Abstract
Development of sustainable technologies for the production of 3-hydroxypropionic acid (3HP) as a platform chemical has recently been gaining much attention owing to its versatility in applications for the synthesis of other specialty chemicals. Several proposed biological synthesis routes and strategies for producing 3HP from glucose and glycerol are reviewed presently. Ten proposed routes for 3HP production from glucose are described and one of which was recently constructed successfully in Escherichia coli with malonyl–Coenzyme A as a precursor. This resulted in a yield still far from the required level for industrial application. On the other hand, strategies employing engineered E. coli and Klebsiella pneumoniae capable of producing 3HP from glycerol are also evaluated. The titers produced by these recombinant strains reached around 3 %. At its current state, it is evident that a bulk of engineering works is yet to be done to acquire a biosynthesis route for 3HP that is acceptable for industrial-scale production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The balance between environmental and economic aspects has been one of the primary considerations in developing alternative cleaner technologies. This includes the development of bio-based production processes that utilize cheaper and renewable biomass feedstocks (Saxena et al. 2009). Herein biomass is converted to desired products such as platform chemicals and biofuels. This conversion makes use of novel chemical or biological transformation technologies such as green synthesis of platform chemicals from biomass-derived sugars and conversion of C5 and C6 sugars to fuels, platform chemicals, and biopolymers using metabolically engineered microorganisms (Malihan et al. 2012; Lee et al. 2012; Liu et al. 2012a,b; Jang et al. 2012; Park et al. 2012, 2013). In 2004, the US Department of Energy created a priority list containing the names of the platform chemicals suitable for the development of alternative renewable production methods with cheap and viable processes. These compounds were selected based on the involved processes, economics, industrial viability, market size, and their versatility as bridge molecules for the production of more industrially relevant chemicals. The final list provides a guideline for product identification in research, which includes a range of C2–C6 platform chemicals, one of which is 3-hydroxypropionic acid (3HP) (Werpy et al. 2004).
3-Hydroxypropionic acid is a C3 non-chiral hydroxycarboxylic acid. It is a structural isomer of 2-hydroxypropionic acid (lactic acid), which contains a hydroxyl and carboxyl functional groups. This bifunctionality gives 3HP the versatility in several chemical transformations to produce other specialty chemicals (Della Pina et al. 2011) (Fig. 1). For example, reduction of the carboxyl functional group produces 1,3-propanediol (13PDO), a precursor for the production of polyesters such as polytrimethylene terephthalate. On the other hand, dehydration of the hydroxyl functional group leads to the formation of acrylic acid, while its oxidation product is malonic acid. Condensation of 3HP will also produce biopolymers such as poly(3-hydroxypropionate). Other derivatives of 3HP can be obtained through lactonization, amination, and esterification (Della Pina et al. 2011).
Currently, no viable process is available for the production of 3HP at industrial scale. However, several studies have investigated the development of green synthesis routes for the production of 3HP as discussed in a recent review by Della Pina et al. (2011) (Fig. 1). The review discussed seven known chemical conversion routes for the synthesis of 3HP using different starting materials such as ketene, acrylic acid, β-hydroxypropionitrile, 13PDO, 3-hydroxypropionaldehyde (3HPA), vinyl acetate, and allyl alcohol. These routes must overcome the constraints of starting material, reaction intermediate formation and catalyst toxicity, substrate cost and process selectivity. Such limitations can be addressed through biosynthetic means of producing 3HP (Della Pina et al. 2011). Development of efficient 3HP biosynthesis using cheap renewable substrates such as carbohydrates has been a challenge during the past years.
This review discusses recent progress in the development of engineered microorganisms for 3HP production. First, the presence of 3HP as a metabolic intermediate in some microorganisms is discussed, and several potential key enzymes are identified for biotechnological applications. Next, several pathways for the conversion of glucose and other unrelated carbon source to 3HP are evaluated, highlighting their potentials and constraints. The conversion of glycerol to 3HP is also thoroughly reviewed with emphasis on the metabolic engineering efforts done to improve the host strain. At its current state, it is evident that a bulk of engineering works is yet to be done to acquire a biosynthesis route for 3HP that is acceptable for industrial-scale production. More efforts must be given on the development of economically feasible processes employing metabolically engineered microorganisms for the synthesis of 3HP from abundant and cheap substrates.
3-Hydroxypropionate as a metabolic pathway intermediate
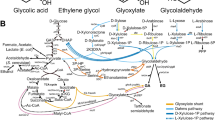
Several microorganisms have been identified to have 3HP as an intermediate in certain metabolic pathways. 3HP as a metabolic intermediate was first observed in thermophilic phototrophic eubacterium Chloroflexus aurantiacus. It is secreted into the culture medium at levels of 1.5 μM to 0.35 mM and was suggested to be an intermediate in the autotrophic assimilation of CO2 (Holo 1989). This was later proven through 13C-NMR studies and enzyme assays, from which the term “3-hydroxypropionate cycle” (3HP cycle) for this CO2 fixation mechanism was derived (Strauss et al. 1992). This cycle was reported to exist in several microorganisms such as Acidianus brierleyi, Metallosphaera sedula, Acidianus ambivalens, Sulfolobus sp. strain VE6, and Sulfolobus metallicus (Hügler et al. 2003a).
After two decades of studies on 3HP cycle, the bicyclic 3HP cycle in C. aurantiacus was completely elucidated (Zarzycki et al. 2009) along with the 3-hydroxypropionate/4-hydroxybutyrate (4HB) cycle in M. sedula (Berg et al. 2007) (Fig. 2a, b). The first cycle of the bicyclic 3HP cycle in C. aurantiacus starts with acetyl–Coenzyme A (acetyl–CoA) and the fixation of two bicarbonate molecules to form glyoxalate. The second cycle starts with propionyl–CoA and glyoxalate, forming pyruvate and acetyl–CoA without any redox step or any CoA dissociation (Zarzycki et al. 2009). On the other hand, the 3HP/4HB cycle in M. sedula fixes CO2 using a promiscuous acetyl–CoA/propionyl–CoA carboxylase. Through this enzyme, two molecules of bicarbonate are reductively converted to succinyl–CoA and then to 4HB. Two acetyl–CoA molecules are derived from 4HB after several transformation steps (Berg et al. 2007). The key feature in these two cycles is the formation of 3HP from acetyl–CoA via a two-step enzymatic reaction: carboxylation of acetyl–CoA by the acetyl–CoA carboxylase to form malonyl–CoA (Menendez et al. 1999; Hügler et al. 2003b) and then further reduction of malonyl–CoA to 3HP via NADPH-dependent malonyl–CoA reductase (Hügler et al. 2002; Alber et al. 2006).
