Abstract
Elimination of acetate overflow in aerobic cultivation of Escherichia coli would improve many bioprocesses as acetate accumulation in the growth environment leads to numerous negative effects, e.g. loss of carbon, inhibition of growth, target product synthesis, etc. Despite many years of studies, the mechanism and regulation of acetate overflow are still not completely understood. Therefore, we studied the growth of E. coli K-12 BW25113 and several of its mutant strains affecting acetate-related pathways using the continuous culture method accelerostat (A-stat) at various specific glucose consumption rates with the aim of diminishing acetate overflow. Absolute quantitative exo-metabolome and proteome analyses coupled to metabolic flux analysis enabled us to demonstrate that onset of acetate overflow can be postponed and acetate excretion strongly reduced in E. coli by coordinated activation of phosphotransacetylase-acetyl-CoA synthetase (PTA-ACS) and tricarboxylic acid (TCA) cycles. Fourfold reduction of acetate excretion (2 vs. 8 % from total carbon) at fastest growth compared to wild type was achieved by deleting the genes responsible for inactivation of acetyl-CoA synthetase protein (pka) and TCA cycle regulator arcA. The Δpka ΔarcA strain did not accumulate any other detrimental by-product besides acetate and showed identical μ max and only ~5 % lower biomass yield compared to wild type. We conclude that a fine-tuned coordination between increasing the recycling capabilities of acetate in the PTA-ACS node through a higher concentration of active acetate scavenging Acs protein and downstream metabolism throughput in the TCA cycle is necessary for diminishing overflow metabolism of acetate in E. coli and achieving higher target product production in bioprocesses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acetate is the major by-product in bioprocesses using Escherichia coli and glucose as the carbon source. Acetate excretion or overflow during fast growth/glucose uptake aerobic fermentations of E. coli has been studied for a long time, and it is known to lead to decreased biomass formation, inhibition of recombinant protein and low molecular weight compound production (Mazumdar et al. 2010; Shiloach et al. 2010; Wolfe 2005).
Acetate overflow is generally believed to be caused by an imbalance between substrate uptake and anabolic/catabolic throughput of downstream pathways, and many theories about different possible limitations in metabolism have been proposed: limitation in tricarboxylic acid (TCA) cycle (Majewski and Domach 1990; Veit et al. 2007), respiratory chain (Han et al. 1992; Paalme et al. 1997; Varma and Palsson 1994) or glyoxylate shunt (GS) activity (Shin et al. 2009; Waegeman et al. 2011) and competition for membrane space (Zhuang et al. 2011). Several process level and genetic manipulations have been tried to mitigate acetate overflow (reviewed in De Mey et al. 2007). It has been shown that disruption of the main acetate synthesis pathway—phosphotransacetylase-acetate kinase (PTA-ACKA)—reduces acetate excretion but results in lower specific growth rate (μ), biomass yield (Y xs) and substantially elevated lactate and formate excretion (Castaño-Cerezo et al. 2009; Contiero et al. 2000; Dittrich et al. 2005; El-Mansi and Holms 1989; Yang et al. 1999), while deletion of the second major route for acetate production through pyruvate oxidase (PoxB) causes loss of aerobic growth efficiency (Abdel-Hamid et al. 2001). These results demonstrate that acetate overflow cannot be simply avoided by removing the main synthesis pathways without any negative side effects. Furthermore, none of the theories or process/genetic efforts has been able to unequivocally explain the mechanism of acetate overflow.
Recently, a new perspective on the long-studied phenomenon has emerged as it is proposed by several groups that E. coli actually synthesises acetate constantly at all μ under aerobic conditions and no acetate overflow occurs at low μ since acetate is fully recycled in the phosphotransacetylase-acetyl-CoA synthetase (PTA-ACS) node (Fig. 1) (Renilla et al. 2011; Shin et al. 2009; Valgepea et al. 2010). Furthermore, we have proposed a new theory for the cause of acetate overflow: acetate excretion in fast growing cells is closely related to the loss of acetate-glucose co-utilisation capability and triggered by carbon catabolite repression-mediated down-regulation of acetyl-CoA synthetase (Acs) leading to disruption of the PTA-ACS node (Valgepea et al. 2010).
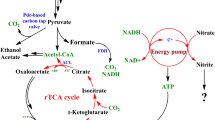
Simplified overview of glucose metabolism, PTA-ACS node and regulation of Acs by Pka and CobB in E. coli. Acetate is mainly generated from: (a) acetyl-CoA by Pta and AckA enzymes via the high energy intermediate acetyl-phosphate (acetyl-P); (b) acetyl-P during different cellular regulation processes (see Wolfe 2005 and Valgepea et al. 2010 for details); (c) pyruvate by PoxB. Acetate in the PTA-ACS node is reactivated by recycling it back to acetyl-CoA by the high-affinity Acs enzyme. Activity of Acs is essential for functioning of the PTA-ACS node: protein lysine acetyltransferase (Pka) inactivates Acs by acetylation (AcsAc), while the NAD+-dependent protein deacetylase CobB reactivates Acs by deacetylating it. Pta phosphotransacetylase, AckA acetate kinase, Acs acetyl-CoA synthetase, PoxB pyruvate oxidase, TCA tricarboxylic acid cycle, GS glyoxylate shunt
Known regulators involved in the control of acs expression in E. coli are cAMP receptor protein (CRP), factor for inversion stimulation (Fis), integration host factor (IHF) (Wolfe 2005) and possibly the dual transcriptional regulator for anoxic redox control (ArcA) (Covert et al. 2004). Besides transcriptional regulation, Acs protein activity in bacteria is regulated by the Pat/CobB system which was first described in Salmonella enterica (Starai et al. 2002; Starai and Escalante-Semerena 2004). An identical system is present in E. coli: protein lysine acetyltransferase (Pka) inactivates Acs by acetylation (Castaño-Cerezo et al. 2011), while the NAD+-dependent regulator protein deacetylase CobB releases Acs from repression by deacetylating it (Zhao et al. 2004) (Fig. 1). Interestingly, increased Acs activity together with derepression of GS enzymes decreases acetate accumulation in E. coli batch cultures (Shin et al. 2009) pointing to the importance of Acs and its active/inactive pools in acetate metabolism.
As regulation of Acs and PTA-ACS node seems to have an important role in acetate overflow, the aim of this work was to gain further understanding of the relevance of the PTA-ACS node in overflow metabolism possibly leading to reduction of acetate overflow in aerobic E. coli cultivations. For this, we studied the growth of E. coli K-12 BW25113 and several of its mutant strains affecting acetate-related pathways in continuous cultures (accelerostat (A-stat); Paalme et al. 1995) at various specific glucose consumption rates (q glc) using absolute quantitative exo-metabolome and proteome analyses coupled to metabolic flux analysis (MFA). Furthermore, as repression of the high-affinity acetate scavenging enzyme Acs seems to be central in acetate overflow regulation through the loss of acetate-glucose co-utilisation capability (Valgepea et al. 2010), we focused this study on Acs-related inhibition of acetate overflow by improving acetate recycling capabilities.
