Abstract
Canthaxanthin is a natural diketo derivative of β-carotene primarily used by the food and feed industries. Mucor circinelloides is a β-carotene-accumulating zygomycete fungus and one of the model organisms to study the carotenoid biosynthesis in fungi. In this study, the β-carotene ketolase gene (crtW) of the marine bacterium Paracoccus sp. N81106 fused with fungal promoter and terminator regions was integrated into the M. circinelloides genome to construct stable canthaxanthin-producing strains. Different transformation methods including polyethylene glycol-mediated transformation with linear DNA fragments, restriction enzyme-mediated integration and Agrobacterium tumefaciens-mediated transformation were tested to integrate the crtW gene into the Mucor genome. Mitotic stability, site of integration and copy number of the transferred genes were analysed in the transformants, and several stable strains containing the crtW gene in high copy number were isolated. Carotenoid composition of selected transformants and effect of culturing conditions, such as temperature, carbon sources and application of certain additives in the culturing media, on their carotenoid content were analysed. Canthaxanthin-producing transformants were able to survive at higher growth temperature than the untransformed strain, maybe due to the effect of canthaxanthin on the membrane fluidity and integrity. With the application of glucose, trehalose, dihydroxyacetone and l-aspartic acid as sole carbon sources in minimal medium, the crtW-expressing M. circinelloides strain, MS12+pCA8lf/1, produced more than 200 μg/g (dry mass) of canthaxanthin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Canthaxanthin is a dark red, natural diketo derivative of β-carotene (β,β-carotene-4,4′dione; Fig. 1). It is used as food colorant and feed additive, primarily in aquacultures and poultry industries (Bhosale and Bernstein 2005; Dufossé 2006). This carotenoid has stronger antioxidant activity than β-carotene (Palozza and Krinsky 1992), and several beneficial effects have been attributed to it. Canthaxanthin induced apoptosis in human cancer cell lines (Kumaresan et al. 2008; Palozza et al. 1998), reduced the risk of several types of cancer in animal models (Mayne and Parker 1989) and effectively stimulated immune defences compared to other carotenoid species (Jyonouchi et al. 1996; Okai and Higashi-Okai 1996). Although several bacteria, such as Corynebacterium michiganense (Saperstein and Star 1954), Micrococcus roseus (Cooney et al. 1966), Brevibacterium sp. strain KY 4313 (Nelis and De Leenheer 1989) and Gordonia jacobaea MV-1 (De Miguel et al. 2001; Veiga-Crespo et al. 2005), and some microalgae, such as Chlorella pyrenoidosa (Czygan 1964) and Chlorella zofingiensis (Pelah et al. 2004), have been reported as canthaxanthin producers, compounds for the food and feed industries are primarily produced by chemical synthesis (Ernst 2002). The canthaxanthin market is growing but hampered by the lack of appropriate microbial sources (Bhosale and Bernstein 2005).
Mucor circinelloides is a β-carotene-producing filamentous fungus. Besides Phycomyces blakesleeanus and Blakeslea trispora, this species has been involved in the study of the molecular background of the carotene biosynthesis in zygomycetes. Consequently, the biosynthetic process is well resolved, and several structural and regulatory genes participating in or related with the pigment production have been isolated and characterized (Navarro et al. 2001; Papp et al. 2006; Velayos et al. 2000a, b; 2003). Although the main carotenoid product in M. circinelloides is β-carotene, it also has a poor β-carotene hydroxylase activity; thus, the fungus is able to produce β-cryptoxanthin and zeaxanthin in small amounts (Álvarez et al. 2006; Papp et al. 2006). We previously reported the heterologous expression of a β-carotene ketolase (crtW) from the marine bacterium, Paracoccus sp. N81106 (formerly Agrobacterium aurantiacum) in M. circinelloides (Csernetics et al. 2011; Papp et al. 2006). In those studies, crtW was introduced into the fungus in autonomously replicating plasmids, and the resulting transformants were able to produce astaxanthin and canthaxanthin. However, the copy number of the plasmids (0.07–1 per host genome) and the amount of the new carotenoid products remained low. Therefore, the aim of the present study was the construction of M. circinelloides strains, which harbour the bacterial crtW gene integrated in the genome and able to effectively express it to produce canthaxanthin in significant amount.
It is fairly difficult to obtain stable transformants in zygomycetes. The transferred DNA rarely integrates into the genome but often form autonomously replicating, occasionally rearranged and/or concatenated structures (Michielse et al. 2004; Ibrahim and Skory 2007; Papp et al. 2010). Such transformants generally display mitotic instability losing their extrachromosomal elements under nonselective conditions. Integration can be forced by transformation with linear fragments holding extensive homologous regions at their termini to direct homologous recombination and gene replacement. For this purpose, polyethylene glycol (PEG)-mediated transformation is traditionally used (Navarro et al. 2001; Silva et al. 2006), but the integration frequency is generally low in this system. Agrobacterium tumefaciens-mediated transformation (ATMT) has been established for some zygomycetes. However, stability of the transformants remained problematic in case of Mucor (Monfort et al. 2003; Nyilasi et al. 2005). Modification of the integrated DNA resulting in rearrangements, excision and recircularization may also occur in the integrative transformation systems of zygomycetes (Michielse et al. 2004; Ibrahim and Skory 2007).
In this study, different methods, such as transformation with linear DNA fragments, restriction enzyme-mediated integration (REMI) (Maier and Schäfer 1999) and ATMT, were tested to integrate the bacterial gene into the Mucor genome. Carotenoid composition and effect of some culturing conditions, such as temperature, carbon sources and application of certain additives, on the carotenoid content were analysed in the resulting transformants.