3HP is also known to be a product in the degradation of dimethylsulfoniopropionate by Alcaligenes faecalis M3A (Ansede et al. 1999) and the degradation of uracil in yeast and bacteria (Loh et al. 2006; Kim et al. 2010) (Fig. 2c, d).
Certain key enzymes involved in the pathways enumerated have high potential for biotechnological applications (Ishii et al. 2004). These include the acetyl–CoA carboxylase (with propionyl–CoA carboxylase activity) from M. sedula, malonyl–CoA reductase from C. aurantiacus, l-malyl–CoA lyase (with β-methylmalyl–CoA lyase activity), and the trifunctional propionyl–CoA synthase. The multi-functionality of some of these enzymes might find their way into biorefinery applications, wherein a single enzyme is designed to carry out several reactions. Recently, a portion of the 3HP cycle has been constructed in Escherichia coli for the synthesis of polyketides by recruiting five enzymes from M. sedula and Sulfolobus tokadii (Yuzawa et al. 2012). These heterologous enzymes convert malonyl–CoA to propionyl–CoA, which is then further converted to (2S)-methylmalonyl–CoA by the propionyl–CoA carboxylase from Streptomyces coelicolor. A portion of the 3HP cycle was also used in E. coli or Ralstonia eutropha for the production of biopolymers (Wang et al. 2012; Fukui et al. 2009). Malonyl–CoA reductase, with or without the 3HP–CoA synthase domain, was used for the development of a host strain capable of producing polyhydroxyalkanoate (PHA) containing 3HP monomer (Wang et al. 2012; Fukui et al. 2009).
Metabolic engineering of microorganisms for the production of 3HP from glucose
Metabolic engineering of host strains for the production of target products usually aims one of the following aspects: improving the titer, productivity and yield of the product and the overall performance of the host strain; enabling consumption of a wider substrate range; deletion or reduction of undesired by-product accumulation; and introduction of new pathways to produce new products. To achieve this, metabolic engineers have utilized strategies employing systems metabolic engineering (Kern et al. 2007; Lee et al. 2012). This enabled engineers to improve the production of natural metabolites as well as to design microorganisms capable of producing any desired molecules (Lee et al. 2012). Novel pathways have been proposed (both in patents and scientific reports) for the production of important bioproducts including 3HP using a combination of synthetic biology, systems/computational biology, evolutionary engineering, and metabolic engineering (Henry et al. 2010; Brunk et al. 2011; Lee et al. 2012).
Suggested novel 3HP production pathways from glucose and other unrelated carbon sources
Several patents (Gokarn et al. 2002; Lynch 2011; Burk and Osterhout 2010) and pathway prediction methods (Brunk et al. 2011; Bar-Even et al. 2012; Cho et al. 2010; Henry et al. 2010) have proposed novel biosynthetic pathways for the production of 3HP. These designed pathways are based on an extension of an existing metabolic pathway (e.g., lactate and succinate production), the diversion of a glycolysis or a tricarboxylic acid (TCA) cycle intermediate (e.g., acetyl–CoA, oxaloacetate), or further conversion of an amino acid (e.g., β-/l-alanine). Ten proposed pathways are summarized in Table 1.
Pathway 1 requires further conversion of lactate from any lactate-producing microorganisms (Gokarn et al. 2002; Cho et al. 2010; Henry et al. 2010; Bar-Even et al. 2012). This pathway involves the attachment of CoA to lactate to produce lactoyl–CoA which is then further converted to acryloyl–CoA and finally to 3HP. The enzymes recruited for this route includes a CoA transferase, two CoA dehydratases, and a CoA hydrolase. A major obstacle for this pathway is the thermodynamics of the dehydration reaction employed (formation of acryloyl–CoA), wherein the ΔG° for the reactionFootnote 1 is about +9.9 kJ mol−1. In addition, the dehydration of lactoyl–CoA to acryloyl–CoA is similar to the known dehydration reaction of (R)-2-hydroxyglutaryl–CoA to glutaconyl–CoA (Buckel 1996). This type of dehydration reaction is not activated by any electron-withdrawing group. It instead follows an oxygen-sensitive and reversible reaction mechanism (Bar-Even et al. 2012; Buckel 1996).
The second pathway makes use of the conversion of the TCA cycle intermediate succinate to propionyl–CoA (Table 1; Bar-Even et al. 2012; Cho et al. 2010; Gokarn et al. 2002). Here, the overexpression of phosphoenolpyruvate carboxylase (encoded by pcc gene) in a pfl and ldhA double mutant E. coli strain greatly improved succinic acid production at the expense of reduced cell growth and excess pyruvate secretion. Succinic acid can be converted to propionic acid by expression of the scpC (ygfH) gene from E. coli, coding for succinate–CoA transferase (Haller et al. 2000). The E. coli prpE, gene encoding for propionyl–CoA synthase, may be used to attach CoA to propionic acid (Zhang et al. 2010). Further conversion of propionyl–CoA to acryloyl–CoA is challenging in this proposed pathway. The reduction potential of the acryloyl–CoA/propionyl–CoA couple is reported to be unusually high (E° = +0.069 V) as compared with other enoyl–CoA/acyl–CoA redox couples, which has E° < 0 (Sato et al. 1999). By virtue of thermodynamic principles, a higher reduction potential is needed for the oxidizing agent (in this case, NAD(P)+/NAD(P)H) in order to form propionyl–CoA from acryloyl–CoA. However, the NAD(P)+/NAD(P)H couple has a relatively lower reduction potential than the acryloyl–CoA/propionyl–CoA couple (E° = −0.32 V) (Unden and Bongaerts 1997). This implies that the reaction is not energetically feasible. In addition, the only known enzyme that can catalyze this reaction is the acryloyl–CoA reductase (recruited either from Clostridium propionicum or C. aurantiacus) and is known to be irreversible and prefers the formation of propionyl–CoA (Kuchta and Abeles 1985; Hetzel et al. 2003; Teufel et al. 2009).
