Abstract
In search of effective nitrogen-fixing strains for inoculating Leucaena leucocephala, we assessed the symbiotic efficiency of 41 rhizobial isolates from root nodules of L. leucocephala growing in the arid–hot river valley area in Panxi, China. The genetic diversity of the isolates was studied by analyzing the housekeeping genes 16S rRNA and recA, and the symbiotic genes nifH and nodC. In the nodulation and symbiotic efficiency assay, only 11 of the 41 isolates promoted the growth of L. leucocephala while the majority of the isolates were ineffective in symbiotic nitrogen fixation. Furthermore, one fourth of the isolates had a growth slowing effect on the host. According to the 16S rRNA and recA gene analyses, most of the isolates were Ensifer spp. The remaining isolates were assigned to Rhizobium, Mesorhizobium and Bradyrhizobium. The sequence analyses indicated that the L. leucocephala rhizobia had undergone gene recombination. In contrast to the promiscuity observed as a wide species distribution of the isolates, the results implied that L. leucocephala is preferentially nodulated by strains that share common symbiosis genes. The symbiotic efficiency was not connected to chromosomal background of the symbionts and isolates carrying a similar nifH or nodC showed totally different nitrogen fixation efficiency.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rhizobia are well known for their ability to carry out symbiotic nitrogen fixation by forming nodules on the roots or occasionally on the stems of legumes. Owing to this ability, leguminous plants can grow in arid, infertile soils, acting as pioneer plants that stabilize soil and enhance fertility, thereby preventing water loss, soil erosion and desertification (Howieson et al. 2008; Sprent and Gehlot 2010).
Panxi (Panzhihua–Xichang) in the southwest of Sichuan province, China, is at the transitional zone from Qinhai–Tibet and Yunnan–Guizhou plateaus to Sichuan basin. The arid–hot river valley area in Panxi, characterized with xerothermic climate and abundant photothermal source, is the northernmost place with the Southeast Asia tropical climate (Zhao et al. 2011). Because of overcutting and shortage of fuel wood, Panxi area suffers from soil erosion and environmental deterioration. Therefore, improving environment and preserving water and soil by reforestation are imperative long-term tasks in this area.
Leucaena leucocephala is a fast-growing, tropical leguminous tree species in the subfamily Mimosoideae. It is referred as the miracle tree because of its exceptional capacity to produce biomass and protein (Somasegaran and Martin 1986). It is also used in agroforestry, soil improvement, preventing soil erosion, land reclamation and for firewood, timber, forage, and green manure (Moawad and Bohlool 1984; Somasegaran and Martin 1986). In its region of origin, Mexico, L. leucocephala is nodulated by a diverse selection of rhizobia, including Ensifer (Sinorhizobium), Rhizobium, and Mesorhizobium (Wang et al. 1999). The amount of nitrogen fixed by L. leucocephala rhizobium symbiosis is similar to or higher than that of crop legumes such as peanut and soybean, and most of the fixed nitrogen is returned to the soil in the leaf litter (Peoples et al. 1995). L. leucocephala was introduced into Panxi in the 1980s and 1990s as a pioneer species of planted forests. The planted L. leucocephala were not inoculated with rhizobia, and the indigenous Leucaena rhizobia in this area have not yet been investigated and described. We became aware that inoculants would be able to promote L. leucocephala development. Consequently, the first objective of this investigation was to isolate Panxi area Leucaena rhizobia and to find strains effective in plant growth promotion for inoculation. The second objective was to investigate the genetic diversity, the phylogeny and the taxonomic position of these isolates based on housekeeping genes 16S rRNA and recA, and on symbiosis related genes nifH and nodC.
Materials and methods
Isolation of nodule bacteria
Nodules were collected from lateral roots of L. leucocephala growing in 20 sites in the arid–hot river valley area in Panxi, China (Table 1). Bacteria were isolated from surface-sterilized nodules using the standard procedure and yeast extract–mannitol agar (YMA) medium (Vincent 1970). Single colonies were picked up and checked for purity by repeatedly streaking on YMA medium (Vincent 1970), and verified by colony morphology, absorption of Congo red (25 mg ml−1) and Gram reaction. All the isolates were incubated on YMA slants at 28 °C and maintained at 4 °C for temporary storage and in 20 % glycerol at −70 °C for long-term storage.
