbiol 60:4303–4309
Meyer V (2008) Genetic engineering of filamentous fungi—progress, obstacles and future trends. Biotechnol Adv 26:177–185
Moreira PR, Almeida-Vara E, Sena-Martins G, Polonia I, Malcata FX, Duarte JC (2001) Decolourisation of Remazol Brilliant Blue R via a novel Bjerkandera sp. strain. J Biotechnol 89:107–111
Moreira PR, Almeida-Vara E, Malcata FX, Duarte JC (2007) Lignin transformation by a versatile peroxidase from a novel Bjerkandera sp. strain. Int Biodeter Biodegr 59:234–238
Nicolaou SA, Gaida SM, Papoutsakis ET (2010) A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng 12:307–331
Palma C, Martínez AT, Lema JM, Martínez MJ (2000) Different fungal manganese-oxidizing peroxidases: a comparison between Bjerkandera sp. and Phanerochaete chrysosporium. J Biotechnol 77:235–245
Pérez-Boada M, Doyle WA, Ruiz-Dueñas FJ, Martínez MJ, Martínez AT, Smith AT (2002) Expression of Pleurotus eryngii versatile peroxidase in Escherichia coli and optimisation of in vitro folding. Enzym Microb Technol 30:518–524
Punekar NS, Suresh-Kumar SV, Jayashri TN (2003) Isolation of genomic DNA from acetone-dried Aspergillus mycelia. Fungal Genet Newsl 50:15–16
Ruiz-Dueñas FJ, Martínez MJ, Martínez ÁT (1999) Heterologous expression of Pleurotus eryngii peroxidase confirms its ability to oxidize Mn2+ and different aromatic substrates. Appl Environ Microbiol 65:4705–4707
Ruiz-Dueñas FJ, Morales M, Garcia E, Miki Y, Martínez MJ, Martínez AT (2009) Substrate oxidation sites in versatile peroxidase and other basidiomycete peroxidases. J Exp Bot 60:441–452
Salame TM, Knop D, Levinson D, Mabjeesh SJ, Yarden O, Hadar Y (2014) Inactivation of a Pleurotus ostreatus versatile peroxidase-encoding gene (mnp2) results in reduced lignin degradation. Environ Microbiol 16:265–77
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Harbor Press, Cold Spring Harbor, NY
Singh D, Chen S (2008) The white-rot fungus Phanerochaete chrysosporium: conditions for the production of lignin-degrading enzymes. Appl Microbiol Biotechnol 81:399–417
Sollewijn Gelpke MD, Mayfield-Gambill M, Lin Cereghino GP, Gold MH (1999) Homologous expression of recombinant lignin peroxidase in Phanerochaete chrysosporium. Appl Environ Microbiol 65:1670–1674
Stewart P, Kersten P, Vanden Wymelenberg A, Gaskell J, Cullen D (1992) Lignin peroxidase gene family of Phanerochaete chrysosporium: complex regulation by carbon and nitrogen limitation and identification of a second dimorphic chromosome. J Bacteriol 174:5036–5042
Tien M, Kirk T (1988) Lignin peroxidase of Phanerochaete chrysosporium. Methods Enzymol 161:238–249
Tokuoka M, Tanaka M, Ono K, Takagi S, Shintani T, Gomi K (2008) Codon optimization increases steady-state mRNA levels in Aspergillus oryzae heterologous gene expression. Appl Environ Microbiol 74:6538–6546
Tsukihara T, Honda Y, Sakai R, Watanabe T, Watanabe T (2006) Exclusive overproduction of recombinant versatile peroxidase MnP2 by genetically modified white rot fungus, Pleurotus ostreatus. J Biotechnol 126:431–439
Wariishi H, Valli K, Gold MH (1992) Manganese (II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators. J Biol Chem 267:23688–23695
Acknowledgments
NCL and DMO are indebted to CONACYT for doctoral fellowships 219893 and 219950.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Coconi-Linares, N., Magaña-Ortíz, D., Guzmán-Ortiz, D.A. et al. High-yield production of manganese peroxidase, lignin peroxidase, and versatile peroxidase in Phanerochaete chrysosporium . Appl Microbiol Biotechnol 98, 9283–9294 (2014). https://doi.org/10.1007/s00253-014-6105-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6105-9