The limitations of the first and second pathways may also apply to pathways 3 and 4, where acryloyl–CoA is a common intermediate. The moderate toxicity of acryloyl–CoA should also be considered (Bar-Even et al. 2012) as well as the thermodynamic favorability of the whole pathway (Table 1). All four pathways give a positive net Gibbs free energy change indicating an inhibited forward flow of reaction and are therefore thermodynamically unfavorable. Specific reaction steps found in each pathway have positive Gibbs free energy change of reaction, such as the transfer of the amino group from β-alanine to α-ketoglutarate in pathway 3 with ΔG° of +9.8 kJ mol−1 and the conversion of β-alanyl–CoA to acryloyl–CoA in pathways 3 and 4 with ΔG° of +19 kJ mol−1 (ΔG° values were calculated using eQuilibrator; Flamholz et al. 2011). In addition, a particular trait of pathways 4 and 6 is the use of a novel amino mutase enzyme, which converts l-alanine to β-alanine. This enzyme was screened and engineered to mediate the conversion of l-alanine to β-alanine as described by Cargill, Inc. (Liao et al. 2003). However, no relevant biochemical data were disclosed in the patent.
Pathways 5 and 6 both have a similar downstream reaction: the conversion of β-alanine to malonate semialdehyde and then to 3HP. This distinctive reaction step uses a β-alanine aminotransferase, which has been found in Saccharomyces kluyveri (Schnackerz et al. 2008). If this enzyme is recruited along with an engineered amino mutase enzyme (construction of pathway 6), the bacterial host can be expected to produce high titers of 3HP from glucose (Henry et al. 2010). With regards to the reaction thermodynamics, both pathways are feasible, having net ΔG° < 0 (Table 1). However, cellular energetic considerations will result in a zero net NADH for both pathways (Jiang et al. 2009). Employing these two pathways for 3HP production may cause some cellular redox imbalance in the cell since there are also other redox reactions occurring within the cell for growth and maintenance.
The last four pathways use malonyl–CoA or oxaloacetate as a precursor for 3HP. Pathway 7 is based on the 3HP cycle found in several archaeal bacteria (Hügler et al. 2003a). The key steps of this route are the formation of malonyl–CoA by the acetyl–CoA carboxylase and the reduction of malonyl–CoA to 3HP catalyzed by the malonyl–CoA reductase. The thermodynamics of this pathway is favorable due to the individual carboxylation and the reduction reaction with ΔG° values of −5.7 and −8.8 kJ mol−1, respectively (ΔG° values calculated using eQuilibrator; Flamholz et al. 2011). One of the bottlenecks for this pathway is probably the reductase step since the recruitable enzymes known to catalyze this reaction are likely to have optimum activities at temperatures ≥ 50 °C. It can be expected that these enzymes would have lower activities at lower temperature culture conditions. When the mcr gene encoding for the malonyl–CoA reductase from C. aurantiacus was cloned, highest purified enzyme activity of 10–11 U mg−1 protein was obtained at 55–60 °C (Hügler et al. 2002; Kroeger et al. 2011). When this pathway was constructed in recombinant E. coli BL21 strain, 3HP concentration reached up to 0.193 g L−1 in a shake flask culture and the reductase activity of the crude enzyme extract was only at 0.008 U mg−1 protein (Rathnasingh et al. 2012). On the other hand, pathway 8 involves the recruitment of a 2-keto acid decarboxylase that is active on oxaloacetate to produce malonate semialdehyde. There is no known enzyme which catalyzes this reaction and therefore enzyme engineering of a 2-ketoglutarate decarboxylase is necessary. The reaction catalyzed by this enzyme is favorable considering that the ΔG° for this single reaction is −14.1 kJ mol−1 (values calculated using eQuilibrator; Flamholz et al. 2011). Pathway 9 involves recruiting an engineered CoA-dependent oxaloacetate dehydrogenase, which catalyzes the conversion of oxaloacetate to malonyl–CoA. No known microorganism has been reported to naturally involve this unique reaction step, which means that enzyme engineering should be employed to develop novel enzymes. Subsequently, malonyl–CoA is further converted to 3HP by employing malonyl–CoA reductase from C. aurantiacus or M. sedula, similar to pathway 7. This designed synthetic pathway is thermodynamically favorable with net ΔG° of −34.6 kJ mol−1 (Table 1). The strategy for pathway 10 is to employ a malate decarboxylase enzyme which can catalyze the decarboxylation of malate at the carbon adjacent to the hydroxyl-functionalized carbon to produce 3HP. Similar to the CoA-dependent oxaloacetate dehydrogenase from pathway 9, this malate decarboxylase has not yet been reported to exist, thereby also necessitating enzyme engineering. On the other hand, this pathway is the most thermodynamically promising among the pathways described (Table 1) as it has a net ΔG° of −53.9 kJ mol−1.
Pathway potential and engineering strategies
Among the ten pathways reviewed earlier, pathway 7 has been implemented in E. coli BL21 strain (Rathnasingh et al. 2012). So far, the highest titer was achieved using recombinant E. coli BL21 strain expressing C. aurantiacus malonyl–CoA reductase, E. coli acetyl–CoA carboxylase encoded by the accADBC genes, E. coli biotinilase encoded by the birA gene, and E. coli nicotinamide nucleotide transhydrogenase encoded by the pntAB genes. Further improvement of this strain was made by removing reactions that compete for the available acetyl–CoA and diverting the carbon flow towards malonyl–CoA formation. The strategy involved the disruption of lactate and acetate formations by deleting the ldhA and pta-ackA genes in the E. coli host chromosome, respectively (Fig. 3a). The resulting mutant strain exhibited a drastic decrease in lactate and acetate formations without any significant change in the 3HP titer. The improvement of intracellular malonyl–CoA concentration might not be enough to drive the formation of 3HP. This may have been caused by its usual low intracellular concentration of 0.01–0.23 nmol mg−1 dry cell weight (DCW) or 4–90 μM and by the direct participation of malonyl–CoA intermediate in the biomass formation (Takamura and Nomura 1988). Several metabolic engineering strategies are available to increase the intracellular levels of malonyl–CoA. In a study by Fowler et al. (2009), an in silico metabolic engineering approach that integrates an evolutionary algorithm within constraint-based modeling (termed CiED) was applied to identify gene deletion strategies to improve malonyl–CoA pools. Results of the CiED showed that a single deletion of the sdhCDAB (encoding succinate dehydrogenase) gene which is involved in the conversion of succinate to fumarate in the TCA cycle is expected to increase acetyl–CoA pools. Compared with other deletions in the TCA cycle, this deletion has the lower impact on cellular growth. However, disruption of the sdhCDAB genes in the chromosome of E. coli resulted in a lower 3HP concentration along with decreased biomass yield (Rathnasingh et al. 2012). This approach may not be enough owing to the complexity of the metabolic networks related with the acetyl–CoA and malonyl–CoA synthesis and consumption. Other additional gene disruptions predicted by the CiED applicable to 3HP production included mutation of the adhE (acetaldehyde dehydrogenase), citE (citrate lyase), and brnQ (branched-chain amino acid transporter) genes (Fig. 3a). Employment of all four mutations along with the overexpression of the acetyl–CoA carboxylase and biotinilase has resulted in an intracellular malonyl–CoA concentration of 2.6 nmol mg−1 DCW (Fowler et al., 2009). In a different study by Xu et al. (2011), OptForce in silico metabolic engineering was performed to predict gene manipulations that would lead to an increase in malonyl–CoA pools. This software was designed to identify the relevant changes in the metabolic flux by comparing the maximum range of flux variability of the wild-type strain against a strain having a pre-specified overproduction target (Xu et al. 2011). Mutations in the fumC (fumarase) and sucC (succinate dehydrogenase) genes involved in the TCA cycle, along with the overexpression of the pgk (phosphoglycerate kinase), pdh (pyruvate dehydrogenase), and accADBC (acetyl–CoA carboxylase) genes, led to an increase of intracellular malonyl–CoA concentration up to 10.5 nmol mg−1 DCW (Xu et al. 2011). Using this approach in 3HP production will probably increase the product titer and yield.
