Abstract
Traditional Chinese medicinal plants are sources of biologically active compounds, providing raw material for pharmaceutical, cosmetic and fragrance industries. The endophytes of medicinal plants participate in biochemical pathways and produce analogous or novel bioactive compounds. Panxi plateau in South-west Sichuan in China with its unique geographical and climatological characteristics is a habitat of a great variety of medicinal plants. In this study, 560 endophytic actinomycetes were isolated from 26 medicinal plant species in Panxi plateau. 60 isolates were selected for 16S rDNA-RFLP analysis and 14 representative strains were chosen for 16S rDNA sequencing. According to the phylogenetic analysis, seven isolates were Streptomyces sp., while the remainder belonged to genera Micromonospora, Oerskovia, Nonomuraea, Promicromonospora and Rhodococcus. Antimicrobial activity analysis combined with the results of amplifying genes coding for polyketide synthetase (PKS-I, PKS-II) and nonribosomal peptide synthetase (NRPS) showed that endophytic actinomycetes isolated from medicinal plants in Panxi plateau had broad-spectrum antimicrobial activity and potential natural product diversity, which further proved that endophytic actinomycetes are valuable reservoirs of novel bioactive compounds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Actinomycetes produce a diverse range of secondary metabolites including various antibiotics, antitumor and immunosuppressive agents and plant growth hormones [9, 26, 31] that play an important role in pharmaceutical industry. However, the dispersion of drug resistance in bacteria and the appearance of life-threatening viruses promote the search for new and useful metabolites. Endophytes are microorganisms that for the whole or part of their life history live inside plant tissues by symbiotic, parasitic or mutualistic means without causing immediately overt negative effects [28]. As a result of these long-held associations, endophytic microorganisms and plants have developed good information transfer [29]. Medicinal plants synthesize chemical substances, providing raw material for pharmaceutical, cosmetic and fragrance industries [15]. Endophytes of medicinal plants probably participate in metabolic pathways of medicinal plants and produce analogous or novel bioactive compounds, for example taxol [30]. Endophytic actinomycetes are considered as potential sources of novel bioactive compounds and various bioactive compounds have been isolated from them until now [4, 11, 32].
Species distribution and biological diversity are significantly influenced by ecological environment [10, 27]. In previous studies, the diversity and antibiotic activity of endophytic actinomycetes isolated from medicinal plants growing in tropical regions have been reported exclusively [2, 17, 23], while the medicinal plants from Panxi plateau, China, have not gained research attention before our study. The Panxi plateau is in the southwest of Sichuan province, located between the Qinghai-Tibet plateau, Yunnan-Guizhou plateau and Sichuan basin. The weather in this plateau is characterised with abundant rainfall and sunshine and with wet and dry seasons. Panxi plateau is the northernmost place with Southeast Asian tropical climate. Xerothermic climate is another characteristic of this area. The unique geographic position, favourable environment, complicated tectonic geographic condition, special climate and various agrotypes all contribute to the peculiar landscape and biodiversity in the area. About 2,400 species of wild medicinal plants are found in this area, with many of them having a long history of application by local people [14]. While the microbial diversity of Panxi plateau has been investigated by our laboratory in the past years [18], the actinomycetes and especially the endophytic actinomycetes of this plateau have not been studied until now.
In this study, we screened the endophytic actinomycetes of medicinal plants from Panxi plateau based on the medicinal function of the plants to identify their potential as biocontrol agents for phytopathogens and bacteria. Because polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) pathways synthesize a large number of biologically active molecules and exist widely in the genomes of actinomycetes, we used degenerate primers of PKS-I, PKS-II and NRPS genes to analyse the potential capacity of the endophytic actinomycetes to synthesize secondary metabolites. The phylogenetic diversity of isolates was assessed using 16S rDNA RFLP analysis and sequencing. This is the were first report on the diversity and bioactivity of endophytic actinomycetes of medicinal plants from Panxi plateau where we hope to bioprospect endophytic actinomycetes resources possessing potential to be applied in pharmacy and agriculture field.
Materials and Methods
Sampling of Wild Medicinal Plants
According to the ethnobotanical properties, healthy medicinal plant samples with antitumor, antibacterial and antiviral properties were collected from Panxi plateau in South-west Sichuan, China in July 2008. In order to ensure the endophytic nature of the isolates, the cut ends of root and stem samples were sealed with wax. All the samples were screened for endophytic actinomycetes within 48 h.