Materials and methods
Strains, media and growth conditions
MS12, a leuA − and pyrG − mutant (Benito et al. 1992) derived from the wild-type M. circinelloides strain CBS277.49, was used in the transformation experiments; this strain is auxotrophic for leucine and uracil but wild type for the carotene biosynthesis. The A. tumefaciens strain GV3101 containing the pMP90 helper plasmid was used in the ATMT experiments; this strain harbours rifampicin and gentamicin resistance markers in the bacterial genome and the helper plasmid, respectively. GV3101 was grown on LB medium (Sambrook et al. 1989) containing 25 μg mL−1 gentamicin and 100 μg mL−1 rifampicin at 28 °C. Induction medium (IM) for ATMT was prepared as described by Bundock and Hooykaas (1996). Escherichia coli strain DH5α was used in all cloning experiments and plasmid amplifications; it was cultivated on LB medium containing 50 μg mL−1 ampicillin at 37 °C. For both nucleic acid and carotenoid extraction, M. circinelloides strains were cultured on solid minimal medium (YNB, 10 g glucose, 0.5 g yeast nitrogen base without amino acids, 1.5 g (NH4)2SO4, 1.5 g sodium glutamate and 20 g agar/L) supplemented with leucine or uracil (0.5 mg mL−1) as required. To test the mitotic stability of the transformants, malt extract agar (MEA, 10 g glucose, 5 g yeast extract, 10 g malt extract and 20 g agar/L) was used as a complete medium. Fungal cultures were grown for 4 days under continuous light at 25 °C. Temperature dependence of the carotenoid production was tested cultivating the fungal strains on YNB at 20, 25, 30 35 and 37 °C. To examine the effect of different carbon sources on the carotenoid production, glucose was replaced with mannose, trehalose, fructose, galactose, cellobiose, maltose, ethanol, glycerol, glycerol-l-monoacetate, dihydroxyacetone and l-aspartic acid in a final concentration of 1 % in YNB. When the effect of some chemical additives was analysed, 20 mM H2O2, 1 % palm oil or 1 mM FeSO4, CuSO4 or CoCl2 were added to YNB containing 1 % glucose.
Molecular techniques
General procedures for plasmid DNA preparation, cloning, transformation of E. coli and Southern blotting were performed by following standard methods (Sambrook et al. 1989). Genomic DNA was prepared from mycelia disrupted with a pestle and mortar in liquid nitrogen. DNA was isolated using a method described earlier (Iturriaga et al. 1992) or with the DNeasy Plant Mini Kit (Qiagen). DNA sequencing was performed by LGC Genomics (Berlin, Germany). For Southern hybridizations, probes were labelled with the digoxigenin-based PCR DIG Probe Synthesis Kit (Roche), and the DIG Nucleic Acid Detection Kit (Roche) was used for immunological detection of the nucleic acid blots, following the instructions of the manufacturer.
Construction of plasmids
Plasmids used in the present study are summarized in Table 1. In all constructions, either the leuA gene encoding the α-isopropylmalate isomerase or the pyrG gene encoding the orotidine-5′-monophosphate decarboxylase was used as a selection marker; these genes complement the leucine and the uracil auxotrophy of the MS12 strain, respectively. The crtW cassette containing the β-carotene ketolase gene (crtW) of Paracoccus sp. N81106 fused with the promoter (gpd1P) and terminator (gpd1T) regions of the glyceraldehyde-3-phosphate dehydrogenase 1 gene (gpd1) of M. circinelloides was derived from the pPT51 plasmid (Papp et al. 2006). Map of the plasmids constructed in this study is presented in Online Resource 1.
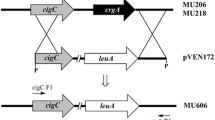
The plasmid pCA8 was constructed by placing the crtW cassette between the leuA gene and its 5′ adjacent region, as follows: leuA cut from the pAVB107 plasmid (Velayos 2000) with XbaI and PstI enzymes was placed between the corresponding sites of pUC18 (Fermentas) giving rise pCA7; then, the 4.01-kb SmaI-ScaI fragment of pCA7 and the 8.67-kb NaeI-ScaI fragment derived from pPT51 were ligated to form pCA8.
To construct pCA9, the crtW casette and pyrG were built between the fragments of the 18S and 28S ribosomal RNA genes (rDNA) of M. circinelloides. The 18S and 28S rDNA fragments were amplified from the genomic DNA of M. circinelloides using the RIB1–RIB2 and RIB3–RIB4 primer pairs (Table 2), respectively, and they were cloned into pUC18 (Fermentas) giving rise the p18S and p28S plasmids, respectively. These plasmids were digested with the enzymes SalI and BamHI, and the 1.41-kb fragment of p28S was ligated with the 4.5-kb fragment of p18S resulting in p18S-28S. At the same time, pCA2 was constructed by ligating the 5.01- and 1.32-kb fragments obtained by the XbaI and PstI digestion of pPT51 and pEPM901 (Benito et al. 1992), respectively. Finally, p18S-28S and pCA2 were digested with ClaI and KpnI, and the arising 5.65-kb fragment of p18S-28S and the 3.45-kb fragment of pCA2 were used to create pCA9. The pCA15 plasmid used in the ATMT experiments was derived from the pPK2 Agrobacterium binary vector (Covert et al. 2001) by the replacement of the original hph cassette with the modified crtW and the pyrG genes (Papp et al. 2012).