Metabolic engineering of microorganisms for 3HP production from glycerol
Glycerol has been intensively examined as a promising alternative substrate for the production of 3HP. Since both glycerol and 3HP are three-carbon compounds, the reaction steps involved in the 3HP production from glycerol are expected to be shorter compared with the six-carbon substrate glucose. Conversion of glycerol into 3HP only takes two reactions: dehydration of glycerol to 3-hydroxypropionaldehyde followed by the further reduction of the terminal aldehyde group into a hydroxyl group to produce 3HP (Fig. 3b). The first step is catalyzed by a coenzyme B12-dependent glycerol dehydratase found in several gram-positive and gram-negative microorganisms including Klebsiella and Citrobacter (Daniel et al. 1999). The second step is catalyzed by an NAD(P)+-dependent aldehyde dehydrogenase, which can be recruited from E. coli, Azospirillum brasilense, and Klebsiella pneumoniae (Jo et al. 2008; Rathnasingh et al. 2009; Ashok et al. 2011). Recently, coenzyme B12-independent glycerol dehydratase that is only functional in complete anaerobic condition has also been identified in Clostridium butyricum and used for the construction of recombinant microorganisms for the production of 1,3-propanediol (13PDO) and PHA containing 3HP monomer (Andreessen et al. 2010; Raynaud et al. 2003; Tang et al. 2009)
3HP production from glycerol employing K. pneumoniae as host strain
The glycerol metabolism of K. pneumoniae consists of two branches. The oxidative branch begins with the conversion of glycerol to dihydroxyacetone (DHA) along with the formation of NADH. This is catalyzed by the glycerol dehydrogenase (DhaD) encoded by the dhaD gene. The dihydroxyacetone kinase (DhaK) encoded by the dhaK gene further phosphorylates DHA while consuming one ATP. The product dihydroxyacetone-phosphate (DHAP) is then converted to other necessary metabolic intermediates for cell growth and for the formation of other metabolites. The ATP consumed is regenerated upon conversion of DHAP to phosphoenolpyruvate. Meanwhile, the reductive branch starts with the dehydration of glycerol to 3HPA catalyzed by glycerol dehydratase (DhaB), which is encoded by the dhaB gene. This step requires the participation of coenzyme B12 (Forage and Foster 1982). 3HPA is further converted to 13PDO by an NADH-dependent 1,3-propanediol dehydrogenase encoded by the dhaT gene. This step is responsible for immediate regeneration of the NAD+ generated from the first step of the oxidative branch.
Since 3HP is not a natural metabolite of K. pneumoniae, an aldehyde dehydrogenase efficient in converting 3HPA to 3HP has not been extensively examined until recently. An NAD+-dependent aldehyde dehydrogenase (AldH) in K. pneumoniae was identified to be active on 3HPA (Luo et al. 2011b). When AldH was overexpressed in a K. pneumoniae strain with mutations in its oxidative branch, 17 g L−1 13PDO and 6.8 g L−1 3HP were produced. Although the results in this study suggested that AldH plays a role in 3HP formation, chromosomal deletion of this gene in K. pneumoniae did not completely eliminate 3HP formation. This supports the hypothesis that other aldehyde dehydrogenases are also present and have activity towards 3HPA (Luo et al. 2011b). In a more recent study, the propanediol utilization protein (PduP), encoded by the pduP gene from Lactobacillus reuteri, was characterized and confirmed to be active towards 3HPA (Luo et al. 2011a). This enzyme showed broad substrate specificity for aliphatic aldehydes and uses both NAD+ and NADP+ as coenzymes. The role of PduP in 3HP formation was further investigated and showed that this enzyme converts 3HPA to 3HP through a CoA-dependent mechanism (Luo et al. 2012). A separate investigation of other known aldehyde dehydrogenases was done by recruiting dehydrogenases from Zymomonas mobilis, Lactobacillus collinoides, E. coli, and K. pneumoniae (Huang et al. 2012). Among the 14 different aldehyde dehydrogenases cloned and overexpressed, the AldH from E. coli expressed in K pneumoniae exhibited the highest 3HP accumulation of 1.13 g L−1 (Huang et al. 2012).
Another aldehyde dehydrogenase, the NAD+-dependent γ-glutamyl-γ-aminobutyraldehyde dehydrogenase (PuuC), encoded by the puuC gene of K. pneumoniae was suggested to have considerable activity towards 3HPA despite the undetectable 3HP accumulation in K. pneumoniae (Raj et al. 2010). Overexpression of PuuC in K. pneumoniae under microaerobic conditions resulted in the production of 1.1 g L−1 3HP, which is at about half of the 13PDO concentration. Disruption of the dhaT in K. pneumoniae is expected to support the balanced formation of 3HP and 13PDO. This resulted in the production of equal amounts of 3HP and 13PDO at 3.6 g L−1 (Raj et al. 2010). Even though this mutation improved the titer and yield of 3HP from glycerol, an increase in NADH level might have inhibited the carbon flux through the oxidative branch. This in turn lowered the growth of the mutant strain (Ashok et al. 2011). To overcome the unfavorable effects of the overexpression of PuuC and dhaT mutation, nitrate was used as an electron acceptor to regenerate NAD+. Oxygen cannot be used as the terminal electron acceptor due to its inhibitory effect on certain enzymes involved in the synthesis of coenzyme B12. This strategy, along with the mutation of the glpK to divert more glycerol to 3HP production, resulted in the final concentration of 22.5 g L−1 3HP (Ashok et al. 2013a). Other strategies to increase 3HP and decrease 13PDO formation involve the disruption of both of the dhaT and yqhD genes and the overexpression of PuuC and DhaB. A fed-batch culture of recombinant K. pneumoniae strain developed with this strategy resulted in the highest titer of 3HP at 28.0 g L−1 (Ashok et al. 2013b). Although the elimination of 13PDO formation is expected upon disruption of both the dhaT and yqhD genes, a considerable amount of 13PDO was still detected. Aeration also is determined to be the critical factor in minimizing 13PDO accumulation, noting that its formation is favored under anaerobic conditions. Complete aeration may minimize 13PDO accumulation but will result in growth inhibition due to low flux towards coenzyme B12 synthesis (Ashok et al. 2013b).