Surface Sterilization
The procedure of surface sterilization was from Johannes et al. [13] with some modifications. Samples were dried at room temperature for 48 h before being thoroughly washed with running tap water and graded by size and surface appearance. All the visibly damaged material was excluded. Samples were washed by sonification for 10 min at 150 w to dislodge soil and organic matter. The surface sterilization included the following steps: tissue pieces were rinsed in 0.1% Tween 20 for 30 s, sequentially immersed in 75% ethanol for 5 min and in 2% sodium hypochlorite for 5 min and rinsed with 10% NaHCO3 for 10 min to inhibit fungal growth. After each treatment, samples were rinsed three times in sterile water. Finally, the samples were thoroughly dried in a laminar flow chamber. To confirm the success of the sterilization process, aliquots of the sterile distilled water from the final rinse were inoculated on ISP2 medium plates. The surface-sterilized samples were aseptically dissected into small pieces, plated onto selective media and incubated for about 1 month at 28°C. The actinomycetes isolates were stored on ISP4 slope medium at 4°C.
Isolation Media
The seven isolation media, all supplemented with 1.8% agar, were as follows: E1: Low-Nutrient Mineral Salts-agar (LNMS) [6]; E2: Type Water Yeast Extract agar (TWYE) [8]; E3: Oatmeal agar [24]; E4: Humic-vitamin agar (HV) [3]; E5: Histidine-Raffinose agar [37]; E6: Modified Gause No.1 [22]; E7: Chitin agar [21]. To inhibit the growth of nonactinomycetes, the isolation media were supplemented with nalidixic acid and K2Cr2O7, both to a final concentration of 50 μg/ml.
Identification of Actinomycetes
The isolates were dereplicated by cultural and morphological characteristics, including morphology and colour of aerial mycelium, characteristics of colonies on the plate, spore mass colour, colour of diffusible pigments and sporophore and spore chain morphology. Visual observation using light microscopy and Gram-straining were performed for further identification [40].
16S rRNA Gene Restriction Fragment Length Polymorphism Analysis (ARDRA)
The total genomic DNA of isolates were extracted and amplified as described earlier [7]. The PCR products were, respectively, digested with restriction endonucleases Hae-III and Rsa-I. The 10-μl reactions included 1× buffer, 0.1-μg/μl BSA (Bovine Serum Albumin), 10-U Hae-III/Rsa-I (TaKaRa, China) and 3-μl PCR product. The digestions were performed at 37°C for 6 h. The digested fragments were separated by gel electrophoresis and visualised with a UV transilluminator. Isolates were grouped based on the combined amplified rDNA restriction analysis patterns using the approaches described [10].
16S rDNA Sequencing and Analysis
According to the results of 16S rRNA-RFLP, representative isolates were chosen for 16S rRNA gene sequencing carried by Shengong Biotechnology Ltd. (Shanghai, China). Sequences were compared with GenBank database using BlastN for searching the closest match sequence. The sequences were pairwise aligned using Clustal X [34]. A phylogenetic tree was constructed under the Kimura two-parameter model and bootstrap analyses with 1,000 resamplings were performed with MEGA 4.0. [33].
Evaluation of Antimicrobial Activity
11 indicator organisms (Verticillium dahliae Kleb. [SAUM 0110], Fusarium oxysporum f. sp. vasinfectum [SAUM 2312], Aspergillus niger [SAUM 1275], Fusarium oxysporum f. sp. Niveum [SAUM0674], Colletotrichum orbiculare [SAUM 0321], Fusarium graminearum [SAUM 2912], Exerohilum turcicum [SAUM 0218], Curvularia lunata [SAUM 1373], Botrytis cinerea [SAUM 1294], Staphylococcus aureus [ATCC 25923], Escherichia coli [ATCC 35218]) were used in the screening tests of antimicrobial activity as described earlier [35]. The antagonism was detected by formation of an inhibition zone in 3 days.