Transformation
Protoplasts of M. circinelloides were prepared as described earlier (Papp et al. 2006). In all transformation experiments, transformants were selected on the basis of auxotrophy complementation and colour change cultivating them on YNB, supplemented with leucine or uracil if required. Transformation with linear fragments and REMI was carried out via PEG-mediated transformation of protoplasts, which was performed according to van Heeswijck and Roncero (1984). Map of DNA fragments and molecules for transformation are shown in Online Resource 1 and 2. To force gene replacement, the linear DNA molecules pCA8lf and pCA9lf obtained by the ClaI-AatII and AatII-NheI digestion of pCA8 and pCA9, respectively, were used to transform MS12. For REMI, the SalI-SacI fragment of pCA8 (pCA8′R), the ClaI-KpnI and PstI-KpnI fragments of pCA9 (pCA9′R1 and pCA9′R2, respectively) and the NaeI-NdeI, NaeI-ClaI and SalI-ScaI fragments of pPT51 (pPT51′R1, pPT51′R2 and pPT51′R3, respectively) were introduced into the recipient MS12 strain. REMI experiments were performed according to the PEG-mediated method except that the restriction enzymes used previously to digest the plasmid were added to the protoplasts together with the linear DNA, and the solution was incubated for 0.5 h on ice and 1.5 h at 33 °C.
For ATMT, pCA15 was introduced into the A. tumefaciens strain GV3101 by electroporation at 2.5 kV and 25 μF; LB medium supplemented with 25 μg mL−1 gentamicin, 100 μg mL−1 rifampicin and 40 μg mL−1 kanamycin was used to select the cells that contain pPCA15 and pMP90. ATMT was performed according to Papp et al. (2012). Co-cultivation of A. tumefaciens cells and M. circinelloides protoplasts was performed on cellophane sheets placed on the surface of IM supplemented with 0.8 M sorbitol and 200 μM acetosyringone at 28 °C for 3 days. Following the incubation, the cellophane sheets were transferred to selection medium (YNB) and incubated at room temperature for 4–10 days. Putative transformants were then transferred to fresh YNB plates to obtain monosporangial (i.e. colonies from single spores) cultures.
Molecular analysis of the transformants
Real-time quantitative PCR (qPCR) was used to determine the copy number of the transferred gene in the total DNA samples. The qPCR experiments were performed in iQ5 and CFX96 real-time PCR detection systems (Bio-Rad), using the iQTM SYBR Green Supermix (Bio-Rad). The primers used in the qPCR amplifications are shown in Table 2. The amplification conditions were as follows: an initial denaturing step of 3 min at 95 °C was followed by 40 cycles of 15-s denaturation (95 °C) and 30-s annealing and extension (69 °C). The relative quantification of the copy number was performed using the 2-ΔCt method (Livak and Schmittgen 2001). Copy number of the crtW was referred to those of the carG and isoA genes in a given DNA sample; the latter genes exist in one copy in the M. circinelloides genome (Csernetics et al. 2011).
The inverse PCR (IPCR) method (Ochman et al. 1988) was used to analyse the integration site of the transferred DNA. About 1 μg genomic DNA of the tested transformants was digested with one of the restriction enzymes SalI, ScaI, BstXI, ClaI, SmaI or NheI overnight and precipitated with two volumes of ethanol at −20 °C. Then, the samples were washed with 70 % ethanol, dried under vacuum and resuspended in 10 μl distilled water. The digested samples were ligated at 8 °C overnight and incubated at 65 °C for 5 min to inactivate the ligase. The ligation samples were precipitated and washed again and dissolved in 20 μl distilled water. These samples (20–50 ng) served as templates in the IPCR experiments using different combinations of the primers shown in Table 2. Amplifications were performed with the Pfu polymerase (ZenonBio), and the PCR products were cloned using the CloneJET PCR Cloning Kit (Thermo Scientific) and sequenced. To determine the sites of integration, the resulting sequences were tested in BLAST searches against the M. circinelloides genome database (DoE Joint Genome Institute; M. circinelloides CBS277.49 v2.0; http://genome.jgi-psf.org/Mucci2/Mucci2.home.html). The IPCR strategy is shown in Online Resource 2.
Plasmid rescue experiments were performed as follows. E. coli DH5α cells were transformed with 3 μg genomic DNA of the MS12+pCA8′R transformants. The bacterium transformation protocol followed general procedures (Sambrook et al. 1989). As pCA8′R fragments contain the appropriate bacterial gene, bacterial transformants were selected on the basis of their resistance to ampicillin. The plasmids were purified from the transformants using the Mini Plus Ultrapure Plasmid DNA kit (Viogene) and then sequenced with the primers designed to the ampicillin resistance gene (Table 2). Location of these primers on the pCA8′R fragment is shown in Online Resource 2.
Carotenoid extraction and analysis
Carotenoid extraction was performed as described earlier (Papp et al. 2006). For high-performance liquid chromatography (HPLC), samples were analysed by using a modular Shimadzu low-pressure gradient HPLC system. The dried samples were re-dissolved in 100 μL tetrahydrofuran supplemented with butylated hydroxytoluene (100 μg mL−1), and 2 μL was subjected to HPLC analysis on a Prodigy ODS-3 (4.6 × 150, ODS 3 μm) column (Phenomenex). The separation was performed with a gradient (min/volume of solvent A%/volume of solvent B% was 0/99/1; 8/60/40; 13/46/54; 15/0/100; 18/0/100; 21/99/1; and 25/99/1), 4 % water-96 % methanol being used as solvent A and 100 % methyl-tert-butyl ether as solvent B, at a flow rate of 1 mL min−1. The detection wavelength was 450 nm. The following standards were used to identify the carotenoids: astaxanthin, lycopene and β-carotene (Sigma), β-cryptoxanthin, zeaxanthin and canthaxanthin (Carl Roth) and echinenone (DHI Water and Environment); γ-carotene was purified by HPLC from Mucor azygosporus.
Results
Transformation, transformation frequency and mitotic stability
Transformation with linear fragments and REMI was performed via PEG-mediated protoplast transformation. All these experiments resulted in transformant colonies except REMI using the two pCA9′R fragments; the transformation frequencies were relatively low, one to four colonies per 105 protoplasts using 10–15 μg linear DNA. During the optimization of the transformation protocol, application of the DNA for transformation in higher amounts did not lead to further increase in the number of the transformants. In case of the REMI, application of 30 U of the restriction enzymes and incubation of transformation mixture containing the protoplasts, the DNA fragment and the restriction enzymes for 30 min on ice and subsequently for 1.5 h at 33 °C proved to be suitable.