A different bioconversion technique has been reported by Kumar et al. (2012), wherein resting cells of recombinant K. pneumoniae expressing the α-ketoglutaric semialdehyde dehydrogenase (KgsadH, encoded by kgsadH gene) from A. brasilense were used for the co-production of 3HP and 13PDO from glycerol. This system achieved 11.3 g L−1 3HP. When KgsadH was overexpressed in a dhaT mutant K. pneumoniae, 16 g L−1 of 3HP was produced, higher than that obtained by the overexpression of PuuC (Ko et al. 2012). However, impeded cell growth and glycerol assimilation found in the recombinant K. pneumoniae strain should be overcome to improve 3HP production. The growth inhibition and glycerol assimilation might have been caused by an increase in intracellular NADH concentration as it was previously reported that a high NADH ratio inhibited some metabolic pathways involved in biomass precursor synthesis (Ashok et al. 2011).
The 3HP production obtained by the different recombinant K. pneumoniae strains is summarized in Table 2. Currently, the highest 3HP titer achieved is 28 g L−1 with a yield of 0.39 g 3HP g−1 glycerol consumed and a productivity of 0.58 g L−1 h−1 obtained from a dhaT and yqhD mutant K. pneumoniae strain overexpressing the DhaB and PuuC (Ashok et al. 2013b). On the other hand, highest 3HP productivity of 1.02 g L−1 h−1 was attained by overexpression of AldH from E. coli in K. pneumoniae under anaerobic culture conditions (Huang et al. 2012). By combining these two strategies, a high 3HP titer and yield can be expected.
3HP production from glycerol employing E. coli as host strain
A two-step pathway for 3HP production was constructed in recombinant E. coli by recruiting the necessary genes (Table 2; Fig. 3). The first prototype strain capable of producing 3HP from glycerol was constructed by recruiting the K. pneumoniae DhaB and the E. coli AldH (Raj et al. 2008). This strain accumulated 0.58 g L−1 3HP in aerobic culture conditions using a medium containing glycerol as carbon source. The 3HP formation is attributed to the high activity of the AldH towards 3HPA at 38.1 U mg−1 protein (Jo et al. 2008). Process parameters such as pH, inducer concentration, aeration, and substrate concentration were further optimized using this strain and resulted in a 3HP concentration of 31 g L−1 (Raj et al. 2009). Despite the improved 3HP titer, the enzyme activities of DhaB and AldH were unbalanced and unstable. This instability is more pronounced on DhaB and may have been a result of oxygen-rich environment and intracellular 3HPA accumulation (Raj et al. 2009). The instability problem is attributed to its coenzyme B12 dependency (Maervoet et al. 2011) and its mechanism-based inactivation during catalysis (Toraya et al. 1976). These factors make this step appear to be rate-limiting. Therefore, additional expression of a glycerol dehydratase reactivase (Gdr) enzyme complex encoded by the gdrAB gene was employed (Tobimatsu et al. 1999; Toraya and Mori 1999; Rathnasingh et al. 2009). This approach, combined with the usage of two compatible isopropyl-thio-β-galactoside (IPTG)-inducible plasmid systems, allowed a balanced expression of both enzymes. This also prevented the accumulation of the toxic intermediate 3HPA. The final recombinant E. coli strain expressing KgsadH from A. brasilense along with Gdr and DhaB from K. pneumoniae resulted in a final 3HP concentration of 38.73 g L−1 (Rathnasingh et al. 2009). This value is so far the highest concentration obtained for 3HP. However, further improvements are still needed to address the issues on low productivity, enzyme activity loss, and redox balance, all of which significantly affect 3HP production using recombinant E. coli as host strain.
Host comparison for 3HP production: E. coli vs. K. pneumoniae
There are several limiting factors unique to each host strain used for 3HP production specifically from glycerol (Table 3). The inability of E. coli to synthesize its own coenzyme B12 is a major disadvantage; expensive coenzyme B12 must be added to the culture medium to produce 3HP. On the other hand, K. pneumoniae is able to make its own coenzyme B12. However, recruitment of the coenzyme B12 synthesis pathway from K. pneumoniae and its construction in E. coli can make E. coli a more competitive strain for 3HP production. Alternatively, a B12-independent glycerol dehydratase from C. butyricum may be employed (Raynaud et al. 2003; O’Brien et al. 2004), but this enzyme is oxygen sensitive; hence, strict anaerobic condition is needed for 3HP production. The issue of the stability of DhaB from K. pneumoniae is also encountered when expressed in E. coli. As described earlier, this instability is caused by an instantaneous deactivation of the DhaB upon dehydration of glycerol. In K. pneumoniae, the dehydratase is reactivated by the Gdr enzyme complex endogenous to the strain. When this enzyme complex is recruited to E. coli along with the glycerol dehydratase, the stability is improved but only to a certain extent.
The by-products formed during 3HP production in K. pneumoniae are mainly 13PDO, lactic acid, and acetic acid, along with traces of ethanol and 2,3-butanediol. The accumulation of 13PDO in K. pneumoniae is difficult to prevent since 13PDO is originally the main product of glycerol metabolism (Maervoet et al. 2011). Most of the glycerol is usually diverted to 13PDO formation and has become a competing pathway in 3HP production. Although genetic manipulations have been done to lower the 13PDO accumulation, substantial amount of 13PDO is still observed, probably due to some other unknown 13PDO dehydrogenases present (Ashok et al. 2011, 2013a, b; Ko et al. 2012). In contrast, acetic acid and lactic acid are the only major by-products of 3HP production in E. coli, with trace amounts of ethanol. Synthesis of these by-products can be easily prevented through simple standard genetic manipulations in E. coli. The availability of these genetic manipulation tools makes E. coli simpler and easier to handle.