In the restriction fragment length polymorphism (RFLP) analysis, 14 reference strains were included, as follows: Bradyrhizobium japonicum USDA6T, Bradyrhizobium yuanmingense CCBAU10071T, Bradyrhizobium liaoningense USDA3622T; Rhizobium mongolense USDA1844T, Rhizobium hainanense CCBAU 57015T and Rhizobium huautlense LMG18254T; Ensifer/Sinorhizobium fredii USDA205T, Ensifer adhaerens/Sinorhizobium morelense LMG21331T and Ensifer/Sinorhizobium kostiense HAMBI1489T; Mesorhizobium ciceri USDA3383T and Mesorhizobium plurifarium LMG11892T; and Agrobacterium rubi IAM13569T, Agrobacterium vitis IAM14140T and, Allorhizobium undicola LMG11875T.
Nodulation and symbiotic efficiency assays
The nodulation ability and the symbiotic efficiency of the isolates were tested by plant inoculation assay on their original host plant (Table 1). L. leucocephala seeds were surface sterilized, placed on sterile Whatman filter paper, and germinated in sterile water in Petri dishes for 3–5 days. The seedlings were transplanted into glass bottles containing Jensen’s solution with 0.003 % of Ca(NO3)2 as starter nitrogen in all inoculation assays, including the uninoculated controls. The fast- and slow-growing isolates were grown in YEM broth (Vincent 1970) for 5 and 7 days, respectively. The seedlings were inoculated with 1.5 ml of the culture containing ca 1010 bacterial cells per milliliter and grown under a 16 h light and 8 h dark regime at 25 °C (Chen et al. 2003). All inoculation assays were done in triplicate. After 56 days, the plants were harvested, and the nodule numbers and plant shoot dry weights were determined. The data was analyzed by one-way analysis of variance, with the mean values compared using SPSS15.0 least significant difference (LSD) analysis (P = 0.05).
DNA extraction and PCR-RFLP analysis of the 16S rRNA gene
The subsequent DNA analyses were done on the 41 isolates that successfully nodulated L. leucocephala (Table 2). Total genomic DNA was obtained after lysozyme–sodium dodecyl sulfate lysis, followed by phenolchloroform extraction and ethanol precipitation as described by Li et al. (2009).
16S rRNA gene fragments were amplified using 20 pmol of each primer P1 and P6 (Li et al. 2009) in 50 μl of amplification buffer (250 μM of each dNTP, 10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, 50 mM KCl and 2.5 U Taq DNA polymerase), with about 50 ng of total DNA as template. The amplification was performed using a Bio-RAD MyCycler™ with the following temperature profile: an initial denaturation at 92 °C for 3 min; 30 cycles of denaturation at 94 °C for 3 min, annealing at 58 °C for 1 min and extension at 72 °C for 2 min; and a final extension at 72 °C for 6 min, with a final soak at 4 °C. The size of the amplification products was verified by electrophoresis in 1 % agarose gel.
Aliquots (5 μl) of 16S polymerase chain reaction (PCR) products were individually digested with restriction endonucleases MspI, HinfI, HaeIII, and TaqI (MBI, Fermentas; 5 U per reaction) as specified by the manufacturer in a total volume of 10 μl. The fragments were separated by electrophoresis in 2.0 % agarose at 120 V for 3 h. Gels were stained with ethidium bromide and photographed with Gel Document System. The band patterns of RFLP analysis were converted into a two-dimensional binary matrix through a binary scoring system and the similarities were evaluated by simple matching coefficient. Dendrograms were constructed from the distance matrix by UPGMA clustering algorithm in NTSYS-pc (2.1) program (Rohlf 1990). Isolates sharing the same RFLP patterns were classified as an rRNA genotype.
16S rRNA, recA, nifH, and nodC gene sequencing
Based on the symbiotic effectiveness of the isolates and on the PCR-RFLP, 15 representative isolates were chosen for sequencing of the 16S rRNA, recA, nifH, and nodC genes. The 16S rRNA gene was amplified as for 16S RFLP. The recA gene was amplified as described using primers recAF1 and recAR1, with the exception of SCAU209 and SCAU211 whose recA was amplified using recAF2 and recAR2 (Gaunt et al. 2001). The nifH gene was amplified using primers nifHF and nifHI as described (Laguerre et al. 2001). Five forward primers, nodCF, nodCFu, nodCF2, nodCF4, and nodCFn, and the reverse primer nodCR were used for amplification and sequencing of the nodC gene (Laguerre et al. 2001). The amplification products of 16S rRNA, recA and nodC genes were directly purified and sequenced (Invitrogen, Shanghai, China). The purified nifH gene fragments were cloned into a pMD19-T vector (Takara, Dalian, China) transformed into competent Escherichia coli DH5α cells and examined with M13 primers according to the manufacturer’s instructions before sequencing at Invitrogen, Shanghai, China. The quality of the sequences was verified by sequencing both strands.