Detection and Analysis of PKS-I, PKS-II and NRPS
Three sets of degenerate primers PKS-I-A: 5′-GCS ATG GAY CCS CAR CAR CGS VT-3′, PKS-I–B: 5′-GTS CCS GTS CCR TGS SCY TCS AC-3′ [25]; PKS-II-A: 5′-TSG CST GCT TGG AYG CSA TC-3′, PKS-II-B: 5′-TGG AAN CCG CCG AAB CCG CT-3′ [20]; NRPS-A: 5′-GCS TAC SYS ATS TAC ACS TCS GG-3′, NRPS-B: 5′-SAS GTC VCC SGT SCG CTAS-3′ [1] targeting genes encoding polyketide synthases (PKS-I, PKS-II) and nonribosomal peptide synthetase (NRPS) were used to screen the biosynthetic potential of the isolates. Amplication and sequencing of the genes and sequence analysis were performed as described previously [12].
Results
In total, we sampled 26 species healthy medicinal plants. No colonies emerged from the final rinses of the sterilization procedure showing that the surface sterilization was effective and the subsequent isolates were endophytes. We isolated 560 strains that showed morphological characteristics of actinomycetes after 1–1.5-month incubation. Most of the strains were isolated from root (58.2%), secondly from stem (27.8%) and least from leaves (14%). According to their colour on media and sporing structure, we selected 60 isolates for further analysis.
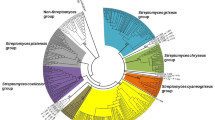
We digested the 16S rRNA genes of 60 isolates by restriction endonucleases Hae-III and Rsa-I and assigned the isolates to 12 groups at the similarity level of 90% (Table 1). The isolates formed two dominant groups of 19 and 31 strains, named groups B and D, respectively. Each of the other ten groups contained one strain only. There was no direct correlation between the isolation sources and the RFLP grouping. For example, strains SAUK6024, SAUK6102 and SAUK6117, isolated from Potentilla discolor, Pteris dactylina Hook and Senecio declouxii Dunn, respectively, were all in RFLP group D, and strains SAUK6012 and SAUK6015, isolated from Ainsliaea henryi Diels, were in groups F and H, respectively. Based on 16S rRNA-RFLP analysis and morphological characteristics, three strains were selected from RFLP group D and one from each of the other groups for sequence analyses. The 16S rRNA of those strains were compared with their closest relatives in GenBank and a phylogenetic tree of the 16S rRNA sequences were constructed by combining the sequences of the 14 representative strains with their closest relatives (Table 1; Fig. 1). The seven strains representing the 51 strains in RFLP-groups A–E were most similar to Streptomyces species. The other seven strains were members of the genera Micromonospora, Nonomuraea, Oerskovia, Promicromonospora and Rhodococcus.
Neighbor-joining phylogenetic tree analysis of 16S rDNA of endophytic actinomycete The numbers at the nodes indicate the levels of bootstrap support (%) based on 1000 resampled data sets. Only values above 50% are given. The scale bar corresponds to 0.02 substitutions per nucleotide position. Numbers in parentheses indicate accession numbers in Genbank. Bacillus pumilus DSM 27 T was used as an outgroup
In our study, 59 out of the 60 strains showed antagonistic activity against at least one of the 11 indicator organisms (Table 1). The 15 strains that inhibited the growth of S. aureus were considerable candidates for screening for new antibiotics (Table 1). Over half of the strains, altogether 38, inhibited the growth of at least five indicator organisms (Table 1). All of them belonged to the genus Streptomyces, as assigned by morphological characteristics and DNA analyses. The rare actinomycetes strains were more limited in their antimicrobial scope. However, SAUK6015, a strain most similar to Nonomuraea roseola, was a very potent inhibitor of pathogens Exerohilum turcicum and Curvularia lunata, as well as SAUK6030, a strain most similar to Micromonospora chokoriensis was a potent inhibitor of Curvularia lunata. Among the tested endophytic strains, there were isolates from 18 plant species with known antibacterial activity (Supplemental material 2), and at least one of the isolates from 9 out of 18 plant species showed antibacterial activity (Table 1).
In this study, we assessed the biosynthetic potential of the 60 isolates by amplifying the genes encoding polyketide synthases (PKS-I, PKS-II) and nonribosomal peptide synthetase (NRPS) using three sets of degenerate primers. 56 out of the 60 strains carried at least one of the biosynthetic enzyme genes (Table 1). The number of strains with PKS-I, PKS-II and NRPS gene fragments matching the anticipated length were 32 (53%), 49 (82%) and 32 (53%), respectively. Sequence analysis of the amplified products of PKS-I, PKS-II and NRPS genes of the 14 representative isolates confirmed that all the partial sequences encoded parts of biosynthetic enzymes. SAUK6023 carried all the biosynthetical genes, yet it did not have antimicrobial activity against any of tested pathogens. Four strains SAUK6113, SAUK6114, SAUK6013 and SAUK6033 showed antimicrobial activity even though we were not able to amplify any of the PKS or NPRS genes from them (Table 1).