We chose A-stat (Paalme et al. 1995) for the cultivation method since it allows accurate detection of metabolic switch points (e.g. start of acetate overflow) and high-resolution description of metabolism dynamics (e.g. acetate accumulation) which are both instrumental for a system-wide study of acetate overflow regulation in E. coli (Valgepea et al. 2010). A-stat experiments produce comparable results with chemostats at the level of major growth characteristics in E. coli and Lactococcus lactis (Lahtvee et al. 2011; Valgepea et al. 2010, 2011) and at transcriptome (Valgepea et al. 2010) and proteome (Nahku 2012) levels in E. coli.
In this work, we show that acetate overflow in aerobic E. coli cultivations can be remarkably reduced by coordinated activation of PTA-ACS and TCA cycles which was achieved by deleting the Acs-inactivating gene pka and TCA cycle regulator arcA. We conclude that a fine-tuned coordination between increasing the recycling capabilities of acetate in the PTA-ACS node through a higher concentration of active acetate scavenging Acs protein and downstream metabolism throughput in the TCA cycle is necessary for diminishing overflow metabolism of acetate in E. coli.
Materials and methods
Bacterial strains, growth medium and continuous culture conditions
Bacterial strains used in this work were E. coli K-12 BW25113 wild type (WT) and its single-gene knockouts obtained from the Keio collection (Baba et al. 2006) or constructed using homologous recombination. Strains overexpressing acs (acsOE and Δpka acsOE) were constructed by swapping the native acs-yjcH-actP promoter region of the parent strain (WT and Δpka, respectively) with a Ptet promoter sequence (Veit et al. 2007). The Ptet promoter sequence was first annealed with a kanamycin resistance gene from a pCP13 plasmid (Datsenko and Wanner 2000) by PCR after which this construct was used to swap the native promoter region using homologous recombination as described by Datsenko and Wanner (2000). For the construction of Δpka ΔarcA double knockout strain, arcA gene was deleted from the Δpka strain using a homologous recombination method that uses pSIM6 plasmid instead of pKD46 (Sharan et al. 2009). The strains constructed in this work are deposited in the public strain collection of the Collection of Environmental and Laboratory Strains (University of Tartu, Estonia) with reference numbers shown in Table 1. All strains used in this work with their abbreviations are listed in Table 1.
All A-stat experiments were carried out using a defined minimal medium supplemented with glucose (4.5 g/L) as described before by Valgepea et al. (2010). Glucose-limited cultures were grown in 1.25 L BioBundle bioreactors (Applikon Biotechnology B.V., Schiedam, the Netherlands) controlled by an ez-Control biocontroller (Applikon Biotechnology B.V.) and a cultivation control program “BioXpert XP” (Applikon Biotechnology B.V.). The system was equipped with OD, pH, pO2, off-gas O2 and CO2 and temperature sensors.
A-stat experiments were carried out at 37 °C, pH 7 and aerobic conditions ensured by agitation of 800 rpm and air flow rate of 150 mL/min. Glucose-limited cultures were stabilised in chemostat at dilution rate (D) 0.1 or 0.2 h−1 after which A-stat with a continuous increase of D (acceleration of 0.01 h−2) was initiated. The control algorithm for A-stat was: D = D 0 + a D × t, where D 0 is the initial D of chemostat (h−1), a D is the acceleration of the A-stat phase (h−2) and t is the time from the start of A-stat (h).
WT and ΔarcA, Δpka ΔarcA strains were cultivated in four and two independent A-stat experiments, respectively. Other strains were cultivated in single A-stat experiments. Growth characteristics in A-stat were calculated using the formulas described in a previous study (Kasemets et al. 2003) on the basis of total volume of medium pumped out from bioreactor (L), biomass (gram dry cellular weight (gDCW)/L) and organic acid concentrations (mM) in culture broth and CO2 concentration in the off-gas (mM). We note that in CO2 production calculations, also the CO2 and HCO3 − dissolved in the culture broth were taken into account as described by Taymaz-Nikerel et al. (2013).
Analytical methods
Exo-metabolome analysis for a more complete carbon balance was carried out as described previously in Valgepea et al. (2011). In short, samples of culture broth were centrifuged (14,000 × g for 5 min), and the supernatant was collected and analysed for glucose and organic acids by HPLC (Alliance2795 system, Waters Corporation, Milford, MA, USA) using either a Bio-Rad HPX-87H Aminex ion-exclusion (Bio-Rad Laboratories, Inc., Hercules, CA, USA) or Agilent Hi-Plex H (Agilent Technologies, Santa Clara, CA, USA) column connected to RI and UV detectors (35 °C, flow rate 0.6 mL/min). The column was eluted with 4.1 mM sulphuric acid for glucose, carbamoyl-aspartate, lactate and orotate and with 26.5 mM formic acid for acetate, dihydroorotate and acetyl-aspartate analysis.
Biomass concentration (X) in the experiments with WT, ΔarcA and Δpka ΔarcA was determined gravimetrically and expressed in gram dry cell weight per liter. Approximately 15 ml of culture broth was collected, weighed and transferred onto a pre-weighed glass fiber filter (S-Pak Membrane Filter White gridded 0.45 μm, EMD Millipore Corporation, Billerica, MA, USA) which was mounted on a vacuum filtration device. Special care was taken to ensure that all the culture broth was transferred onto the filter. Next, the filter was heated in an oven at 105 °C for >24 h after which the filter with dried biomass was cooled in a desiccator for >1 h and finally weighed to determine the dry biomass weight. Coefficient of variation for the determination of X with this method between technical replicates was <1 % (three technical replicates in 27 sample points). The developed method was used to determine the correlation factor (K) for all μ values between X and optical density (at 600 nm) of culture broth (OD), (K = X / OD). The obtained linear μ-dependent relationship of K = −0.2309 × μ + 0.4534 for WT was used to calculate X for the experiments with strains ΔcobB, Δacs, Δpka, acsOE and Δpka acsOE based on measured OD values.
Metabolic flux analysis
MFA using strain-dependent growth characteristics and exo-metabolome data for ΔarcA, Δpka ΔarcA and WT was carried out as described previously (Valgepea et al. 2011) except that pyruvate instead of oxaloacetate was set as the metabolite characterising carbon outflow that was not identified experimentally. In short, the simplified metabolic network contained the main central carbon metabolism pathways—glycolysis, TCA cycle, pentose phosphate pathway (PPP)—one pyrimidine synthesis pathway (to include the excreted carbamoyl-aspartate, orotate and dihydroorotate) and the acetyl-aspartate synthesis reaction. The reconstructed network converted into a fully determined and calculable stoichiometric matrix consisting of 24 metabolites and 50 fluxes (24 unknown, 1 measured inflow, 7 outflow and 18 calculated fluxes based on biomass composition and stoichiometries of anabolic pathways). The same metabolic network, stoichiometric matrix and other MFA parameters were used for all the three analysed strains, since, to the best of our knowledge, neither arcA nor pka deletion should affect MFA characteristics compared to WT.