The sequences of representative isolates and the reference sequences were aligned using ClustalW as implemented in Mega4.0 (Tamura et al. 2007). Phylogenetic trees were constructed using the Neighbor-joining method in MEGA program version 4.0 (Tamura et al. 2007), and confidence level determined by 1,000 replicates bootstrapping. The percentage similarity of the genes was estimated using DNAman (version 6.0; Woffelman 1994). Isolates were assigned to genus or species with the highest sequence similarity with the type strain. The sequences obtained in this study were submitted to the GenBank sequence database under the accession numbers JQ362359-JQ362389, JF330102-JF330106, JF330109-JF330110, HQ538614, HQ538616, HQ538622, HQ538618-HQ538620, and JX073914-JX073927.
Results
Symbiotic effectiveness of L. leucocephala rhizobia
When searching for effective strains for inoculating L. leucocephala in Panxi, altogether 45 isolates were obtained from L. leucocephala root nodules. From them, 41 isolates that were able to nodulate L. leucocephala plants were called Leucaena rhizobia. Four isolates were not considered as rhizobia because they did not form nodules on their original host (Table 2). However, one of the four non-nodulating isolates, isolate SCAUnon4 increased plant shoot dry weight significantly (Table 2), indicating that some endophytic non-nodulating bacteria are able to promote plant growth.
Plant shoot dry weight was consistent with plant shoot fresh weight (data not shown), and correlation (r = 0.96**) was significant at the 0.01 level (two-tailed). Only 11 of 41 Leucaena rhizobia promoted the growth of L. leucocephala significantly (56.1–75.6 mg of shoot dry weight/plant) as compared to the uninoculated control (45.8 mg of shoot dry weight/plant; Table 2). Hence, the 11 isolates were good candidates and among them isolates SCAU223 and SCAU221 the best candidates for inoculating L. leucocephala in this area (Table 2). Over two thirds of the tested rhizobial isolates were ineffective in symbiotic nitrogen fixation. Strikingly, 10 Leucaena rhizobial isolates could be considered as parasitic because plant shoot dry weight decreased significantly (22.1–35.3 mg of shoot dry weight/plant) as compared to the uninoculated control. On the average, both the effective nitrogen fixing and parasitic isolates formed from two to 17 nodules per plant (Table 2).
RFLP analyses of the 16S rRNA gene
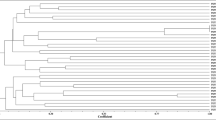
To preliminary estimate the taxonomic positions of isolates, their 16S rRNA RFLP restriction profiles were compared with the restriction profiles of 14 reference strains that represented different rhizobial species and genera (Table 2). A dendrogram was constructed by UPGMA algorithm based on the combined restriction patterns (Fig. 1). This analysis resulted in six, seven, five, and five different restriction patterns of 41 Leucaena rhizobia for HaeIII, HinfI, MspI, and TaqI, respectively. Restriction patterns were arbitrarily identified by letters and used to assign the isolates into RFLP groups (Table 2). For the 41 isolates analyzed, 10 distinct genotypes were distinguished. Genotype I comprising 28 isolates was the most frequent 16S rRNA genotype (Table 2). According to the 16S RFLP analysis, Leucaena rhizobia were distributed into four genera. Ensifer (32 isolates) was the dominant genus. The remaining isolates were assigned as Rhizobium (four isolates), Mesorhizobium (three isolates), and Bradyrhizobium (two isolates).
UPGMA dendrogram constructed based on the RFLP analysis of PCR-amplified 16S rRNA genes of the isolates and reference strains using four restriction endonucleases (MspI, HinfI, HaeIII, and TaqI). Genotype I to X (indicated on the right) are described in Table 2. Numbers in parentheses show the number of isolates with the same 16S rRNA genotype
Phylogenetic analyses of 16S rRNA and recA genes
To get more detailed information on the diversity of the L. leucocephala rhizobia, two housekeeping genes (16S rRNA and recA) of 15 representative isolates were sequenced. Amplification of the 16S rRNA and recA genes was successful, resulting in a single fragment of about 1,500 and 500 bp, respectively (data not shown).