ATMT experiments had a similar transformation frequency resulting in one to eight transformant colonies per experiment. All stable transformants obtained in this study were deposited in the culture collection of the University of Szeged (Szeged Microbiology Collection, Szeged, Hungary). After the second cultivation, the presence of the crtW gene could be detected clearly in the transformants by PCR (data not shown). Southern hybridization analysis verified the integration in the majority of the transformants (Fig. 2).
Examples of Southern analyses performed to prove the presence of the crtW gene and its integration into the genome of the M. circinelloides transformants using the digoxigenin-labelled crtW gene as a hybridization probe. Panels: the fragment pCA8lf and the DNAs of three MS12+pCA8lf strains digested with SmaI (a) and PvuII (b), the fragment pCA9lf and the DNAs of three MS12+pCA9lf strains digested with ClaI and KpnI (c), the fragment pCA8′R and the DNAs of three MS12+pCA8′R strains digested with SmaI (d) and ClaI (e), the fragment pPT51′R and the DNAs of three MS12+pPT51′R strains digested with SmaI (f) and the plasmid pCA15 and the DNAs of three MS12+pCA15 strains digested with XhoI (g)
To analyse their mitotic stability, transformants were passed several times onto selective and non-selective media. Strains transformed with the linear fragments, pCA8lf or pCA9lf, were stable after more than 15 consecutive cultivation steps under non-selective conditions (i.e. on MEA plates). Out of the REMI constructed strains, the strains transformed with pCA8′R and pPT51′R3 were found to be stable. ATMT transformants proved to be unstable losing the transferred DNA during the first three cultivation cycles.
Molecular analysis of the mitotically stable transformants
The site of integration was analysed only in the stable transformants by using the IPCR technique. In all tested MS12+pCA8lf and MS12+pCA8′R transformants, integration occurred by gene replacement in the homologous site and was directed by the leuA gene and its 5 adjacent region. IPCR analysis detected ectopic integration in the tested MS12+pCA9lf and MS12+pPT51′R3 transformants in different genomic regions (the genomic regions revealed by the IPCR experiments are presented in Online Resource 3). In several REMI transformants, IPCR analysis suggested multiple integrations and/or rearrangements in the transferred DNA and the adjacent host regions. Sequences of the fragments amplified by PCR from the genomic DNA of these transformants using the inverse primers also suggested the existence of such multicopy structures.
Copy number of the crtW gene in the transformants was analysed by using real-time qPCR (Table 3). Mucor species generally produce multinucleate protoplasts and spores and form coenocytic mycelia. As a consequence, primary integrative transformants are heterokaryotic to the transferred DNA. In order to obtain homokaryotic strains, it is necessary to isolate monosporangial colonies and to perform some consecutive cultivation cycles on the selection medium with them. Copy number increased in all tested transformants in the consecutive generations. In several transformants, such as in MS12+pCA8lf/1-2, MS12+pCA9lf/1 and MS12+pPT51′R3/2, the copy number detected after the 13th cultivation cycle was very high (Table 3). In the cases of the MS12+pCA8lf/1 and MS12+pCA9lf/1, IPCR also suggested the presence of multiple copies of the introduced DNA. In line with the increasing copy number, the primary yellowish colony colour of these transformants changed to orange-red indicating the accumulation of keto-carotenoids in consequence of the expression of the transferred crtW gene (see Online Resource 4). In some transformants, the copy number of the crtW gene did not reached one copy per genome even after more than ten generations showing that these strains remained heterokaryotic to the transferred DNA (Table 3).
To reveal the potential existence of autonomously replicating plasmids came from the excision and recircularization of the integrated DNA fragments, plasmid rescue experiments were performed with the MS12+pCA8′R transformants. Circular plasmids with sizes significantly lower than the originally transferred DNA (2,000–5,500 bp) could be recovered. Sequencing of these plasmids revealed rearranged and truncated fragments of the transferred DNA, but most of them did not contain the crtW gene or its parts.
Carotenoid production of the transformants
Expression of the bacterial crtW gene led to the production of keto derivatives of β-carotene, mainly canthaxanthin and echinenone and very small amounts of astaxanthin in the transformants. Detailed carotenoid composition of some transformants is shown in Table 3. During the first three cultivation cycles, concentration of the keto carotenoids was low, except in some MS12+pPT51′R3 transformants, which had red-orange colour directly after the transformation. After several cultivation cycles (>10), the amount of canthaxanthin and echinenone increased significantly in most of the transformants corresponding to the elevated copy number of the crtW gene. At the same time, the REMI constructed MS12+pCA8′R transformants produced only small amounts of the keto derivatives independently from the number of the cultivation cycles. Out of the good canthaxanthin-producing strains, MS12+pCA8lf/1 and MS12+pCA9lf/1 were selected to examine the effect of the culturing temperature, the carbon source and certain chemical additives on the carotenoid content and composition.
Effect of the culturing temperature on the carotenoid production
Carotenoid content of the two selected transformants in comparison with that of the MS12 strain was examined after cultivating the fungi on YNB at 20, 25, 30, 35 and 37 °C. Figure 3 shows the average β-carotene and canthaxanthin content of the three strains measured at the different temperatures. The canthaxanthin and echinenone production was the highest at 20 °C and decreased at the higher cultivation temperatures. At the same time, the highest total carotenoid and β-carotene levels were detected at 35 °C. Interestingly, transformants produced significantly higher amounts of β-carotene at this temperature than MS12. The maximum growth temperature of M. circinelloides is 36 °C. In accordance with this, the original MS12 strain did not grow at 37 °C. However, the tested transformants were able to survive and produce carotenoids at this temperature.