Proteome analysis
Sample preparation
One milliliter of WT, Δpka, ΔarcA and Δpka ΔarcA culture broth was harvested at μ = 0.4 h−1 and centrifuged (14,000 × g for 1 min at 4 °C), and the pellet was washed once with phosphate buffered saline (PBS), flash frozen with liquid N2 and kept at −80 °C until further processing. Frozen pellets were melted on ice after which 100 μg of the sample biomass was mixed with 100 μg of stable isotope labeling by amino acids in cell culture (SILAC)-labeled (Ong et al. 2002) E. coli biomass (internal standard) and frozen at −80 °C. Cell pellets were suspended in 100 μL SDS lysis buffer (4 % SDS/100 mM Tris-HCl pH 8/100 mM DTT) and heated at 95 °C for 5 min. Cell lysates were sonicated with ultrasound for a few pulses and pelleted by centrifugation. Cell lysates were digested with trypsin according to the Filter Aided Sample Preparation protocol (FASP) (Wiśniewski et al. 2009) and purified with C-18 StageTips (Rappsilber et al. 2007).
Sample analysis
Sample analysis was done as in Arike et al. (2012) with the exception of running one technical replicate for WT cells (triplicate for mutant strains). In short, LC-MS/MS analysis was performed using an Agilent 1200 series nanoflow system (Agilent Technologies) connected to an LTQ Orbitrap mass spectrometer (Thermo Electron, San Jose, CA, USA) equipped with a nanoelectrospray ion source (Proxeon, Odense, Denmark). Peptides were separated with a 240-min gradient from 2 to 40 % B (A—0.5 % acetic acid, B—0.5 % acetic acid/80 % acetonitrile) using a flow rate of 200 nL/min. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaíno et al. 2013) with the dataset identifier PXD000556.
Data analysis
Data analysis of raw MS files was performed by the MaxQuant software package version 1.3.0.5 (Cox and Mann 2008). Peak lists were searched using the Andromeda search engine (built into MaxQuant) against an E. coli database (downloaded on September 5, 2013 from http://www.uniprot.org/) which was supplemented with common contaminants (e.g. human keratin, trypsin). Full tryptic specificity, a maximum of two missed cleavages and a mass tolerance of 0.5 Da for fragment ions, was specified in the MaxQuant search. Carbamidomethylation of cysteine was set as a fixed modification, and methionine oxidation and protein N-terminal acetylation were set as variable modifications. The required false discovery rate was set to 1 % for both peptide and protein levels and the minimum required peptide length was set to seven amino acids. “Match between runs” option with a time window of 2 min was allowed. To increase peptide identification by “Match between runs” function, additional 26 WT E. coli raw proteome MS files were supplemented to the MaxQuant search.
Relative and absolute proteome quantification
Since each sample was mixed with equal amounts of SILAC-labeled E. coli internal standard, relative protein expression differences for >1,000 proteins between mutants and WT could be calculated from expression differences between each sample and the internal standard. All protein ratios were normalised assuming a median expression change of 1 since equal amounts of labeled internal standard and unlabeled sample were mixed together.
Absolute proteome quantification to determine intracellular protein concentrations (protein copies/fL of biomass) was performed using a label-free protein quantification method termed the total protein approach (Arike et al. 2012; Wiśniewski et al. 2012). This method assumes that most of the proteins are detected by MS and this is usually the case as the top 1,000 proteins make up more than 90 % of the protein mass (Milo 2013; Valgepea et al. 2013). For each protein, its relative fraction from the total proteome can be derived from the ratio of its individual LFQ intensity (reported by MaxQuant) to the total MS signal. This ratio can be converted into protein concentration (copies/fL) by dividing with the protein’s molecular weight and multiplying with the Avogadro constant and the protein concentration in a cell (g protein/mL cell volume) (Milo 2013). We determined the protein concentration using measured biomass concentration and protein fraction of dry biomass, cell buoyant density of 1.1 g/mL (Baldwin et al. 1995; Loferer-Krössbacher et al. 1998) and dry fraction of wet biomass of 30 % (Cayley et al. 1991). As SILAC-labeling is more sensitive and reproducible than label-free quantification (Asara et al. 2008; Hendrickson et al. 2006), we determined protein concentrations using the above-described method only for a WT sample from the A-stat experiment at μ = 0.4 h−1. All the reported protein concentrations in this work were calculated by multiplying the values of the latter sample with the relative protein expression differences between each sample and the SILAC-labeled E. coli internal standard to maintain the high accuracy of SILAC labeling for relative comparison of strains. Protein concentrations instead of abundances per cell were calculated because cell size can vary between conditions and strains (Milo 2013; Valgepea et al. 2013).
Gene expression analysis
Real-time PCR for acs expression analysis was carried out as described before (Nahku et al. 2010) except that the primer concentration used in this work was 20 μM.
Results
Glucose-limited cultures were first stabilised in a chemostat at either D = 0.1 or 0.2 h−1 to achieve steady-state conditions after which the A-stat phase with a continuous increase of D was started using an acceleration of 0.01 h−2. This yielded steady-state representative growth until cells could not keep up with the rising D after achieving μ max, resulting in culture washout. E. coli BW25113 wild-type (WT) and ΔarcA, Δpka ΔarcA strains were cultivated in four and two independent A-stat experiments, respectively. Other strains were analysed in single experiments since the data of strains cultivated in multiple replicates confirmed the high reproducibility of A-stat data (Lahtvee et al. 2011; Valgepea et al. 2010). We note that growth characteristics of different strains are compared relative to q glc instead of μ in this study since glucose uptake is known to regulate the magnitude of carbon catabolite repression (Görke and Stülke 2008; Narang 2009) possibly triggering the down-regulation of Acs and acetate overflow (Valgepea et al. 2010).
Effect of Acs deletion and overexpression on acetate metabolism
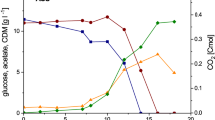
We first wanted to verify the hypothesis that E. coli synthesises acetate also at low μ using WT with deleted acs (Δacs) which should lead to disruption of the PTA-ACS node and loss of acetate recycling. Indeed, as expected, the PTA-ACS node disruption in Δacs led to acetate excretion at all q glc values studied, while no acetate accumulated in WT up to q glc ~ 4.2 mmol gDCW−1 h−1 (Fig. 2a). This phenotype of Δacs is consistent with previous observations (Renilla et al. 2011; Shin et al. 2009) and confirms the hypothesis that acetate is constantly synthesised at all μ (Renilla et al. 2011; Shin et al. 2009; Valgepea et al. 2010). As Δacs constantly directed more carbon to acetate, a lower maximum q glc (Fig. 2a) and on average 5 % lower Y xs (0.42 ± 0.00 vs. 0.44 ± 0.01 gDCW/g glucose; average of the q glc range ± standard deviation) were observed compared to WT.