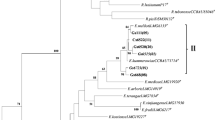
The assignments of the 15 representative isolates as defined by 16S rRNA gene were consistent with 16S RFLP at the genus level (Figs. 1 and 2). In the phylogenetic analyses of the 16S RNA gene, the 15 representative strains were grouped into four main groups corresponding to Ensifer, Rhizobium, Mesorhizobium, and Bradyrhizobium or six clades in detail (Fig. 2). Most isolates (11/15) were assigned to genus Ensifer, and were distributed over three distinct clades. Four isolates clustered together with Ensifer xinjiangense LMG 17930T, E. fredii USDA205T, Ensifer americanum CFNEI 156T, and Ensifer kummerowiae CCBAU 71714T, and represented a distinct phylogenetic branch (clade E1). Six isolates clustered together with Ensifer mexicanus HAMBI 2910T with similarities ranging from 99.1 to 99.7 % (clade E2; Table 1, Fig. 2). SCAU224 clustered with Ensifer meliloti LMG 6133T, Ensifer medicae USDA 1037T, and Ensifer arboris HAMBI 1552T (clade E3). In the Mesorhizobium group, SCAU202A and SCAU215 each shared 99.6 % similarity and clustered together with M. plurifarium LMG 11892T (Table 1, Fig. 2; clade M). SCAU209 formed a distinct clade (clade R) with Rhizobium huautlense LMG18254T (99.9 % sequence similarity) in the Rhizobium group. SCAU211 clustered together with Bradyrhizobium japonicum USDA6T (99.6 % sequence similarity) in Bradyrhizobium group (clade B; Table 1, Fig. 2).
Phylogenetic tree of 16S rRNA gene sequences showing the relationships between the representative isolates and the reference strains for defined rhizobial species. Bootstrap values >50 % are given at the branching points. The scale bar indicates the number of substitutions per site. Sequence accession numbers are given in parentheses. The representative isolates are shown in bold. ↑*, ↓*, and ns indicates an increase, a decrease or a non-significant difference in plant shoot dry weight according to the LSD test (at *p = 0.05 level), respectively
In the phylogenetic analysis of the recA gene, the 15 representative isolates were grouped into four main groups Ensifer, Rhizobium, Mesorhizobium, and Bradyrhizobium or six clades in detail as in the 16S rRNA tree (Figs. 2 and 3). The recA phylogeny was consistent with the 16S rRNA phylogeny at the genus and species level in Rhizobium and Mesorhizobium (Figs. 2 and 3, Table 1). However, the topology of the recA tree was different from that of the 16S rRNA tree for the Ensifer group. In the recA clade E1, three isolates had 100 % recA sequence similarity and formed a sister clade to E. xinjiangense LMG 17930T and E. fredii USDA205T. SCAU218, a member of 16S rRNA clade E1, had 100 % recA sequence similarity with the six isolates from 16S rRNA clade E2. Together, they formed recA clade E2 with the closest type strain E. americanum LMG CFNEI 156T. SCAU224 had 92.2 % similarity with the closest type strain E. americanum species and was the most divergent branch in the genus. Like in the 16S rRNA tree, SCAU202A and SCAU215 clustered together with M. plurifarium, and SCAU209 with R. huautlense LMG18254T (Fig. 3). SCAU211 formed a clade (clade B) with the nearest type strain B. liaoningense USDA 3622T (95.1 %; Table 1, Fig. 3).
Phylogenetic tree of recA gene sequences showing the relationships between the representative isolates and the reference strains for defined rhizobial species. Bootstrap values >50 % are given at the branching points. The scale bar indicates the number of substitutions per site. Sequence accession numbers are given in parentheses. The representative isolates are shown in bold. ↑*, ↓*, and ns indicates an increase, a decrease or a non-significant difference in plant shoot dry weight according to the LSD test (at *p = 0.05 level), respectively
Phylogenetic analysis of nifH and nodC genes
To characterize the in vivo observed symbiotic properties on gene level, two symbiotic genes (nifH and nodC) of 15 representative isolates were sequenced. The nifH and nodC genes from SCAU209, assigned as R. huautlense in the 16S rRNA and recA phylogeny, failed to be amplified. For the other 14 representative isolates, amplification of the nodC using the primer pair nodCF/nodCR resulted in a single fragment of about 900 bp (data not shown). The 737 bp sequences of the nifH gene were obtained after removal of the primer sequences. The topology of the nodC phylogenetic tree (Fig. 4) was very similar to that of the nifH tree (Fig. 5). As in the 16S rRNA and recA trees, the isolates grouped with Ensifer, Mesorhizobium, and Bradyrhizobium in both the nifH and nodC trees. Even though the 11 Ensifer isolates formed three clades in both the 16S rRNA and recA trees, they formed only one clade in both the nifH and nodC trees, suggesting that they harbored very similar or same symbiotic genes.