Effect of different carbon sources and chemical additives on the carotenoid production
This experiment was performed with strains passed more than 20 times after the transformation. MS12, MS12+pCA8lf/1 and MS12+pCA9lf/1 were cultured on solid YNB, where glucose was replaced to different compounds as the sole carbon sources (Table 4). Some chemical additives given to the glucose containing minimal medium were also tested. Canthaxanthin content of the two transformants significantly differed on glucose: MS12+pCA8lf/1 contained 33 % canthaxanthin and 7 % β-carotene compared to the total carotenoid content; in contrast, these proportions were 13 and 36 %, respectively, in the case of MS12+pCA9lf/1. Other carbon sources and the tested additives also exerted different effects on their carotenoid composition. Although application of fructose, trehalose and mannose enhanced the total carotenoid level in both transformants, the canthaxanthin–β-carotene ratio of the strains changed only on fructose. l-aspartic acid significantly stimulated the canthaxanthin production of the MS12+pCA8lf/1 strain, while glycerine had a similarly positive effect on that of MS12+pCA9lf/1. Dihydroxyacetone increased the canthaxanthin content in both transformants. Although the other tested carbon sources and additives did not change or even decreased the total carotenoid level, several compounds increased the proportion of the keto derivatives of β-carotene, such as H2O2 and CuSO4 in the case of MS12+pCA8lf/1 or glycerol and ethanol in the case of MS12+pCA9lf/1. Online Resource 4 shows the colony colours of the MS12+pCA8lf/1 strain cultured on the different carbon sources.
Discussion
In the present study, attempts to integrate the crtW gene of Paracoccus sp. N81106 encoding a β-carotene ketolase into the M. circinelloides genome were carried out using different techniques. Transformation systems that allow stable integration of the transferred DNA into the host genome are essential for the genetic modification of the organisms as well as for the functional analysis of genes. It is well known that transformation of zygomycetes with circular plasmids generally results in strains that maintain the introduced plasmids episomally without any integration event (Ibrahim and Skory 2007; Papp et al. 2010). Such transformants generally prove to be mitotically unstable because of the poor segregation of their plasmids into the spores (Appel et al. 2004). However, at least in the case of M. circinelloides, integration has been achieved by using linear DNA fragments for transformation, which harbour homologous sequences at their termini to direct the double-crossing over gene replacement (Navarro et al. 2001; Silva et al. 2006), and several zygomycetes have been successfully transformed with the ATMT method (Michielse et al. 2004; Monfort et al. 2003; Nyilasi et al. 2005; 2008), which generally led to single-copy integration. Successful REMI transformation has not yet been reported in zygomycetes. Despite the fact that several transformation methods are established for zygomycetes, construction of stable transformants, which maintain and express a heterologous (especially a bacterial) gene integrated into the genome, has remained a great challenge (Ibrahim and Skory 2007; Michielse et al. 2004; Obraztsova et al. 2004; Papp et al. 2010).
In our study, all tested methods, including PEG-mediated protoplast transformation with linear DNA fragments, REMI and ATMT, resulted in transformants. However, ATMT and certain REMI transformants proved to be unstable losing the transferred DNA during the first few cultivation cycles. In a previous experiment, ATMT of M. circinelloides with a bacterial gene (hygromycin B phosphotransferase) also led to the formation of unstable transformants (Nyilasi et al. 2005), and a similar instability was found in the ATMT of other Mucorales, such as Rhizomucor miehei and Backusella lamprospora (Monfort et al. 2003; Nyilasi et al. 2008). Stable transformants were obtained by ATMT from Rhizopus oryzae but only if the integrated gene was endogenous or originated from a closely related organism (Michielse et al. 2004; Ibrahim and Skory 2007). REMI method generally leads to random integration where the applied enzymes and the site of integration are important factors of the stability of the integrated DNA.
Although majority of the transformants produced by PEG-mediated protoplast transformation with linear DNA fragments and REMI retained the transferred DNA even under non-selective cultivation conditions, rearrangements to large concatemers and/or excision and recircularization of the integrated linear fragments can be suggested in several strains based on the results of the IPCR and the plasmid rescue experiments. Besides the multiple integration of the transformed DNA, such processes also may explicate the extremely high copy number of the transferred DNA detected in certain transformants (Table 3). Rearrangements of the transferred DNA have often observed in zygomycetes (Yanai et al. 1990; Takaya et al. 1996; Papp et al. 2010). Several authors proposed the existence of a genome defence mechanism that eliminates the heterologous DNA via these DNA rearrangements and deletions (Ibrahim and Skory 2007; Michielse et al. 2004; Nyilasi et al. 2005; Obraztsova et al. 2004; Papp et al. 2010). However, this was not the case in our transformants, where rearrangements did not eliminate the bacterial gene. In contrast, copy number of the crtW gene increased, sometimes dramatically, during the consecutive cultivation cycles. Higher copy number of crtW generally led to increased canthaxanthin and echinenone production, but this relation was not directly proportional in the different transformants (Table 3) indicating that the site of integration, possible DNA rearranges and/or other factors, such as gene regulation or silencing, may also affect the efficacy of the heterologous gene expression as reported in other organisms (Verdoes et al. 1995; Liang et al. 1996; Lubertozzi and Keasling 2009). Over a certain expression level of the exogenous gene, the amount of the available precursor, i.e. β-carotene, could also limit the synthesis of canthaxanthin.
As linear DNA molecules used in the PEG-mediated transformations held homologous sequences at their termini to direct gene replacement, integration was expected in the corresponding homologous sites. This could be proven only in the transformants produced by using the fragment pCA8lf and REMI with pCA8′R. In the other cases, ectopic integration occurred. As restriction enzymes digest the genomic DNA at several sites, it is well known that REMI frequently causes random integration (Turgeon et al. 2010). Although we expected gene replacement with the pCA9lf fragment, it also integrated ectopically. Maybe the extensions of the homologous regions were not sufficient to direct the double-crossing over or integration into the ribosomal cluster caused defects in the host genome.