Specific glucose consumption rate (q glc)-dependent carbon flow to acetate in E. coli K-12 BW25113 wild type and its Acs-related mutants. The percentage of carbon flow from glucose to acetate: a in wild type (blue solid line), acsOE (pink dashed line), Δacs (green dotted line); b in WT (blue solid line), ΔcobB (pink dashed line), Δpka (green dotted line), Δpka acsOE (yellow dash-dotted line). Lines for each strain are best-fit splines of 9–36 acetate and glucose concentration measurements in each experiment. Error bars represent standard deviation of four independent A-stat experiments
As one of the proposed reasons for acetate overflow is the repression of Acs (Valgepea et al. 2010), we tested the effect of increasing Acs expression on acetate excretion. Hence, acs was overexpressed (acsOE) by swapping the native promoter of the acs-yjcH-actP operon by a strong constitutive promoter Ptet (Veit et al. 2007) to maintain recycling of acetate in the PTA-ACS node also at faster growth. A 114 ± 10.3- and 9 ± 0.6-fold higher expression of acs messenger RNA (mRNA) in acsOE compared to WT was measured by real-time PCR at μ = 0.5 h−1 (q glc ~ 6 mmol gDCW−1 h−1) and μ = 0.2 h−1 (q glc ~ 3 mmol gDCW−1 h−1), respectively. However, acetate overflow started at a lower q glc in acsOE, and the strain also excreted more carbon as acetate than WT at all the q glc studied (Fig. 2a). Similar to Δacs, lower maximum q glc and μ max were achieved in acsOE compared to WT. The fact that higher expression of the acetate-consuming gene acs led to earlier onset of acetate overflow and higher carbon wasting to acetate could possibly be a result of either the negative effects of too high protein overexpression (see “Discussion”) and/or due to most of the Acs protein pool being inactive. Hence, we next concentrated on the effects regulating Acs protein activity.
Protein acetylation plays an important role in acetate metabolism
Two enzymes in E. coli are known to regulate the activity of Acs protein: Pka inactivates Acs by acetylation (Castaño-Cerezo et al. 2011), while CobB can release Acs from repression by deacetylating it (Zhao et al. 2004) (Fig. 1). Moreover, it has been shown in vitro that Acs can autoacetylate itself in the presence of acetate and ATP (Barak et al. 2004), further demonstrating the importance of acetylation in determining Acs protein activity. Hence, to investigate the effects of altered protein acetylation on acetate metabolism, we analysed growth of Pka (Δpka) and CobB (ΔcobB) single-gene knockout mutants of E. coli.
ΔcobB cells, which cannot reactivate Acs by deacetylation, excreted more acetate (Fig. 2b) and showed on average 28 % lower Y xs (0.32 ± 0.01 vs. 0.44 ± 0.01 gDCW/g glucose) compared to WT at all studied q glc values. Similar to the Δacs strain, a relatively constant fraction from the carbon balance was directed to acetate up to q glc ~ 4 mmol gDCW−1 h−1 (Fig. 2b). This further confirms the importance of the state of the Acs protein pool and an active PTA-ACS node in acetate metabolism.
Next, we analysed the growth of a strain lacking Pka (Δpka), known to inactivate Acs in S. enterica (Starai and Escalante-Semerena 2004) and E. coli (Castaño-Cerezo et al. 2011), which should eliminate or at least substantially decrease the fraction of inactivated Acs protein in the cell, possibly leading to higher recycling of acetate in the PTA-ACS node and postponed acetate overflow. Indeed, Δpka showed a postponed start of acetate overflow at q glc = 5.5 compared to ~4.2 mmol gDCW−1 h−1 in WT (Fig. 2b). We wanted to verify if the diminished acetate overflow in Δpka was indeed caused by higher Acs activity in vivo. However, this hypothesis cannot be unfortunately tested since, to the best of our knowledge, there exists no assay to unequivocally determine the in vivo activity of Acs in E. coli cells expressing proteins of both acetate-consuming pathways (ACKA-PTA and Acs). Since all the strains studied in this work express the three enzymes of the acetate-consuming pathways (AckA, Pta and Acs) at considerable concentrations as can be seen from the absolute quantitative proteome data described below, in vivo activity of Acs cannot be unequivocally determined in any of the strains.
As we saw a positive effect of pka deletion on diminishing acetate metabolism, we next introduced pka deletion into the acsOE background (Δpka acsOE) to test if the lack of a positive effect of acs overexpression on mitigating acetate accumulation could have arisen from a substantial part of the Acs protein pool being inactive in the acsOE strain (see above). This was not the case as acetate overflow was not postponed in Δpka acsOE compared to WT (Fig. 2b). Therefore, we searched for other means to further postpone the onset and decrease the magnitude of acetate overflow.
Strong reduction of acetate overflow by higher levels of active Acs and TCA cycle throughput
Our results showed that strong overexpression of acs had no positive effects on relieving acetate overflow. However, since we saw diminished overflow of acetate in Δpka which altered the levels of active Acs, we surmised that acetate overflow could be further reduced in Δpka if Acs levels would be slightly higher, but not too high as in Δpka acsOE (see “Discussion”). Furthermore, it could be plausible that even if the Δpka cells could recycle more acetate due to higher levels of active Acs, downstream pathways (e.g. TCA cycle) could still be limiting in processing the recycled acetyl-CoA to divert carbon away from acetate. Hence, we also looked for ways to increase downstream throughput for the recycled acetate.
Deleting ArcA induces expression of TCA cycle genes (Covert et al. 2004; Vemuri et al. 2006) and flux throughput (Perrenoud and Sauer 2005). Interestingly, Covert et al. (2004) also detected ~2-fold higher expression of acs in the arcA deletion mutant compared to WT. Hence, it seemed that an arcA deletion would hit two birds with one stone for us, and therefore, we deleted arcA in the Δpka background (Δpka ΔarcA) in order to concurrently increase the levels of active Acs and downstream throughput of the TCA cycle to further diminish acetate overflow. Remarkably, the double-knockout strain showed even further reduced acetate overflow as the onset of acetate accumulation was postponed until q glc ~ 6 mmol gDCW−1 h−1 (q glc ~ 4.2 mmol gDCW−1 h−1 in WT), and more importantly, carbon wasting into acetate in Δpka ΔarcA was more than 4-fold lower compared to WT (2 vs. 8 % of total carbon) at maximal q glc (Fig. 3a).