Future perspectives
Currently, all the proposed metabolic routes for the production of 3HP utilize glucose or glycerol as starting materials. The highest and only reported 3HP titer from glucose is from an engineered E. coli constructed by recruiting several key enzymes from the 3HP cycle found in carbon-fixating bacteria. Further genetic manipulation and engineering is needed to increase 3HP titer to a level acceptable for industrial-scale production. The other proposed plausible pathways using glucose as a carbon source have not yet been explored thoroughly. On the other hand, 3HP biosynthesis using glycerol as the substrate is still at its optimization stage and its highest titer and yield are approaching the level of industrial viability. Between the two hosts compared for this route, E. coli is preferred since it is widely accepted for industrial use, unlike K. pneumoniae. Other industrial hosts may also be explored using this route.
Several challenges should be addressed in order to increase the titer and yield of 3HP derived from glucose. Thermodynamic favorability, enzyme availability, and intermediate toxicity must all be considered in selecting the best pathway to be constructed in a prototype microbial platform. Among the discussed pathways, the most promising route is through the malonyl–CoA and oxaloacetate intermediate if the thermodynamics of each pathway is considered. However, the enzymatic activity and availability will be its next challenge since some of the enzymes proposed for these pathways either have low activity or are yet to be discovered.
A different set of challenges is present when 3HP production via glycerol is employed. First, the co-factor dependence of the first enzymatic step must be dealt with, including its instability. Second, the use of an appropriate and industrially viable host must be selected. Lastly, a balance in the expression of the key enzymes to prevent accumulation of the toxic intermediate, 3HPA, must be achieved. These limitations have been addressed as described above. However, further optimizations of the strain and the process are needed in order to raise the titer and yield to an industrial scale.
The next step after achieving this is to broaden the substrate specificity of the host, wherein aside from glucose and glycerol, other carbohydrates from biomass can also be used as substrates. Utilization of the engineered strains in biorefinery applications is another challenge and will be dealt with in the near future.
It is now possible to produce 3HP from renewable resources by microbial fermentation. However, the bulk of engineering works is yet to be done in order to reach the full potential of commercial 3HP production. Several engineering tools are available and are continually advancing, allowing engineers and scientists to hasten the pace of developing the most efficient and robust microbial platform for 3HP production.
Notes
From here on, all the Gibbs free energies mentioned in the text are calculated using the eQuilibrator online program (http://milolab.webfactional.com/) as described in the report by Flamholz et al. (2011).
References
Alber B, Olinger M, Reider A, Knockelkorn D, Jobst B, Hügler M, Fuchs G (2006) Malonyl–Coenzyme A reductase in the modified 3-hydroxypropionate cycle for autotrophic carbon fixation in archaeal Metallosphaera and Sulfolobus spp. J Bacteriol 188:8551–8559
Andreessen B, Lange AB, Robenek H, Steinbüchel A (2010) Conversion of glycerol to poly(3-hydroxypropionate) in recombinant Escherichia coli. Appl Environ Microbiol 76:622–626
Ansede JH, Pellechia PJ, Yoch DC (1999) Metabolism of acrylate to β-hydroxypropionate and its role in dimethylsulfoniopropionate lyase induction by a salt marsh sediment bacterium, Alcaligenes faecalis M3A. Appl Environ Microbiol 65:5075–5081
Ashok S, Raj SM, Rathnasingh C, Park S (2011) Development of recombinant Klebsiella pneumoniae ΔdhaT strain for the co-production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol. Appl Microbiol Biotechnol 90:1253–1265
Ashok S, Raj SM, Ko Y, Sankaranarayanan M, Zhou S, Kumar V, Park S (2013a) Effect of puuC overexpression and nitrate addition on glycerol metabolism an anaerobic 3-hydroxypropionic acid production in recombinant Klebsiella pneumoniae ΔglpK ΔdhaT. Metab Eng 15:10–24
Ashok S, Sankaranarayanan M, Ko Y, Jae K-E, Ainala SK, Kumar V, Park S (2013b) Production of 3-hydroxypropionic acid from glycerol by recombinant Klebsiella pneumoniae ΔdhaTΔyqhD which can produce vitamin B12 naturally. Biotechnol Bioeng 110:511–524
Bar-Even A, Flamholz A, Noor E, Milo R (2012) Rethinking glycolysis: on biochemical logic of metabolic pathways. Nat Chem Biol 8:509–517
Berg IA, Kockelkorn D, Buckel W, Fuchs G (2007) A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318:1782–1786
Brunk E, Neri M, Tavernelli I, Hatzimanikatis V, Rothlisberger U (2011) Integrating computational methods to retrofit enzymes to synthetic pathways. Biotech Bioeng 109:572–582
Buckel W (1996) Unusual dehydrations in anaerobic bacteria: considering ketyls (radical anions) as reactive intermediates in enzymatic reactions. FEBS Lett 389:20–24
Burk MJ, Osterhout RE (2010) Methods and organisms for production of 3-hydroxypropionic acid. USA Patent US 2010/0021978 A1
Cho A, Yun H, Park JH, Lee SY, Park S (2010) Prediction of novel synthetic pathways for the production of desired chemicals. BMC Syst Biol 4:35
Daniel R, Bobik TA, Gottschalk G (1999) Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes. FEMS Microbiol Rev 22:553–566
Della Pina C, Falletta E, Rossi M (2011) A green approach to chemical building blocks. The case of 3-hydroxypropionic acid. RSC Green Chem 13:1624–1632
Flamholz A, Noor E, Bar-Even A, Milo R (2011) eQuilibrator—the biochemical thermodynamics calculator. Nucleic Acids Res 40:D770–D775
Forage RG, Foster MA (1982) Glycerol fermentation in Klebsiella pneumoniae: functions of the coenzyme B12-dependent glycerol diol dehydratases. J Bacteriol 149:413–419