After the 13th cultivation step, some transformants produced more than 100 μg g−1 (dry mass) canthaxanthin on glucose-containing minimal medium (Table 3), which is a similar amount to that measured previously in Paracoccus sp. N81106 by Yokoyama and Miki (1995). Interestingly, total carotenoid content of the crtW-harbouring transformants also proved to be higher than that of the untransformed MS12 strain suggesting that formation and/or presence of the keto derivatives stimulate the β-carotene biosynthesis (Table 3). It is known that β-carotene itself and its derivatives produced by the fungus have feedback effect on the β-carotene biosynthesis (Lampila et al. 1985; Fraser et al. 1996; Bhosale 2004). It is possible that the β-carotene level, which decreased radically in consequence of its conversion to canthaxanthin, affected the activity of the carotenoid biosynthesis genes. On the other hand, canthaxanthin may also be able to stimulate the carotenogenic pathway since it holds keto groups on the β-ionone rings, which was previously found to be essential in the positive feedback effect of several chemical regulators of the pathway, such as trisporic acids (Bhosale 2004). Previously, crtW was introduced into the same M. circinelloides strain using a circular plasmid (Csernetics et al. 2011; Papp et al. 2006). Under the same culturing conditions, the canthaxanthin content of those transformants was much lower [6–13 μg g−1 (dry mass)], maybe in consequence of the low copy number and the unequal distribution of the plasmids in the mycelium and the spores. Similarly to previous studies (Papp et al. 2006; Csernetics et al. 2011), the astaxanthin level remained low in consequence of the low activity of the endogenous β-carotene hydroxylase.
As an important environmental factor, temperature affects several biosynthetic pathways including carotenoid biosynthesis (Bhosale 2004). In our experiments, both the transformed and the original M. circinelloides strains produced the highest amounts of β-carotene at 35 °C (Fig 3). A similar effect of the temperature to the carotenoid production was previously observed in other Mucor species, such as in Mucor rouxii (Mosqueda-Cano and Gutierez-Corona 1995). Contrarily, canthaxanthin and echinenone formation of the transformants was higher at lower cultivation temperature, maybe due to the temperature requirements of the heterologous β-carotene ketolase. Interestingly, crtW containing transformants were able to survive at higher temperature than the untransformed fungus. Canthaxanthin has the ability to interact with and incorporate in the plasma membrane, where it exerts significant influence on the structural and dynamic properties of the membrane even in very small concentration (Sujak et al. 2007). Recently, Kumaresan et al. (2008) also reported thermotolerance of a mutant Aspergillus carbonarius strain connected with its canthaxanthin content. The effect of canthaxanthin on membrane fluidity and structure may contribute in the higher thermotolerance of the transformants. Moreover, at higher cultivation temperatures, canthaxanthin may protect against the increased endogenous generation of reactive oxygen species. Indeed, canthaxanthin proved to be more effective against free radicals than β-carotene in membrane models (Palozza and Krinsky 1992).
Effects of different carbon sources and chemical additives on the carotenoid production of two selected transformants were also tested. Glucose, mannose, trehalose and fructose had a positive effect on the whole carotenoid content of both transformants tested. Previously, glucose, cellobiose and maltose enhanced the carotenogenesis of M. rouxii in the highest degree compared to other carbon sources (Mosqueda-Cano and Gutierez-Corona 1995). In the case of MS12+pCA8lf/1, application of dihydroxyacetone and l-aspartic acid resulted in that β-carotene almost completely converted to its keto derivatives, mainly to canthaxanthin. We suppose that both compounds affected the keto-carotenoid level through the induction of the gpd1 promoter, which drives the expression of the transferred crtW gene. With the application of glucose (at 20 °C, see Fig. 3), trehalose, dihydroxyacetone and l-aspartic acid (Table 4), MS12+pCA8lf/1 produced more than 200 μg g−1 (dry mass) of canthaxanthin. This amount is comparable with the canthaxanthin content of the wild-type G. jacobaea [200 μg g−1 (dry mass)] (Veiga-Crespo et al. 2005), but lower than those of Brevibacterium KY-4313 [600 μg g−1 (dry mass)] (Nelis and De Leenheer 1989) and Haloferax alexandrinus [700 μg g−1 (dry mass)] (Asker and Ohta 2002).
The aim of this study was to examine the biological requirements of the application of M. circinelloides as a xanthophyll producer and to develop strains and methods, which could be used in the further applied studies. Stable strains expressing a bacterial β-carotene ketolase and producing canthaxanthin in considerable amounts were constructed. Taking into account that the canthaxanthin level reported in this study for MS12+pCA8lf/1 was achieved in minimal medium and the echinenone level remained relatively high in the majority of experiments, we expect that optimization of the growth conditions and improvement of the culturing medium will allow much higher carotenoid production and more complete β-carotene–canthaxanthin conversion. Stable canthaxanthin-producing mutants also can be used as a host for heterologous genes to construct astaxanthin-producing strains as well as for model organisms to study the physiological effect of the canthaxanthin accumulation. Although further studies are needed to increase the canthaxanthin productivity of the modified M. circinelloides strains, our results indicate that they may be promising candidates for further canthaxanthin-producing strain improvement studies.