Specific glucose consumption rate (q glc)-dependent carbon flow through metabolism in E. coli K-12 BW25113 wild type and its acetate overflow-reduced mutants. The percentage of carbon flow from glucose to acetate (a) and CO2 (b). The percentage of carbon flow from glucose through TCA cycle (c) and pentose phosphate pathway (PPP) (d) represented by suc and gnd fluxes, respectively, expressed as the C-molar percentage of flux through the reaction from the consumed carbon. Wild type (blue solid line), ΔarcA (pink dashed line), Δpka ΔarcA (green dotted line). Lines for each strain are best-fit splines of 13–43 glucose and product concentration measurements in each experiment. Error bars represent standard deviation of four and two independent A-stat experiments for wild type and ΔarcA, Δpka ΔarcA, respectively
Carbon balance showed that Δpka ΔarcA primarily produced more CO2 instead of acetate (2–8 % more than WT) (Fig. 3b). In addition, MFA revealed that the higher CO2 production was the result of increased TCA cycle fluxes as expected from an arcA deletion (Fig. 3c; Supplementary Table S1). MFA also showed that while more CO2 was produced in the TCA cycle, less CO2 was produced through PPP in Δpka ΔarcA, demonstrating the relevance of balancing TCA cycle and PPP also in acetate metabolism (Fig. 3d; Supplementary Table S1). Compared to acetate overflow-reduced strains of acetate synthesis pathway deletions (pta + ackA and poxB) which show reduced μ, Y xs and substantially elevated lactate and formate excretion (see above), our Δpka ΔarcA strain with strongly reduced overflow did not accumulate any other detrimental by-product, maintained μ max of WT (Fig. 3a) and exhibited only ~5 % lower Y xs (0.42 ± 0.00 vs. 0.44 ± 0.01 gDCW/g glucose) compared to WT.
The results of the study of Vemuri et al. (2006) indicated that acetate overflow could be postponed by a single arcA knockout in E. coli K-12 MG1655. Hence, we also analysed the growth of ΔarcA in our E. coli K-12 BW25113 background to check if the arcA knockout effect could be strain-dependent and whether the positive effects of Δpka ΔarcA on diminishing acetate overflow could actually arise only from an arcA deletion. It is evident that the arcA deletion alone could not be accounted for the reduced overflow in Δpka ΔarcA in our BW25113 background as acetate overflow started even earlier (q glc ~ 3.5 vs. ~4.2 mmol gDCW−1 h−1) in ΔarcA compared to WT (Fig. 3a). Interestingly, accumulation of acetate was slower and ΔarcA wasted ~4 % less carbon into acetate compared to WT near maximal q glc (Fig. 3a). As expected, ΔarcA diverted less carbon to CO2 compared to Δpka ΔarcA (Fig. 3b; Supplementary Table S1), which is possibly due to an active PTA-ACS node in Δpka ΔarcA that provides the TCA cycle with more acetyl-CoA through higher acetate recycling compared to ΔarcA. The latter points to the importance of Δpka in facilitating higher TCA cycle throughput by supplying it with more acetyl-CoA through higher acetate recycling in the PTA-ACS node.
Proteomic response to different genetic disruptions
We carried out absolute quantitative proteomics analysis of WT and several mutant strains yielding intracellular protein concentrations to find out if the different metabolic flux patterns could be described by proteome expression profiles. For this, we used SILAC-labeled E. coli biomass as an internal standard (see “Materials and methods”) to compare the proteomes in single A-stat experiments of Δpka, ΔarcA and Δpka ΔarcA with WT at μ = 0.4 h−1 (Fig. 4 and Supplementary Table S2). We chose to compare the proteome expression profiles at μ = 0.4 h−1 corresponding to q glc values of 5.55, 5.25, 5.44 and 5.01 mmol gDCW−1 h−1 for Δpka, ΔarcA, Δpka ΔarcA and WT, respectively, since significant differences in acetate overflow characteristics between the strains had occurred by that high μ (q glc). Proteome analysis for WT was carried out also at μ = 0.2 and 0.51 h−1 in two independent A-stat experiments yielding an average coefficient of variation of protein expression changes over the quantified >1,000 proteins to be <7 %. The latter means that protein expression differences >14 % between a mutant strain and WT could be considered significant. Proteome-wide data for protein concentrations and expression changes between strains is in Supplementary Table S2.
Proteome expression of E. coli K-12 BW25113 ΔarcA (left box), Δpka (middle box) and Δpka ΔarcA (right box) compared to wild type at μ = 0.4 h−1. Numbers represent fold change of protein concentration compared to wild type; down-regulation is shown in red and denoted by negative values, up-regulation in blue and by positive values. Colour intensity is proportional to fold change and scaled by the maximum and minimum protein concentration fold change on the figure. Proteins aligned on the grey area are known to be associated with cell membrane. NDH I and ATP synthase represent the average fold change of NADH dehydrogenase and ATP synthase complex protein concentrations, respectively. Arrow heads show the presumed reaction direction in given conditions
Based on the observation that acs expression is increased ~2-fold in ΔarcA compared to WT (Covert et al. 2004), we introduced arcA deletion into Δpka strain with the expectation of increasing the levels of active Acs protein. Indeed, our proteome analysis confirmed the latter as expression of Acs increased ~1.7-fold in response to arcA deletion both in ΔarcA and Δpka ΔarcA strains (Fig. 4; all comparisons relative to WT). This demonstrates that the total Acs protein concentration, most probably together with the levels of its active form, plays a role in acetate metabolism as acetate overflow was strongly reduced in Δpka ΔarcA (see above).
We detected increased carbon fluxes through the TCA cycle resulting in higher CO2 fraction from the carbon balance in Δpka ΔarcA (Fig. 3b, c). This was confirmed also on the translational level as expression of the TCA cycle enzymes was enhanced in Δpka ΔarcA (on average 1.19-fold compared to WT; p ~ 0.001, paired two-tailed t test) (Fig. 4). Notably, although TCA cycle fluxes in ΔarcA were lower compared to Δpka ΔarcA at the proteome data points of q glc ~ 5 mmol gDCW−1 h−1, protein concentrations in the pathway were elevated in ΔarcA (on average 1.25-fold, p < 10−3). The result of lower flux throughput with higher protein concentrations (comparing ΔarcA to Δpka ΔarcA) points to non-constant catalytic rates of enzymes exerted through post-translational regulation. This observation is in line with recent bacterial studies (Adamberg et al. 2012; Valgepea et al. 2013) which demonstrate that post-translational control of fluxes through alteration of catalytic rates of enzymes is the prevalent mode of flux regulation in E. coli and L. lactis.
Deletion of arcA leads to a strong 7-fold expression increase of the TCA cycle and respiratory chain mRNAs in E. coli chemostat cultures at D = 0.4 h−1 (Vemuri et al. 2006). However, protein concentrations in the TCA cycle were increased only 1.50-fold in our ΔarcA strain at the same D (Fig. 4). Moreover, instead of a 7-fold increase, we detected lower expression of cytochromes (CyoABD and CydA) both in ΔarcA and Δpka ΔarcA and small changes in NADH dehydrogenase complexes NDH I and NDH II (Fig. 4). The difference between our results and those of Vemuri et al. (2006) highlights the importance of protein measurements as analysing only mRNA data could lead to equivocal conclusions.
Shin et al. (2009) report that expression of GS in addition to Acs has to be increased for reducing acetate overflow in E. coli batch cultures. However, we detected lower expression of GS proteins AceA and AceB in Δpka ΔarcA (Fig. 4), and yet this strain showed strongly reduced acetate overflow. This discrepancy could possibly be explained by different E. coli strains (W3110 in Shin et al. 2009) and growth conditions of glucose limitation in our case and glucose excess in the experiments by Shin et al. (2009).