Fowler ZL, Gikandi WW, Koffas MA (2009) Increased malonyl–Coenzyme A biosynthesis by tuning the Escherichia coli metabolic network and its application to flavones production. Appl Environ Microbiol 75:5831–5839
Fukui T, Suzuki M, Tsuge T, Nakamura S (2009) Microbial synthesis of poly((R)-3-hydroxybutyrate-co-3-hydroxypropionate) from unrelated carbon sources by engineered Cupriavidus necator. Biomacromolecules 10:700–706
Gokarn RR, Selifona OV, Jessen H, Gort SJ, Selmer T, Buckel W (2002) 3-Hydroxypropionic acid and other organic compounds. US Patent WO 02/42418 A2
Haller T, Buckel T, Rétey J, Gerlt JA (2000) Discovering new enzymes and metabolic pathways: conversion of succinate to propionate by Escherichia coli. Biochem 39:4622–4629
Henry CS, Broadbelt LJ, Hatzimanikatis V (2010) Discovery and analysis of novel metabolic pathways for the biosynthesis of industrial chemicals: 3-hydroxypropanoate. Biotechnol Bioeng 106:462–473
Hetzel M, Brock M, Selmer T, Pierik AJ, Golding BT, Buckel W (2003) Acryloyl–CoA reductase from Clostridium propionicum, an enzyme complex of propionyl–CoA dehydrogenase and electron-transferring flavoprotein. Eur J Biochem 270:902–910
Holo H (1989) Chloroflexus aurantiacus secretes 3-hydroxypropionate, a possible intermediate in the assimilation of CO2 and acetate. Arch Microbiol 151:252–256
Huang Y, Li Z, Shimizu K, Ye Q (2012) Simultaneous production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol by a recombinant strain of Klebsiella pneumoniae. Bioresour Technol 103:351–359
Hügler M, Menendez C, Schägger H, Fuchs G (2002) Malonyl–Coenzyme A reductase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J Bacteriol 184:2404–2410
Hügler M, Huber H, Stetter KO, Fuchs G (2003a) Autotrophic CO2 fixation pathways in archaea (Crenarchaeota). Arch Microbiol 179:160–173
Hügler M, Krieger RS, Jahn M, Fuchs G (2003b) Characterization of acetyl–CoA/propionyl–CoA carboxylase in Metallosphaera sedula. Eur J Biochem 270:736–744
Ishii M, Chuakrutt S, Arai H, Igarashi Y (2004) Occurrence, biochemistry and possible biotechnological application of the 3-hydroxypropionate cycle. Appl Microbiol Biotechnol 64:605–610
Jang Y-S, Kim B, Shin JH, Choi YJ, Choi S, Song CW, Lee J, Park HG, Lee SY (2012) Bio-based production of C2-C6 platform chemicals. Biotechnol Bioeng 109:2437–2459
Jiang X, Meng X, Xian M (2009) Biosynthetic pathways for 3-hydroxypropionic acid production. Appl Microbiol Biotechnol 82:995–1003
Jo J, Raj SM, Rathnasingh C, Selvakumar E, Jung W-C, Park S (2008) Cloning, expression, and characterization of an aldehyde dehydrogenase from Escherichia coli K-12 that utilizes 3-hydroxypropionaldehyde as a substrate. Appl Microbiol Biotechnol 81:51–60
Kern A, Tilley E, Hunter IS, Legiša M, Glieder A (2007) Engineering primary metabolic pathways of industrial microorganisms. J Biotechnol 129:6–29
Kim K-S, Pelton JG, Inwood WB, Andersen U, Kustu S, Wemmer DE (2010) The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems. J Bacteriol 192:4089–4102
Ko Y, Ashok S, Zhou S, Kumar V, Park S (2012) Aldehyde dehydrogenase activity is important to the production of 3-hydroxypropionic acid from glycerol by recombinant Klebsiella pneumoniae. Process Biochem 47:1135–1143
Kroeger JK, Zarzycki J, Fuchs G (2011) A spectrophotometric assay for measuring acetyl–Coenzyme A carboxylase. Anal Biochem 411:100–105
Kuchta RD, Abeles RH (1985) Lactate reduction in Clostridium propionicum, purification and properties of lactyl–CoA dehydratase. J Biol Chem 260:13181–13189
Kumar V, Sankaranarayanan M, Jae K, Dugrapal M, Ashok S, Ko Y, Sarkar R, Park S (2012) Co-production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol using resting cells of recombinant Klebsiella pneumoniae J2B strain overexpressing aldehyde dehydrogenase. Appl Microbiol Biotechnol 96:373–383
Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY (2012) Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nature Chem Biol 8:536–546
Liao HH, Gokarn RR, Gort SJ, Jessen HJ, Selifonova O (2003) Alanine 2,3-aminomutase. WO 03/062173
Liu H, Ramos KRM, Valdehuesa KNG, Nisola GM, Lee W-K, Chung W-J (2012a) Biosynthesis of ethylene glycol in Escherichia coli. Appl Microbiol Biotechnol. doi:10.1007/s00253-012-4618-7
Liu H, Valdehuesa KNG, Nisola GM, Ramos KRM, Chung W-J (2012b) High yield production of d-xylonic acid from D-xylose using engineered Escherichia coli. Bioresour Technol 115:244–248
Loh KD, Gyaneshwar P, Papadimiyriou EM, Fong R, Kim K-S, Parales R, Zhou WI, Kustu S (2006) A previously undescribed pathway for pyrimidine catabolism. Proc Natl Acad Sci 103:5114–5119
Luo LH, Seo J-W, Baek J-O, Oh B-R, Heo S-Y, Hong W-K, Kim D-H, Kim CH (2011a) Identification and characterization of the propanediol utilization protein PduP of Lactobacillus reuteri for 3-hydroxypropionic acid production from glycerol. Appl Microbiol Biotechnol 89:697–703
Luo LH, Seo J-W, Oh B-R, Seo P-S, Heo S-Y, Hong W-K, Kim D-H, Kim CH (2011b) Stimulation of reductive glycerol metabolism by overexpression of an aldehyde dehydrogenase in a recombinant Klebsiella pneumoniae strain defective in oxidative pathway. J Ind Microbiol Biotechnol 38:991–999
Luo LH, Kim CH, Heo S-Y, Oh B-R, Hong W-K, Kim S, Kim D-H, Seo J-W (2012) Production of 3-hydroxypropionic acid through propionaldehyde dehydrogenase PduP mediated biosynthetic pathway in Klebsiella pneumoniae. Bioresour Technol 103:1–6
Lynch MD (2011) Compositions and methods for 3-hydroxypropionate bio-production from biomass. USA Patent US 8,048,624 B1
Maervoet VE, Mey MD, Beauprez J, Maeseneire SD, Soetaert WK (2011) Enhancing the microbial conversion of glycerol to 1,3-propanediol using metabolic engineering. Org Process Res Dev 15:189–202
Malihan LB, Nisola GM, Chung W-J (2012) Brown algae hydrolysis in 1-n-butyl-3-methylimidazolium chloride with mineral acid catalyst system. Bioresour Technol 118:545–552