References
Álvarez V, Rodríguez-Sáiz M, de la Fuente JL, Gudina EJ, Godio RP, Martín JF, Barredo JL (2006) The crtS gene of Xanthophyllomyces denrorhous encodes a novel cytochrome-P450 hydroxylase involved in the conversion of β-carotene into astaxanthin and other xanthophylls. Fungal Genet Biol 43:261–272
Appel KF, Wolff AM, Arnau J (2004) A multicopy vector system for genetic studies in Mucor circinelloides and other zygomycetes. Mol Genet Genom 271:595–602
Asker D, Ohta Y (2002) Production of canthaxanthin by Haloferax alexandrinus under non-aseptic conditions and a simple, rapid method for its extraction. Appl Microbiol Biotechnol 58:743–750
Benito EP, Díaz-Mínguez JM, Iturriaga EA, Campuzano EA, Eslava AP (1992) Cloning and sequence analysis of the Mucor circinelloides pyrG gene encoding orotidine-5′-monophosphate decarboxylase: use of pyrG for homologous transformation. Gene 116:59–67
Bhosale P (2004) Environmental and cultural stimulants in the production of carotenoids from microorganisms. Appl Microbiol Biotechnol 63:351–361
Bhosale P, Bernstein PS (2005) Microbial xanthophylls. Appl Microbiol Biotechnol 68:445–455
Bundock P, Hooykaas PJJ (1996) Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc Nat Acad Sci USA 93:15272–15275
Cooney JJ, Marks HW, Smith AM (1966) Isolation and identification of canthaxanthin from Micrococcus roseus. J Bacteriol 92:342–345
Covert SF, Kapoor P, Lee M, Briley A, Nairn CJ (2001) Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol Res 105:259–264
Csernetics Á, Nagy G, Iturriaga EA, Szekeres A, Eslava AP, Vágvölgyi C, Papp T (2011) Expression of three isoprenoid biosynthesis genes and their effects on the carotenoid production of the zygomycete Mucor circinelloides. Fungal Genet Biol 48:696–703
Czygan FC (1964) Canthaxanthin as a secondary carotenoid in certain green algae. Experientia 20:573–574
De Miguel T, Sieiro C, Poza M, Villa TG (2001) Analysis of canthaxanthin and related pigments from Gordonia jacobaea mutants. J Agric Food Chem 49:1200–1202
Dufossé L (2006) Microbial production of food grade pigments. Food Technol Biotechnol 44:313–321
Ernst H (2002) Recent advances in industrial carotenoid synthesis. Pure Appl Chem 74:1369–1382
Fraser PD, Ruiz-Hidalgo MJ, Lopez-Matas MA, Alvarez MI, Eslava AP, Bramley PM (1996) Carotenoid biosynthesis in wild type and mutant strains of Mucor circinelloides. Biochim Biophys Acta 1289:203–208
Ibrahim AS, Skory CD (2007) Genetic manipulation of zygomycetes. In: Kavanagh K (ed) Medical mycology: cellular and molecular techniques. Wiley, Bognor Regis, pp 305–326
Iturriaga EA, Díaz-Mínguez JM, Benito EP, Álvarez MI, Eslava AP (1992) Heterologous transformation of Mucor circinelloides with the Phycomyces blakesleeanus leu1 gene. Curr Genet 21:215–223
Jyonouchi HS, Sun MM, Gross MD (1996) Effects of various carotenoids on cloned, effector-stage T-helper cell activity. Nutr Cancer 26:313–324
Kumaresan N, Sanjay KR, Venkatesh KS, Kadeppagari R-K, Vijayalakshmi G, Umesh-Kumar S (2008) Partially saturated canthaxanthin purified from Aspergillus carbonarius induces apoptosis in prostate cancer cell line. Appl Microbiol Biotechnol 80:467–473
Lampila LE, Wallen ISE, Bullerman LB (1985) A review of factors affecting biosynthesis of carotenoids by the order Mucorales. Mycopathologia 90:65–80
Liang SH, Skory CD, Linz JE (1996) Characterization of the function of the ver-1A and ver-1B genes, involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ Microbiol 62:4568–4575
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25:402–408
Lubertozzi D, Keasling JD (2009) Developing Aspergillus as a host for heterologous expression. Biotechnol Adv 27:53–75
Maier FJ, Schäfer W (1999) Mutagenesis via insertional or restriction enzyme-mediated-integration (REMI) as a tool to tag pathogenicity related genes in plant pathogenic fungi. Biol Chem 380:855–864
Mayne ST, Parker RS (1989) Antioxidant activity of dietary canthaxanthin. Nutr Cancer 12:225–236
Michielse CB, Salim K, Ragas P, Ram AFJ, Kudla B, Jarry B, Punt J, van den Hondel CAMJJ (2004) Development of a system for integrative and stable transformation of the zygomycete Rhizopus oryzae by Agrobacterium-mediated DNA transfer. Mol Gen Genom 271:499–510
Monfort A, Cordero L, Maicas S, Polaina J (2003) Transformation of Mucor miehei results in plasmid deletion and phenotypic instability. FEMS Microbiol Lett 224:101–106
Mosqueda-Cano G, Gutierez-Corona JF (1995) Environmental and developmental regulation of carotenogenesis in the dimorphic fungus Mucor rouxii. Curr Microbiol 31:141–145
Navarro E, Lorca-Pascual JM, Quiles-Rosillo MD, Nicolas FE, Garre V, Torres-Martinez S, Ruiz-Vazquez RM (2001) A negative regulator of light-inducible carotenogenesis in Mucor circinelloides. Mol Gen Genom 266:463–470
Nelis HJ, De Leenheer AP (1989) Reinvestigation of Brevibacterium sp Strain KY-4313 as a source of canthaxanthin. Appl Environ Microbiol 55:2505–2510
Nyilasi I, Ács K, Papp T, Vágvölgyi C (2005) Agrobacterium tumefaciens-mediated transformation of Mucor circinelloides. Folia Microbiol 50:415–420