Phylogenetic tree of nifH gene sequences showing the relationships between the representative isolates and the reference strains for defined rhizobial species. Bootstrap values >50 % are given at the branching points. The scale bar indicates the number of substitutions per site. Sequence accession numbers are given in parentheses. The representative isolates are shown in bold. ↑*, ↓*, and ns indicates an increase, a decrease or a non-significant difference in plant shoot dry weight according to the LSD test (at *p = 0.05 level), respectively
Phylogenetic tree of nodC gene sequences showing the relationships between the representative isolates and the reference strains for defined rhizobial species. Bootstrap values >50 % are given at the branching points. The scale bar indicates the number of substitutions per site. Sequence accession numbers are given in parentheses. The representative isolates are shown in bold. ↑*, ↓*, and ns indicates an increase, a decrease or a nonsignificant difference in plant shoot dry weight according to the LSD test (at *p = 0.05 level), respectively
Discussion
In search of rhizobial strains for inoculating L. leucocephala, we investigated the symbiotic efficiency of isolates from root nodules of L. leucocephala growing in the arid–hot river valley area in Panxi, China. Due to the rhizobial diversity, effective nitrogen-fixing strains are isolated even from soils with no history of cultivation of the host plant (Cardoso et al. 2012). L. leucocephala is considered as a promiscuous host (Wang et al. 1999, 2006). It is nodulated with both fast- and slow-growing rhizobia, yet again ineffective nodulation is common (Bala and Giller 2001). We found that only 11 out of 41 L. leucocephala rhizobia were efficient in nitrogen fixation (Figs. 1, 2, and 3; Table 2), making them appropriate candidates to be used as inoculants in this area. While the majority of the isolates were ineffective in symbiotic nitrogen fixation, 10 isolates could be considered as parasites with a growth slowing effect on the host. However, it cannot be ruled out that the isolates inefficient in nitrogen fixation do not bring about other benefits, e.g., they might give resistance to plant diseases.
To find explanations for the diversity in nitrogen fixation efficiency, we assessed the genetic diversity of the isolates. The molecular systematics of rhizobia is mainly based on sequence data for the 16S rRNA gene (Binde et al. 2009). Because of the high level of sequence conservation and occasional lateral gene transfer and recombination, it is important to determine the phylogenies of other housekeeping genes, e.g., recA, for rhizobial species assignments (van Berkum et al. 2003). In general, the recA phylogeny is consistent with the 16S rRNA gene phylogeny (Zhao et al. 2010) as it was in this study at genus and species level in Rhizobium and Mesorhizobium (Figs. 2 and 3; Table 1). However, the closest matches of the 16S rRNA and recA of all the Ensifer isolates and the Bradyrhizobium isolate were discordant, indicating that the Ensifer and Bradyrhizobium isolates might represent new species and that lateral transfer of housekeeping genes might occur in interspecies of Ensifer and Bradyrhizobium.
The nodC gene is unique and symbiosis-specific to rhizobia and essential for the synthesis of the Nod factor (Debellé et al. 2001). The nifH gene is essential for nitrogen fixation by rhizobia inside the nodule but also for diazotrophs that fix nitrogen in the free-living state (Raymond et al. 2004). Little polymorphism was found between nifH and nodC genes sequence of L. leucocephala Ensifer symbionts (Figs. 4 and 5). In contrast to the promiscuity observed as a wide species distribution of the isolates, L. leucocephala was nodulated by strains that shared common symbiosis genes. This might reflect host specificity towards this plant by strains that produce essentially identical Nod factors, as has been suggested in other studies (Wei et al. 2009).
L. leucocephala originates from Mexico, and it is most effectively nodulated by Ensifer strains (Bala and Giller 2006). We found that 10 out of 11 effective nitrogen-fixing isolates from Panxi belonged to the genus Ensifer (Figs. 1, 2, and 3; Table 2). The symbiotic efficiency of rhizobia is not connected to the chromosomal background of the symbiont, and strains carrying a similar nifH may show totally different efficiency in nitrogen fixation (Mnasri et al. 2009). These phenomena were observed in our study, too (Table 2; Figs. 2, 3, and 4). For instance, even though the 16S RNA, recA, nifH, and nodC sequences of the six strains in rRNA clade E2 were similar, only two of them were significantly effective in fixing nitrogen. Similarly, only one out of the three strains in rRNA clade E1 was effective. According to a model proposed by Li et al. (2012), nodulating strains are divided into true and sporadic symbionts. Almost all strains of a true symbiont species nodulate effectively, while within the sporadic symbiont species there is large variation in effectiveness and nodulation ability of the strains. Contrary to the promiscuous hosts Glycyrrhiza and Phaseolus vulgaris (Li et al. 2012; Aserse et al. 2012), L. leucocephala seems to be nodulated by sporadic symbionts only. Likewise, genetically close symbionts of Acacia seyal show variation in effectiveness (Diouf et al. 2010), calling upon the questions if these trees should be considered as truly promiscuous hosts and whether this a characteristic feature of other promiscuous tropical leguminous trees, too.