Discussion
Plants growing in areas of great biodiversity are likely to house endophytes with equal or greater biodiversity. Plants with an ethnobotanical history and unusual longevity are expected to be more potent sources of endophytes producing active natural products than other plants [31]. 16S rRNA-RFLP technique is a powerful tool in grouping actinomycetes species and genera when studying diverse collections of microbial isolates [42]. However, there was considerable diversity within the RFLP groups B and D in respect of the presence of PKS-NPRS genes and antagonistic activity of the strains. As assessed by 16S rRNA sequencing, the genus composition of endophytic actinomycetes in our study was more diverse than that of actinomycetes isolated from rainforests [2, 17], wheat root [5], herbaceous and woody plants [35] and Neem tree [36]. Stating that the dominance of the genus Streptomyces noticed in previous reports and in our study reflects the real world situation requires further culture-independent confirmation [2, 17, 36]. We isolated actinomycetes of the genera Promicromonospora and Oerskovia, rare actinomycetes that were first time found to be endophytic by Qin et al. [23] who also studied Chinese medicinal plants. Unlike Qin et al., our isolation methods were not designed to focus on the rare actinomycetes. Since similar isolation media and methods have been applied in studies on other plants, the findings of Qin et al. and the results of our study indicate that medicinal plants in general and the medicinal plants of Panxi plateau in particular host a great diversity of endophytic actinomycetes resources. However, to get more species information of endophytic actinomycetes in Panxi plateau, a wider range of isolation methods should be employed in further studies.
Like in earlier studies, the strains we isolated possessed more antifungal than antibacterial properties [2, 36]. In earlier studies with actinomycetes isolated from nonmedicinal plant species, most of the strains have shown no antimicrobial activity and a small percentage a narrow antimicrobial activity [35]. In our study, only one out of the 60 strains was devoid of antagonistic activity. Streptomyces-like strain SAUK6233 isolated from Mosla dianthera Maxim, a plant with antibacterial activity [38], inhibited the growth of all tested indicator organisms, making it a good candidate for further studies. Also, strains SAUK6159, SAUK6024 and SAUK6185 isolated from Rhizoma arisaematis, Potentilla discolor Bge and Pratia nummularia, respectively, with antioxidant, antitumor, antibacterial and antipyrotic activities [19, 39, 41], which had antimicrobial activity against E. coli and Staphylococcus aureus, deserve further attention. The medicinal plants host numerous Streptomyces strains expected to produce a wide variety of bioactive metabolites [17]. The antagonistic activity seen in our study further suggests that endophytic Streptomyces hosted by medicinal plants are a key source of bioactive compounds. In addition, the rare endophytic actinomycetes offer a novel source for such compounds. Also, our results support the suggestion that the medicinal properties of medicinal plants could be at least partly due to the endophytes the plants host [31].
In recent years, scanning the genes encoding polyketide synthases and nonribosomal peptide synthetases that synthesize most of the biologically active polyketide and peptide compounds have been broadly applied in assessing the biosynthetic potential of culturable microorganisms and culture-independent samples [16, 20]. However, the results from earlier studies and our results suggest that for the culturable actinomycetes, the antimicrobial potential may only be assessed by screening of antimicrobial activity against the desired indicator organisms. The percentages of strains with PKS-I, PKS-II and NPRS and the percentage of strains showing antimicrobial activity do not correlate, and there is an ample amount of examples of strains possessing the functional genes showing no antimicrobial activity and vice versa [17, 23]. Here, the strain SAUK6023, most similar to Streptomyces aurantiacus, did not show any antimicrobial activity, yet we could amplify the PKS-NRPS genes from it. For this phenomenon there are several plausible explanations. Either the antimicrobials of SAUK6023 are effective against pathogens not tested by us or it produced them in quantities too low to inhibit pathogens. It may also be due to the fact that the PKS-NRPS genes of SAUK6023 were silent or the PKS-NRPS gene clusters of the strain were incomplete. Also, four strains showed antimicrobial activity but none of the functional genes was detected in them. Possibly the code genes of antimicrobial products were not PKS or NRPS synthetical genes, or it may be due to the primers amplifying PKS-NRPS genes were not suitable to these strains. Interestingly, SAUK6033 belonged to genus Promicromonospora, in previous report [23] also noted that from the members of that genus the PKS-I, PKS-II and NRPS genes cannot be amplified, yet they possess antimicrobial activity.