Regarding other possibly relevant protein expression changes for reduced acetate overflow, expression of PoxB (converts pyruvate and ubiquinone to acetate and ubiquinol) was considerably lower in all analysed mutants (Fig. 4). Although this could be relevant for lower acetate accumulation at faster growth since PoxB is suggested to be the main enzyme synthesising acetate at high μ (Nahku et al. 2010), the protein concentration of PoxB in all the mutants was low compared to other PTA-ACS node proteins (Supplementary Table S2). In addition, we detected a very strong up-regulation, 2–6-fold in Δpka ΔarcA and 5–30-fold in Δpka and ΔarcA, of proteins related to chemotaxis and flagellar assembly (Supplementary Table S2). However, this has been seen before: highly frequent insertions of insertion sequence elements to the regulatory region of fliD cause expression changes of these genes (Barker et al. 2004; Ishii et al. 2007; Nahku et al. 2011).
Discussion
In this work, we studied the growth of E. coli K-12 BW25113 and several of its mutant strains affecting acetate-related pathways with the aim of further understanding the relevance of the PTA-ACS node in overflow metabolism and mitigating acetate overflow in aerobic E. coli cultivations. Firstly, our results confirm the notion that acetate is synthesised in vivo also at low μ (Renilla et al. 2011; Shin et al. 2009; Valgepea et al. 2010) as strains defective in acetate re-utilisation (Δacs and ΔcobB) excreted acetate during slow growth where WT did not (Fig. 2a, b). This further supports the idea that an active Acs pool prevents acetate overflow during slow growth of WT E. coli since acetate is fully recycled in the PTA-ACS node. Moreover, the PTA-ACS node is also relevant for growth efficiency since the higher acetate excretion in both of the mutants with disrupted PTA-ACS node (Δacs and ΔcobB) leads to lower Y xs compared to WT. Both Δacs and ΔcobB mutations seem to have further negative effects on Y xs in addition to carbon loss to acetate as the increase of carbon flux to acetate cannot solely explain the lower Y xs values observed. For example, the substantially larger fraction of acetate in the carbon balance of ΔcobB compared to WT at high q glc values (Fig. 2b) shows that Acs is not the only protein specifically regulated by CobB, as also shown before (Weinert et al. 2013).
An intermediate molecule of the PTA-ACS cycle—acetyl-phosphate (acetyl-P)—plays a critical role in the acetylation of many proteins (Weinert et al. 2013) and regulation of chemotaxis (Barak et al. 1998; Klein et al. 2007; Mayover et al. 1999), pathogenesis (Anfora et al. 2008), protein degradation (Mizrahi et al. 2009), etc. in E. coli (Fig. 1). Based on the experiment with Δacs, we propose that ~0.8 mmol gDCW−1 h−1 (corresponding to 2 % from the carbon balance in Δacs) of acetate is constantly recycled in the PTA-ACS node of WT E. coli to enable rapid regulation of the acetyl-P pool. This value is higher compared to the calculated value of ~0.3 mmol gDCW−1 h−1 at μ = 0.2 h−1 based on chemotaxis needs and in vivo synthesis of acetate as a by-product during synthesis of cysteine, methionine and arginine (Valgepea et al. 2010), possibly due to the fact that requirement of acetyl-P for protein acetylation was not taken into account in these calculations.
The fact that lack of (active) Acs causes acetate overflow at low μ (Fig. 2a) and the hypothesis that acetate accumulation with increasing q glc is triggered by Acs repression (Valgepea et al. 2010) obviously lead to the idea that higher expression of Acs should reduce acetate overflow. Although acs overexpressing strains have been constructed (Lin et al. 2006; Shin et al. 2009), no steady-state data at different q glc values exists. Moreover, the data in the literature is somewhat contradictory. Shin et al. (2009) showed that both higher activity of GS and acs overexpression are necessary, whereas only higher expression of Acs was concluded to be sufficient by San and coworkers (Lin et al. 2006) to decrease acetate accumulation in E. coli batch cultures. We, however, saw no reduction of acetate overflow in the constructed acsOE strain in our continuous cultures (Fig. 2a). Furthermore, the possibility of inactivation of the higher Acs pool in acsOE was ruled out by introducing deletion of the gene—pka—responsible for Acs inactivation in E. coli (Castaño-Cerezo et al. 2011) in the latter strain since the double mutant strain (Δpka acsOE) accumulated more acetate than WT (Fig. 2b). As both acs overexpression mutants accumulated more acetate than WT and had a lower maximal q glc, we propose that the 114-fold (at high μ) acs overexpression evoked a strong metabolic burden on the cells either by causing the PTA-ACS node to deplete the ATP pool (it is a futile cycle) and/or by significantly altered energy homeostasis through increased energy demands for Acs protein synthesis due to the overexpression. Both of the proposed mechanisms severely hurt the ATP balance which may trigger the cells to excrete acetate at a lower q glc for energy generation purposes (Valgepea et al. 2013).
Our results confirm the hypothesis of Acs repression triggering acetate overflow (Valgepea et al. 2010) as deleting the Acs-inactivating gene pka resulted in postponed acetate overflow (Fig. 2b). Our proteome analysis of Δpka showed lower Acs protein concentration and no major changes in central carbon metabolism compared to WT (Fig. 4). Therefore, it seems most probable that the reduced acetate overflow in the Δpka strain arises from a higher pool of active Acs which enables increased acetate recycling capability in the PTA-ACS node.
The latter results indicated that acetate overflow could probably be further diminished if the level of active Acs would be slightly higher, but not too high as in Δpka acsOE. Approximately 1.8-fold higher expression of Acs in Δpka background was achieved by arcA knockout (Fig. 4), which is consistent with the increased acs expression of E. coli ΔarcA during exponential growth (Covert et al. 2004). Remarkably, the Δpka ΔarcA double-knockout showed strongly reduced acetate overflow by the postponed onset of acetate excretion and 4-fold lower carbon flux to acetate compared to WT at maximal q glc (Fig. 3a). The fact that Δpka ΔarcA synthesised more CO2 due to increased TCA cycle throughput (Fig. 3b) instead of acetate demonstrates an important point: directing higher amounts of recycled acetyl-CoA in the PTA-ACS node further into the TCA cycle is essential for diminishing acetate overflow. Positive effects of arcA knockout on TCA cycle gene expression (Covert et al. 2004; Vemuri et al. 2006) and flux throughput (Perrenoud and Sauer 2005) are expected and were also confirmed on the translational level by our proteome analysis (Fig. 4). It is important to note that the Δpka ΔarcA strain would enable the production of target compounds in the absence of acetate at considerably higher values of q glc, most probably resulting in higher volumetric productivities (~22 % higher gDCW L−1 h−1 compared to WT). Moreover, this strain accumulated no other detrimental by-product and showed identical μ max and only ~5 % lower Y xs compared to WT. This work demonstrates that a simple genetic overexpression does not work in all cases for achieving the desired effects, but the expression level of the relevant fraction of the protein pool (active Acs in this case) has to be fine-tuned together with downstream throughput (TCA). To the best of our knowledge, this is the first successful application of the modification of protein acetylation for metabolic engineering in E. coli.