Introduced legumes are likely to carry their symbionts into the new environment where the symbionts interact with native rhizobia (Weir et al. 2004). In this study, the 16S rRNA gene sequences of all Ensifer isolates had higher similarity to strains not isolated from L. leucocephala than to E. adhaerens LMG21331T, a Mexican L. leucocephala isolate (Fig. 2). Despite the 16S rRNA and recA diversity, the Ensifer isolates harbored similar symbiosis genes (nifH and nodC; Figs. 4 and 5; Table 1). The incongruence indicates possible horizontal transfer of symbiotic genes as previously reported for the Mesorhizobium, Ensifer, Rhizobium, and Bradyrhizobium species (Laranjo et al. 2008; Li et al. 2011; Aserse et al. 2012) and implies that the symbiotic genes which introduced L. leucocephala symbionts carried were transferred into indigenous Ensifer strains.
Invasive Mimosa spp., mimosoid legume woody shrubs and herbs originating in South America, are preferentially nodulated by β-rhizobia (Liu et al. 2012). Liu et al. (2012) isolated Mimosa spp. β-rhizobia in China and found that all the isolates fixed nitrogen effectively, contrary to the small percentage of efficient isolates among the L. leucocephala nodulating rhizobia in this study. The Chinese Mimosa spp. symbionts are closely related to South American strains likely to being brought to China with the host, i.e., to strains with a ca. 50 m years of coexistence with the host (Liu et al. 2012), whereas the L. leucocephala isolates seemed to be indigeneous rhizobia that had received their symbiotic genes laterally during the last 30 years, a relatively short time frame for the partners to adapt to each other. The differences in nitrogen fixation efficiency between the seemingly similar isolates tell us that the isolates possessed diversity outside the characterized gene range. It may be asked whether a promiscuous host like L. leucocephala ever poses such strict requirements for the nodulating strains that an uninoculated plant would host only effective isolates. If the answer is no, it gives further motivation to search for effective inoculant strains.
The species distribution of nodulating strains is different at different sites (Bala and Giller 2006; Liu et al. 2012). Wang et al. (2006) isolated the same rhizobial genera from L. leucocephala growing in subtropical China. However, most of the strains isolated by Wang et al. (2006) were Mesorhizobium spp., whereas in Panxi L. leucocephala had a preference for Ensifer strains. Since soil conditions affect the nodulating species distribution (Bala and Giller 2006; Liu et al. 2012), the dominance of Ensifer strains in arid environments (Romdhane et al. 2006; Benata et al. 2008; Gehlot et al. 2012) and the difference in species distribution between Wang et al. (2006) and our results might be due to the soil properties in the sampling areas. This suggests that when choosing inoculants, strains adapted to local soil conditions should be preferred.
In conclusion, we showed that in the arid–hot river valley area of Panxi L. leucocephala is nodulated by indigineous rhizobia belonging to genera Ensifer, Rhizobium, Mesorhizobium, and Bradyrhizobium. Molecular characterization of the 41 isolates revealed that Ensifer was the predominant genus associated with L. leucocephala in this area. Because the Ensifer isolates had undergone gene recombination, their precise species assignment was not possible. This ambiguity can be clarified by analyzing more housekeeping genes. The search for effectively nitrogen-fixing inoculant strains is justified since most of the indigenous L. leucocephala nodulating isolates were ineffective; furthermore, one fourth of the isolates had a growth slowing effect on the host. As in earlier studies on rhizobia isolated from various legumes (Cardoso et al. 2012; Mnasri et al. 2009), the genetic background of the isolates was diverse and not connected to their symbiotic performance, necessitating the use of traditional plant growth tests in screening effective inoculants.