Menendez C, Bauer Z, Huber H, Gad’on N, Stetter KO, Fuchs G (1999) Presence of acetyl–CoA carboxylase in autotrophic Crenarchaeota and indication for operation of a 3-hydroxypropionate cycle in autotrophic carbon fixation. J Bacteriol 181:1088–1098
O’Brien JR, Raynaud C, Croux C, Girbal L, Soucaille P, Lanzilotta WN (2004) Insight into the mechanism of the B12-independent glycerol dehydratase from Clostridium butyricum: preliminary biochemical and structural characterization. Biochem 43:4635–4645
Park SJ, Kim TW, Kim MK, Lee SY, Lim SC (2012) Advanced bacterial polyhydroxyalkanoates: towards a versatile and sustainable platform for unnatural tailor-made polyesters. Biotechnol Adv 30:1196–1206
Park SJ, Kim EY, Noh W, Park HM, Oh YH, Lee SH, Song BK, Jegal J, Lee SY (2013) Metabolic engineering of Escherichia coli for the production of 5-aminovalerate and glutarate as C5 platform chemicals. Metab Eng 16:42–47
Raj SM, Rathnasingh C, Jo J-E, Park S (2008) Production of 3-hydroxypropionic acid from glycerol by a novel recombinant Escherichia coli BL21 strain. Process Biochem 43:1440–1446
Raj SM, Rathnasingh C, Jung W-C, Park S (2009) Effect of process parameters on 3-hydroxypropionic acid production from glycerol using recombinant Escherichia coli. Appl Microbiol Biotechnol 84:649–657
Raj SM, Rathnasingh C, Jung W-C, Selvakumar E, Park S (2010) A novel NAD+-dependent aldehyde dehydrogenase encoded by the puuC gene of Klebsiella pneumoniae DSM 2026 that utilizes 3-hydroxypropionaldehyde as a substrate. Biotechnol Bioproc Eng 15:131–138
Rathnasingh C, Raj SM, Jo J-E, Park S (2009) Development and evaluation of efficient recombinant Escherichia coli strains for the production of 3-hydroxypropionic acid from glycerol. Biotechnol Bioeng 104:729–739
Rathnasingh C, Raj SM, Lee Y, Catherine C, Ashok S, Park S (2012) Production of 3-hydroxypropionic acid via malonyl–CoA pathway using recombinant Escherichia coli strains. J Biotechnol 157:633–640
Raynaud C, Sarcabal P, Meynial-Salles I, Croux C, Soucaille P (2003) Molecular characterization of the 1,3-propanediol (1,3-PD) operon of Clostridium butyricum. Proc Natl Acad Sci 100:5010–5015
Sato K, Nishina Y, Setoyama C, Miura R, Shiga K (1999) Unusually high standard redox potential of acrylyl–CoA/propionyl–CoA couple among enoyl–CoA/acyl–CoA couples: a reason for the distinct metabolic pathway of propionyl–CoA from longer acyl–CoAs. J Biochem 126:668–675
Saxena RC, Adhikari DK, Goyal HB (2009) Biomass-based energy fuel through biochemical routes: a review. Renew Sustain Energ Rev 13:167–178
Schnackerz KD, Andersen G, Dobritzsch D, Piskur J (2008) Degradation of pyrimidines in Saccharomyces kluyveri: transamination of β-alanine. Nucleos Nucleot Nucleic Acid 27:794–799
Strauss G, Eisenreich W, Bacher A, Fuchs G (1992) 13C-NMR study of autotrophic CO2 fixation pathways in the sulfur-reducing Achaebacterium Thermoproteus neutrophilus and in the phototrophic Eubacterium Chloroflexus aurantiacus. Eur J Biochem 205:853–866
Takamura Y, Nomura G (1988) Changes in the intracellular concentration of acetyl–CoA and malonyl–CoA in relation to the carbon and energy metabolism of Escherichia coli K12. J General Microbiol 134:2249–2253
Tang X, Tan Y, Zhu H, Zhao K, Shen W (2009) Microbial conversion of glycerol to 1,3-propanediol by an engineered strain of Escherichia coli. Appl Environ Microbiol 75:1628–1634
Teufel R, Kung JW, Kockelkorn D, Alber BE, Fuchs G (2009) 3-Hydroxypropionyl–Coenzyme A dehydratase and acryloyl–Coenzyme A reductase, enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in Sulfolobales. J Bacteriol 191:4572–4581
Tobimatsu T, Kajiura H, Yunoki M, Azuma M, Toraya T (1999) Identification and expression of the genes encoding a reactivating factor for adenosylcobalamin-dependent glycerol dehydratase. J Bacteriol 181:4110–4113
Toraya T, Mori K (1999) A reactivating factor for coenzyme B12-dependent diol dehydratase. J Biol Chem 274:3372–3377
Toraya T, Shirakashi T, Kosuga T, Fukui S (1976) Substrate specificity of coenzyme B12-dependent diol dehydratase: glycerol as both a good substrate and a potent inactivator. Biochem Biophys Res Commun 69:475–480
Unden G, Bongaerts J (1997) Alternative respiratory pathways of Escherichia coli: energetic and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta 1320:217–234
Wang Q, Liu C, Mo X, Zhang Y, Zhao G (2012) Biosynthetic pathway for poly(3-hydroxypropionate) in recombinant Escherichia coli. J Microbiol 50:693–697
Werpy, T., Holladay, J., White, J., 2004. Top value added chemicals from biomass, PNNL-14808, Pac. Northw. Nat. Lab., Richland, WA, http://www1.eere.energy.gov/biomass/pdfs/35523.pdf
Xu P, Ranganathan S, Fowler ZL, Maranas CD, Koffas MAG (2011) Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl–CoA. Metab Eng 13:578–587
Yuzawa S, Chiba N, Katz L, Keasling JD (2012) Construction of a part of a 3-hydroxypropionate cycle for heterologous polyketide biosynthesis in Escherichia coli. Biochem 51:9779–9781
Zarzycki J, Brecht V, Müller M, Fuchs G (2009) Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus. Proc Natl Acad Sci 106:21317–21322
Zhang H, Boghigian BA, Pfeifer BA (2010) Investigating the role of native propionyl–CoA and methylmalonyl–CoA metabolism on heterologous polyketide production in Escherichia coli. Biotechnol Bioeng 105:567–573
Acknowledgments
This work was supported by the Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (MEST) (2012-0006693) and the Technology Development Program to Solve Climate Changes (Systems Metabolic Engineering for Biorefineries) from the MEST through the NRF (NRF-2012-C1AAA001-2012M1A2A2026556).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Valdehuesa, K.N.G., Liu, H., Nisola, G.M. et al. Recent advances in the metabolic engineering of microorganisms for the production of 3-hydroxypropionic acid as C3 platform chemical. Appl Microbiol Biotechnol 97, 3309–3321 (2013). https://doi.org/10.1007/s00253-013-4802-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-4802-4