Several explanations for acetate overflow propose limitations in respiratory capacity (Han et al. 1992; Paalme et al. 1997; Varma and Palsson 1994), TCA cycle throughput (Majewski and Domach 1990; Veit et al. 2007) or activity of the GS (Shin et al. 2009; Waegeman et al. 2011). Our results suggest that none of these pathways is limiting and triggering acetate overflow in WT E. coli per se since we detected lower expression of cytochrome and GS proteins in Δpka ΔarcA compared to WT and higher acetate excretion in ΔarcA with up-regulated TCA cycle protein expression compared to Δpka ΔarcA and WT (Figs. 3a and 4). More recently, Zhuang et al. (2011) proposed that acetate overflow could result from the competition for membrane space between respiratory chain enzymes and glucose transporters. However, this hypothesis has been questioned (Huberts et al. 2012), and also our proteomic data of this (Fig. 4) and of a previous study (Valgepea et al. 2010) are not consistent with Zhuang et al. (2011) as expression of respiratory chain and TCA cycle enzymes showed different behaviour. Hence, we conclude that a fine-tuned coordination between increasing the recycling capabilities of acetate in the PTA-ACS node through a higher concentration of active acetate scavenging Acs protein and downstream metabolism throughput in the TCA cycle is necessary for diminishing overflow metabolism of acetate in E. coli. We see this work being a good example for proving the value of systems biology study of metabolism since the strains with potential interest to industrial use reported in this work were created based on the results of previous system-wide studies of E. coli metabolism (Valgepea et al. 2010, 2011, 2013).
References
Abdel-Hamid AM, Attwood MM, Guest JR (2001) Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology 147:1483–1498
Adamberg K, Seiman A, Vilu R (2012) Increased biomass yield of Lactococcus lactis by reduced overconsumption of amino acids and increased catalytic activities of enzymes. PLoS ONE 7(10):e48223
Anfora AT, Halladin DK, Haugen BJ, Welch RA (2008) Uropathogenic Escherichia coli CFT073 is adapted to acetatogenic growth but does not require acetate during murine urinary tract infection. Infect Immun 76(12):5760–5767
Arike L, Valgepea K, Peil L, Nahku R, Adamberg K, Vilu R (2012) Comparison and applications of label-free absolute proteome quantification methods on Escherichia coli. J Proteome 75(17):5437–5448
Asara JM, Christofk HR, Freimark LM, Cantley LC (2008) A label-free quantification method by MS/MS TIC compared to SILAC and spectral counting in a proteomics screen. Proteomics 8(5):994–999
Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008
Baldwin WW, Myer R, Kung T, Anderson E, Koch AL (1995) Growth and buoyant density of Escherichia coli at very low osmolarities. J Bacteriol 177(1):235–237
Barak R, Abouhamad WN, Eisenbach M (1998) Both acetate kinase and acetyl coenzyme A synthetase are involved in acetate-stimulated change in the direction of flagellar rotation in Escherichia coli. J Bacteriol 180(4):985–988
Barak R, Prasad K, Shainskaya A, Wolfe AJ, Eisenbach M (2004) Acetylation of the chemotaxis response regulator CheY by acetyl-CoA synthetase purified from Escherichia coli. J Mol Biol 342(2):383–401
Barker CS, Prüss BM, Matsumura P (2004) Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J Bacteriol 186(22):7529–7537
Castaño-Cerezo S, Pastor JM, Renilla S, Bernal V, Iborra JL, Cánovas M (2009) An insight into the role of phosphotransacetylase (pta) and the acetate/acetyl-CoA node in Escherichia coli. Microb Cell Factories 8:54
Castaño-Cerezo S, Bernal V, Blanco-Catalá J, Iborra JL, Cánovas M (2011) cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol Microbiol 82(5):1110–1128
Cayley S, Lewis BA, Guttman HJ, Record MT Jr (1991) Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J Mol Biol 222(2):281–300
Contiero J, Beatty CM, Kumari S, DeSanti CL, Strohl WR, Wolfe AJ (2000) Effects of mutations in acetate metabolism in high-cell-density growth of Escherichia coli. J Ind Microbiol Biotechnol 24(6):421–430
Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BØ (2004) Integrating high-throughput and computational data elucidates bacterial networks. Nature 429(6987):92–96
Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26(12):1367–1372
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97(12):6640–6645
De Mey M, Lequeux GJ, Beauprez JJ, Maertens J, Van Horen E, Soetaert WK, Vanrolleghem PA, Vandamme EJ (2007) Comparison of different strategies to reduce acetate formation in Escherichia coli. Biotechnol Prog 23(5):1053–1063
Dittrich CR, Bennett GN, San KY (2005) Characterization of the acetate-producing pathways in Escherichia coli. Biotechnol Prog 21(4):1062–1067
El-Mansi EM, Holms WH (1989) Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J Gen Microbiol 135(11):2875–2883
Görke B, Stülke J (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6(8):613–624
Han K, Lim HC, Hong J (1992) Acetic acid formation in Escherichia coli fermentation. Biotechnol Bioeng 39(6):663–671
Hendrickson EL, Xia Q, Wang T, Leigh JA, Hackett M (2006) Comparison of spectral counting and metabolic stable isotope labeling for use with quantitative microbial proteomics. Analyst 131(12):1335–1341
Huberts DH, Niebel B, Heinemann M (2012) A flux-sensing mechanism could regulate the switch between respiration and fermentation. FEMS Yeast Res 12(2):118–128
Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, Hirasawa T, Naba M, Hirai K, Hoque A, Ho PY, Kakazu Y, Sugawara K, Igarashi S, Harada S, Masuda T, Sugiyama N, Togashi T, Hasegawa M, Takai Y, Yugi K, Arakawa K, Iwata N, Toya Y, Nakayama Y, Nishioka T, Shimizu K, Mori H, Tomita M (2007) Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316(5824):593–597
Kasemets K, Drews M, Nisamedtinov I, Adamberg K, Paalme T (2003) Modification of A-stat for the characterization of microorganisms. J Microbiol Methods 55(1):187–200
Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ (2007) The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol 189(15):5574–5581
Lahtvee P-J, Adamberg K, Arike L, Nahku R, Aller K, Vilu R (2011) Multi-omics approach to study the growth efficiency and amino acid metabolism in Lactococcus lactis at various specific growth rates. Microb Cell Factories 10:12
Lin H, Castro NM, Bennett GN, San K-Y (2006) Acetyl-CoA synthetase overexpression in Escherichia coli demonstrates more efficient acetate assimilation and lower acetate accumulation: a potential tool in metabolic engineering. Appl Microbiol Biotechnol 71(6):870–874
Loferer-Krössbacher M, Klima J, Psenner R (1998) Determination of bacterial cell dry mass by transmission electron microscopy and densitometric image analysis. Appl Environ Microbiol 64(2):688–694