References
Aserse AA, Räsänen LA, Assefa F, Hailemariam A, Lindström K (2012) Phylogeny and genetic diversity of native rhizobia nodulating common bean (Phaseolus vulgaris L.) in Ethiopia. Syst Appl Microbiol 35:120–131. doi:10.1016/j.syapm.2011.11.005
Bala A, Giller KE (2001) Symbiotic specificity of tropical tree rhizobia for host legumes. New Phytol 149:495–507. doi:10.1046/j.1469-8137.2001.00059.x
Bala A, Giller KE (2006) Relationships between rhizobial diversity and host legume nodulation and nitrogen fixation in tropical ecosystems. Nutr Cycl Agroecosys 76:319–330. doi:10.1007/s10705-005-2003-y
Benata H, Mohammed O, Noureddine B, Abdelbasset B, Abdelmoumen H, Muresu R, Squartini A, El Idrissi MM (2008) Diversity of bacteria that nodulate Prosopis juliflora in the eastern area of Morocco. Syst Appl Microbiol 31:378–386. doi:10.1016/j.syapm.2008.08.002
Binde DR, Menna P, Bangel EV, Barcellos FG, Hungria M (2009) rep-PCR fingerprinting and taxonomy based on the sequencing of the 16S rRNA gene of 54 elite commercial rhizobial strains. Appl Microbiol Biotechnol 83:897–908. doi:10.1007/s00253-009-1927-6
Cardoso JD, Hungria M, Andrade DS (2012) Polyphasic approach for the characterization of rhizobial symbionts effective in fixing N2 with common bean (Phaseolus vulgaris L.). Appl Microbiol Biotechnol 93:2035–2049. doi:10.1007/s00253-011-3708-2
Chen Q, Zhang XP, Terefework Z, Kaifalainen S, Li DY, Lindström K (2003) Diversity and compatibility of peanut (Arachis hypogaea L.) bradyrhizobia and their host plants. Plant Soil 255:605–617. doi:10.1023/A:1026039503225
Debellé F, Moulin L, Mangin B, Dénarié J, Boivin C (2001) nod genes and Nod signals and the evolution of the Rhizobium legume symbiosis. Acta Biochim Pol 48:359–365
Diouf D, Fall D, Chaintreuil C, Ba AT, Dreyfus B, Neyra M, Ndoye I, Moulin L (2010) Phylogenetic analyses of symbiotic genes and characterization of functional traits of Mesorhizobium spp. strains associated with the promiscuous species Acacia seyal Del. J Appl Microbiol 108:818–830. doi:10.1111/j.1365-2672.2009.04500.x
Gaunt MW, Tumer SL, Rigottier-Gois L, Lloyd-MaeGlip SA, Young JPW (2001) Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int J Syst Evol Microbiol 51(6):2037–2048. doi:10.1099/00207713-51-6-2037
Gehlot HS, Panwar D, Tak N, Tak A, Sankhla IS, Poonar N, Parihar R, Shekhawat NS, Kumar M, Tiwari R, Ardley J, James EK, Sprent JI (2012) Nodulation of legumes from the Thar desert of India and molecular characterization of their rhizobia. Plant Soil. doi:10.1007/s11104-012-1143-5
Howieson JG, Yates RJ, Foster KJ, Real D, Besier RB (2008) Prospects for the future use of legumes. In: Dilworth MJ, James EK, Sprent JI, Newton WE (eds) Nitrogen fixing leguminous symbioses. Springer, Dordrecht, pp 363–393
Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N (2001) Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiol 147:981–993
Laranjo M, Alexandre A, Rivas R, Velázquenz E, Young JPW, Oliveira S (2008) Chickpea rhizobia symbiosis genes are highly conserved across multiple Mesorhizobium species. FEMS Microbiol Ecol 66(2):391–400. doi:10.1111/j.1574-6941.2008.00584.x
Li QF, Zhang XP, Zou L, Chen Q, Fewer DP, Lindström K (2009) Horizontal gene transfer and recombination shape mesorhizobial populations in the gene center of the host plants Astragalus luteolus and Astragalus ernestii in Sichuan, China. FEMS Microbiol Ecol 70:227–235. doi:10.1111/j.1574-6941.2009.00776.x
Li QQ, Wang ET, Zhang YZ, Zhang YM, Tian CF, Sui XH, Chen YW, Chen YX (2011) Diversity and biogeography of rhizobia isolated from root nodules of Glycine max grown in Hebei Province, China. Microb Ecol 61:917–931. doi:10.1007/s00248-011-9820-0
Li L, Sinkko H, Montonen L, Wei G, Lindström K, Räsänen LA (2012) Biogeography of symbiotic and other endophytic bacteria isolated from medicinal Glycyrrhiza species in China. FEMS Microbiol Ecol 79:46–68. doi:10.1111/j.1574-6941.2011.01198.x
Liu XY, Wei S, Wang F, James EK, Guo XY, Zagar C, Xia LG, Dong X, Wang YP (2012) Burkholderia and Cupriavidus spp. are the preferred symbionts of Mimosa spp. in Southern China. FEMS Microbiol Ecol 80:417–426. doi:10.1111/j.1574-6941.2012.01310.x