In conclusion, all but one of our Streptomyces isolates tested displayed antimicrobial activity, with one-third of the strains showing antibacterial activity. In addition to Streptomyces, the medicinal plants carried rare actinomycetes all of which displayed antifungal activity. Our survey suggested that medicinal plants are a potent source of endophytic actinomycetes with wide biological activity against pathogenic fungi as well as Gram-positive and Gram-negative bacteria.
References
Araujo WL, Marcon J, Maccheroni WJ et al (2002) Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol 68:4906–4914
Bascom-Slack CA, Ma C, Moore E et al (2009) Multiple, novel biologically active endophytic actinomycetes isolated from upper Amazonian rainforests. Microb Ecol 58:374–383
Cao L, Qiu Z, You J et al (2004) Isolation and characterization of endophytic Streptomyces strains from surface-sterilized tomato (Lycopersicon esculentum) roots. Lett Appl Microbiol 39:425–430
Castillo UF, Strobel GA, Mullenberg K et al (2006) Munumbicins E-4 and E-5: novel broad-spectrum antibiotics from Streptomyces NRRL 3052. FEMS Microbiol Lett 255:296–300
Coombs JT, Franco CM (2003) Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl Environ Microbiol 69:5603–5608
Crawford DL, Lynch JM, Whipps JM et al (1993) Isolation and characterization of actinomycete antagonists of a fungal root pathogen. Appl Environ Microbiol 59:3899–3905
Cui XL, Mao PH, Zeng M et al (2001) Streptimonospora salina gen nov., sp. nov., a new member of the family Nocardiopsaceae. Int J Syst Evol Microbiol 51:357–363
El-Shatoury S, Abdulla H, El-Karaaly O et al (2006) Bioactivites of endophytic actinomycetes from selected medicinal plants in the world heritage site of saint Katherine, Egypt. Int J Bot 2:307–312
Fiedler HP, Bruntner C, Riedlinger J et al (2008) Proximicin A, B and C, novel aminofuran antibiotic and anticancer compounds isolated from marine strains of the actinomycete Verrucosispora. J Antibiot (Tokyo) 61:158–163
Hou BC, Wang ET, Li Y et al (2009) Rhizobial resource associated with epidemic legumes in Tibet. Microb Ecol 57:69–81
Igarashia Y, Trujillob ME, Martínez-Molinab E et al (2007) Antitumor anthraquinones from an endophytic actinomycete Micromonospora lupini sp. nov. Bioorg Med Chem Lett 17:3702–3705
Jiang SM, Li X, Zhang L et al (2008) Culturable actinobacteria isolated from marine sponge Iotrochota sp. Mar Biol 153:945–952
Johannes H, Gabriele B, Barbara S (2006) Isolation procedures for endophytic microorganisms. Berlin, Springer, pp 299–305
Juan D, Gang F, Yi Z (2007) Current situation and outlook of development and use special biotic resources in Panxi region. Sci Technol Inf Panzh 32:8–11
Karthikeyan B, Jaleel CA, Lakshmanan GM et al (2008) Studies on rhizosphere microbial diversity of some commercially important medicinal plants. Colloids Surf B 62:143–145
Kelly GT, Sharma V, Watanabe CM (2008) An improved method for culturing Streptomyces sahachiroi: biosynthetic origin of the enol fragment of azinomycin B. Bioorg Chem 36:4–15
Li J, Zhao GZ, Chen HH et al (2008) Antitumour and antimicrobial activities of endophytic streptomycetes from pharmaceutical plants in rainforest. Lett Appl Microbiol 47:574–580
Li Q, Zhang X, Zou L et al (2009) Horizontal gene transfer and recombination shape mesorhizobial populations in the gene center of the host plants Astragalus luteolus and Astragalus ernestii in Sichuan, China. FEMS Microbiol Ecol 70:71–79
Matsuura E, Shimomura K, Ishimaru K (2000) Flavonoid and polyacetylene from Pratia nummularia. Nat Med 54:44
Minowa Y, Araki M, Kanehisa M (2007) Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes. J Mol Biol 368:1500–1617
Muscholl-Silberhorn A, Thiel V, Imhoff JF (2008) Abundance and bioactivity of cultured sponge-associated bacteria from the Mediterranean sea. Microb Ecol 55:94–106