Majewski RA, Domach MM (1990) Simple constrained-optimization view of acetate overflow in E. coli. Biotechnol Bioeng 35(7):732–738
Mayover TL, Halkides CJ, Stewart RC (1999) Kinetic characterization of CheY phosphorylation reactions: comparison of P-CheA and small-molecule phosphodonors. Biochemistry 38(8):2259–2271
Mazumdar S, Clomburg JM, Gonzalez R (2010) Escherichia coli strains engineered for homofermentative production of D-lactic acid from glycerol. Appl Environ Microbiol 76(13):4327–4336
Milo R (2013) What is the total number of protein molecules per cell volume? A call to rethink some published values. BioEssays 35(12):1050–1055
Mizrahi I, Biran D, Ron EZ (2009) Involvement of the Pta-AckA pathway in protein folding and aggregation. Res Microbiol 160(1):80–84
Nahku R (2012) Validation of critical factors for the quantitative characterization of bacterial physiology in accelerostat cultures. Dissertation, Tallinn University of Technology, Tallinn
Nahku R, Valgepea K, Lahtvee P-J, Erm S, Abner K, Adamberg K, Vilu R (2010) Specific growth rate dependent transcriptome profiling of Escherichia coli K12 MG1655 in accelerostat cultures. J Biotechnol 145(1):60–65
Nahku R, Peebo K, Valgepea K, Barrick JE, Adamberg K, Vilu R (2011) Stock culture heterogeneity rather than new mutational variation complicates short-term cell physiology studies of Escherichia coli K-12 MG1655 in continuous culture. Microbiology 157:2604–2610
Narang A (2009) Quantitative effect and regulatory function of cyclic adenosine 5′-phosphate in Escherichia coli. J Biosci 34(3):445–463
Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1(5):376–386
Paalme T, Kahru A, Elken R, Vanatalu K, Tiisma K, Vilu R (1995) The computer-controlled continuous culture of Escherichia coli with smooth change of dilution rate (A-stat). J Microbiol Methods 24(2):145–153
Paalme T, Elken R, Kahru A, Vanatalu K, Vilu R (1997) The growth rate control in Escherichia coli at near to maximum growth rates: the A-stat approach. Antonie Van Leeuwenhoek 71(3):217–230
Perrenoud A, Sauer U (2005) Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J Bacteriol 187(9):3171–3179
Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2(8):1896–1906
Renilla S, Bernal V, Fuhrer T, Castaño-Cerezo S, Pastor JM, Iborra JL, Sauer U, Cánovas M (2011) Acetate scavenging activity in Escherichia coli: interplay of acetyl-CoA synthetase and the PEP-glyoxylate cycle in chemostat cultures. Appl Microbiol Biotechnol 93(5):2109–2024
Sharan SK, Thomason LC, Kuznetsov SG, Court DL (2009) Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc 4(2):206–223
Shiloach J, Reshamwala S, Noronha SB, Negrete A (2010) Analyzing metabolic variations in different bacterial strains, historical perspectives and current trends—example E. coli. Curr Opin Biotechnol 21(1):21–26
Shin S, Chang DE, Pan JG (2009) Acetate consumption activity directly determines the level of acetate accumulation during Escherichia coli W3110 growth. J Microbiol Biotechnol 19(10):1127–1134
Starai VJ, Escalante-Semerena JC (2004) Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J Mol Biol 340(5):1005–1012
Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC (2002) Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298(5602):2390–2392
Taymaz-Nikerel H, De Mey M, Baart G, Maertens J, Heijnen JJ, van Gulik W (2013) Changes in substrate availability in Escherichia coli lead to rapid metabolite, flux and growth rate responses. Metab Eng 16:115–129
Valgepea K, Adamberg K, Nahku R, Lahtvee P-J, Arike L, Vilu R (2010) Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase. BMC Syst Biol 4:166
Valgepea K, Adamberg K, Vilu R (2011) Decrease of energy spilling in Escherichia coli continuous cultures with rising specific growth rate and carbon wasting. BMC Syst Biol 5:106
Valgepea K, Adamberg K, Seiman A, Vilu R (2013) Escherichia coli achieves faster growth by increasing catalytic and translation rates of proteins. Mol BioSyst 9(9):2344–2358
Varma A, Palsson BØ (1994) Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl Environ Microbiol 60(10):3724–3731
Veit A, Polen T, Wendisch VF (2007) Global gene expression analysis of glucose overflow metabolism in Escherichia coli and reduction of aerobic acetate formation. Appl Microbiol Biotechnol 74(2):406–421
Vemuri GN, Altman E, Sangurdekar DP, Khodursky AB, Eiteman MA (2006) Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl Environ Microbiol 72(5):3653–3661
Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O’Kelly G, Schoenegger A, Ovelleiro D, Pérez-Riverol Y, Reisinger F, Ríos D, Wang R, Hermjakob H (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41:D1063–D1069
Waegeman H, Beauprez J, Moens H, Maertens J, De Mey M, Foulquié-Moreno MR, Heijnen JJ, Charlier D, Soetaert W (2011) Effect of iclR and arcA knockouts on biomass formation and metabolic fluxes in Escherichia coli K12 and its implications on understanding the metabolism of Escherichia coli BL21 (DE3). BMC Microbiol 11:70
Weinert BT, Iesmantavicius V, Wagner SA, Schölz C, Gummesson B, Beli P, Nyström T, Choudhary C (2013) Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell 51(2):265–272
Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6(5):359–362
Wiśniewski JR, Ostasiewicz P, Duś K, Zielińska DF, Gnad F, Mann M (2012) Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Mol Syst Biol 8:611
Wolfe AJ (2005) The acetate switch. Microbiol Mol Biol Rev 69(1):12–50
Yang YT, Bennett GN, San K-Y (1999) Effect of inactivation of nuo and ackA-pta on redistribution of metabolic fluxes in Escherichia coli. Biotechnol Bioeng 65(3):291–297
Zhao K, Chai X, Marmorstein R (2004) Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J Mol Biol 337(3):731–741
Zhuang K, Vemuri GN, Mahadevan R (2011) Economics of membrane occupancy and respiro-fermentation. Mol Syst Biol 7:500
Acknowledgments
The authors thank Liisa Arike and Andres Maser for their help with absolute protein quantification and fermentation experiments, respectively. The financial support for this work was provided by the European Regional Development Fund project EU29994; Ministry of Education, Estonia, through the grant SF0140090s08; and Estonian Science Foundation through grants G8165 and G9192. The data deposition to the ProteomeXchange Consortium was supported by the PRIDE team.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLS 779 kb)
Rights and permissions
About this article
Cite this article
Peebo, K., Valgepea, K., Nahku, R. et al. Coordinated activation of PTA-ACS and TCA cycles strongly reduces overflow metabolism of acetate in Escherichia coli . Appl Microbiol Biotechnol 98, 5131–5143 (2014). https://doi.org/10.1007/s00253-014-5613-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5613-y