Nyilasi I, Papp T, Csernetics Á, Vágvölgyi C (2008) Agrobacterium tumefaciens-mediated transformation of the zygomycete fungus, Backusella lamprospora. J Basic Microbiol 48:59–64
Obraztsova IN, Prados N, Holzmann K, Avalos J, Cerdá-Olmedo E (2004) Genetic damage following introduction of DNA in Phycomyces. Fung Genet Biol 41:168–180
Ochman H, Gerber AS, Hartl DL (1988) Genetic applications of an inverse polymerase chain reaction. Genetics 120:621–623
Okai Y, Higashi-Okai K (1996) Possible immunomodulating activities of carotenoids in in vitro cell culture experiments. Int J Immunopharmacol 18:753–758
Palozza P, Krinsky NI (1992) Astaxanthin and canthaxanthin are potent antioxidants in a membrane model. Arch Biochem Biophys 297:291–295
Palozza P, Maggiano N, Calviello G, Lanza P, Piccioni E, Ranelletti FO, Bartoli GM (1998) Canthaxanthin induces apoptosis in human cancer cell lines. Carcinogenesis 19:373–376
Papp T, Csernetics Á, Nyilasi I, Ábrók M, Vágvölgyi C (2010) Genetic transformation of zygomycetes fungi. In: Rai M, Kövics G (eds) Progress in mycology, Springer, Scientific Publishers, Jodhpur, pp 75–94
Papp T, Csernetics Á, Nyilasi I, Vágvölgyi C, Iturriaga EA (2012) Integration of a bacterial β-carotene ketolase gene into the Mucor circinelloides genome by the Agrobacterium tumefaciens-mediated transformation (ATMT) method. In: Barredo J-L (ed) Microbial carotenoids: methods and protocols. Methods in molecular biology, vol. 898, Humana Press, Springer, New York, pp 123–132
Papp T, Velayos A, Bartók T, Eslava AP, Vágvölgyi C, Iturriaga EA (2006) Heterologous expression of astaxanthin biosynthesis genes in Mucor circinelloides. Appl Microbiol Biotechnol 69:526–531
Pelah D, Sintov A, Cohen E (2004) The effect of salt stress on the production of canthaxanthin and astaxanthin by Chlorella zofingiensis grown under limited light intensity. World J Microbiol Biotechnol 20:483–486
Sambrook J, Fitsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor
Saperstein S, Star MP (1954) The ketonic carotenoid canthaxanthin isolated from a colour mutant of Corynebacterium michiganense. Biochem J 57:273–275
Silva F, Torres-Martínez S, Garre V (2006) Distinct white collar-1 genes control specific light responses in Mucor circinelloides. Mol Microbiol 61:1023–1037
Sujak A, Strzałka K, Gruszecki WI (2007) Thermotropic phase behaviour of lipid bilayers containing carotenoid pigment canthaxanthin: a differential scanning calorimetry study. Chem Phys Lipids 145:1–12
Takaya N, Yanai K, Horiuchi H, Ohta A, Takagi M (1996) Cloning and characterization of the Rhizopus niveus leu1 gene and its use for homologous transformation. Biosci Biotechnol Biochem 60:448–452
Turgeon BG, Condon B, Liu J, Zhang N (2010) Protoplast transformation of filamentous fungi. In: Sharon A (ed) Molecular and cell biology methods for fungi, series: Methods in molecular biology, vol 638, Humana Press, Springer, New York, pp 3–19
van Heeswijck R, Roncero MIG (1984) High frequency transformation of Mucor with recombinant plasmid DNA. Carlsberg Res Commun 49:691–702
Veiga-Crespo P, Blasco L, dos Santos FR, Poza M, Villa TG (2005) Influence of culture conditions of Gordonia jacobaea MV-26 on canthaxanthin production. Int Microbiol 8:55–58
Velayos A (2000) Carotenogenesis en Mucor circinelloides. PhD Thesis. Universidad de Salamanca, Salamanca, Spain
Velayos A, Blasco JL, Alvarez MI, Iturriaga EA, Eslava AP (2000a) Blue-light regulation of the phytoene dehydrogenase (carB) gene expression in Mucor circinelloides. Planta 210:938–946
Velayos A, Eslava AP, Iturriaga EA (2000b) A bifunctional enzyme with lycopene cyclase and phytoene synthase activities is encoded by the carRP gene of Mucor circinelloides. Eur J Biochem 267:1–12
Velayos A, Papp T, Aguilar-Elena R, Fuentes-Vicente M, Eslava AP, Iturriga EA, Álvarez MI (2003) Expression of the carG gene, encoding geranylgeranyl pyrophosphate synthase, is up-regulated by blue light in Mucor circinelloides. Curr Genet 43:112–120
Verdoes JC, Punt PJ, van den Hondel CAMJJ (1995) Molecular-genetic strain improvement for the overproduction of fungal proteins by filamentous fungi. Appl Microbiol Biotechnol 43:195–205
Yanai K, Horiuchi H, Takagi M, Yano K (1990) Preparation of protoplasts of Rhizopus niveus and their transformation with plasmid DNA. Agric Biol Chem 54:2689–2696
Yokoyama A, Miki W (1995) Composition and presumed biosynthetic pathway of carotenoids in the astaxanthin-producing bacterium Agrobacterium aurantiacum. FEMS Microbiol Lett 128:139–144
Acknowledgments
This work was supported by a grant of the Research and Technology Innovation Fund and the Hungarian Scientific Research Fund (KTIA-OTKA CK 80188) and the Hungarian–French Intergovernmental S&T Cooperation Programme (TÉT_10-1-2011-0747).
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors Tamás Papp and Árpád Csernetics contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 125 kb)
Rights and permissions
About this article
Cite this article
Papp, T., Csernetics, Á., Nagy, G. et al. Canthaxanthin production with modified Mucor circinelloides strains. Appl Microbiol Biotechnol 97, 4937–4950 (2013). https://doi.org/10.1007/s00253-012-4610-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4610-2