Mnasri B, Badri Y, Saїdi S, de Lajudie P, Mhamdi R (2009) Symbiotic diversity of Ensifer meliloti strains recovered from various legume species in Tunisia. Syst Appl Microbiol 32:583–592. doi:10.1016/j.syapm.2009.07.007
Moawad H, Bohlool BB (1984) Competition among Rhizobium spp. for nodulation of Leucaena leucocephala in two tropical soils. Appl Environ Microbiol 48(1):5–9
Peoples MB, Herridge DF, Ladha JK (1995) Biological nitrogen fixation: an efficient source of nitrogen for sustainable agricultural production? Plant Soil 174:3–28. doi:10.1007/BF00032239
Raymond J, Siefert JL, Staples CR, Blankenship RE (2004) The natural history of nitrogen fixation. Mol Biol Evol 21(3):541–554. doi:10.1093/molbev/msh047
Rohlf FJ (1990) NTSYS-pc, Numerical taxonomy and multivariate analysis system, version 2.02. Exeter Software, Setauket, NY
Romdhane SB, Nasr H, Samba-Mbaye R, Neyra M, Ghorbal MH, De Lajudie P (2006) Genetic diversity of Acacia tortilis ssp. raddiana rhizobia in Tunisia assessed by 16S and 16S-23S rDNA genes analysis. J Appl Microbiol 100:436–445. doi:10.1111/j.1365-2672.2005.02765.x
Somasegaran P, Martin RB (1986) Symbiotic characteristics and Rhizobium requirements of a Leucaena leucocephala × Leucaena diversifolia hybrid and its parental genotypes. Appl Environ Microbiol 52(6):1422–1424
Sprent JI, Gehlot HS (2010) Nodulated legumes in arid and semi-arid environments: are they important? Plant Ecol Divers 3:211–219. doi:10.1080/17550874.2010.538740
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi:10.1093/molbev/msm092
van Berkum P, Terefework Z, Paulin L, Suomalainen S, Lindström K, Eardly BD (2003) Discordant phylogenies within the rrn loci of rhizobia. J Bacteriol 185:2988–2998. doi:10.1128/JB.185.10.2988-2998.2003
Vincent JM (1970) A manual for the practical study of root-nodule bacteria. Blackwell, Oxford, UK
Wang ET, Martínez-Romero J, Martínez-Romero E (1999) Genetic diversity of rhizobia from Leucaena leucocephala nodules in Mexican soils. Mol Ecol 8:711–724. doi:10.1046/j.1365-294X.1999.00608.x
Wang FQ, Wang ET, Zhang YF, Chen WX (2006) Characterization of rhizobia isolated from Albizia spp. in comparison with microsymbionts of Acacia spp. and Leucaena leucocephala grown in China. Syst Appl Microbiol 29:502–517. doi:10.1016/j.syapm.2005.12.010
Wei G, Chen W, Zhu W, Chen C, Young JPW, Bontemps C (2009) Invasive Robinia pseudoacacia in China is nodulated by Mesorhizobium and Sinorhizobium species that share similar nodulation genes with native American symbionts. FEMS Microbiol Ecol 68:320–328. doi:10.1111/j.1574-6941.2009.00673.x
Weir BS, Turner SJ, Silvester WB, Park DC, Young JM (2004) Unexpectedly diverse Mesorhizobium strains and Rhizobium leguminosarum nodulate native legume genera of New Zealand, while introduced legume weeds are nodulated by Bradyrhizobium species. Appl Environ Microbiol 70:5980–5987. doi:10.1128/AEM.70.10.5980-5987.2004
Woffelman C (1994) DNAMAN for Windows, version 2.6. Lynon Biosoft, Institute of Molecular Plant Sciences. Leiden University, the Netherlands
Zhao L, Deng Z, Yang W, Yang W, Cao Y, Wang E, Wei G (2010) Diverse rhizobia associated with Sophora alopecuroides grown in different regions of Loess Plateau in China. Syst Appl Microbiol 33:468–477. doi:10.1016/j.syapm.2010.08.004
Zhao K, Penttinen P, Guan T, Xiao J, Chen Q, Xu J, Lindström K, Zhang L, Zhang X, Strobel GA (2011) The diversity and anti-microbial activity of endophytic actinomycetes isolated from medicinal plants in Panxi plateau, China. Curr Microbiol 62:182–190. doi:10.1007/s00284-010-9685-3
Acknowledgments
This research was supported by National Natural Science Foundation of China (31070004), Educational Commission of Sichuan Province of China (09ZB047) and the Specialized Research Found for the Doctoral Program of Higher Education of China (20060626006).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, K.W., Penttinen, P., Chen, Y.X. et al. Symbiotic efficiency and phylogeny of the rhizobia isolated from Leucaena leucocephala in arid–hot river valley area in Panxi, Sichuan, China. Appl Microbiol Biotechnol 97, 783–793 (2013). https://doi.org/10.1007/s00253-012-4246-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4246-2