Peng Y, Jiang Y, Duan S et al (2007) Selective isolation methods of rare actinomycetes. JYNS 29:86–89
Qin S, Li J, Chen HH et al (2009) Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl Environ Microbiol 75:6176–6186
Qiu D, Ruan J, Huang Y (2008) Selective isolation and rapid identification of members of the genus Micromonospora. Appl Environ Microbiol 74:5593–5597
Schirmer A, Gadkari R, Reeves CD et al (2005) Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl Environ Microbiol 71:4840–4849
Schulz D, Nachtigall J, Riedlinger J et al (2009) Piceamycin and its N-acetylcysteine adduct is produced by Streptomyces sp. GB 4-2. J Antibiot (Tokyo) 62:513–518
Sheil D (1999) Tropical forest diversity, environmental change and species augmentation: after the intermediate disturbance hypothesis. J Veg Sci 10:851–860
Stone JK, Charles WB, James FW (2000) An overview of endophytic microbes: endophytism defined. Dekker, New York, pp 3–5
Strobel GA (2003) Endophytes as sources of bioactive products. Microbes Infect 5:535–544
Strobel GA, Miller RV, Martinez-Miller C et al (1999) Cryptocandin, a potent antimycotic from the endophytic fungus Cryptosporiopsis cf quercina. Microbiology 145:1919–1926
Strobel G, Daisy B, Castillo U et al (2004) Natural products from endophytic microorganisms. J Nat Prod 67:257–268
Taechowisan T, Lu C, Shen Y et al (2005) Secondary metabolites from endophytic Streptomyces aureofaciens CMUAc130 and their antifungal activity. Microbiology 151:1691–1695
Tamura K, Dudley J, Nei M et al (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F et al (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Thongchai T, John FP, Saisamorn L (2003) Isolation of endophytic actinomycetes from selected plants and their antifungal activity. World J Microbiol Biotechnol 19:381–385
Verma VC, Gond SK, Kumar A et al (2009) Endophytic actinomycetes from Azadirachta indica A. Juss.: isolation, diversity, and anti-microbial activity. Microb Ecol 57:749–756
Vickers JC, Williams ST, Ross GW (1984) A taxonomic approach to selective isolation of streptomycetes from soil. Academic Press, Orlando, pp 553–561
Wu CP, Lin QQ, Chen MY et al (2006) Studies on the chemical components and antibacterial activity of volatile oil of mosla dianthera maxim. J Fujian Norm Univ (Nat Sci Ed) 22:101–106
Xue PF, Gang L, Wen ZZ et al (2005) Secondary metabolites from Potentilla multifida L. (Rosaceae). Biochem Syst Ecol 33:725–728
Yan XC (1992) Isolation and identification of actinomycete. Science Press, Beijing, pp 45–68
Yang GP, Qian JF (2009) Reviews of research on Arisaematis. Chin J Ethnomed Ethnopharm 18:19–21
Zhang HT, Zhang W, Jin Y et al (2008) A comparative study on the phylogenetic diversity of culturable actinobacteria isolated from five marine sponge species. Antonie Van Leeuwenhoek 93:241–248
Acknowledgements
This research was supported by the foundation of National Science Program of China (Project no. 30570062) and the fund of Sichuan provincal science and technology international cooperative project. We sincerely acknowledge kindness support from the Key Laboratory of Protection and Utilization of Biological Resources, Tarim university and Key Laboratory of Marine Biogenetic Resources, The Third Institute of Oceanography, State Oceanic Administration, when our laboratory was damaged in 2008 Sichuan earthquake.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, K., Penttinen, P., Guan, T. et al. The Diversity and Anti-Microbial Activity of Endophytic Actinomycetes Isolated from Medicinal Plants in Panxi Plateau, China. Curr Microbiol 62, 182–190 (2011). https://doi.org/10.1007/s00284-010-9685-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-010-9685-3