Abstract
In Streptomyces, carbon utilization is of significant importance for the expression of genes involved in morphological differentiation and antibiotic production. Glucose is mainly transported by GlcP, a membrane protein encoded by glcp. In Streptomyces coelicolor, this protein is encoded by sco5578. However, there is little information about the physiology of the GlcP promoter in Streptomyces. The aim of the present work was to clone and perform a functional analysis of the sp7066 promoter (ortholog of sco5578) from Streptomyces peucetius var. caesius. Hydrophobicity and cellular location analysis of the putative amino acid sequence of the cloned gene predicted SP7066 would be a membrane protein with a topology of six plus six transmembrane segments interrupted by a large cytoplasmic loop. In silico analysis of the upstream region of the sp7066 transcription initiation site predicted the sequences 5′-AGGAATAGT-3′ and 5′-TTGACT-3′ for regions -10 and -35 of sp7066 promoter. To reflect sp7066 expression, the promoter sequence was amplified, subcloned, and fused to the egfp reporter gene. Immunoblot analysis revealed that D-glucose and its analog 2-deoxyglucose were able to induce sp7066 expression. This effect was not modified by the presence of equimolar concentrations of D-galactose or N-acetylglucosamine. No expression of egfp was detected with the use of other carbon sources such as L-arabinose, D-fructose, and glycerol. Based on these analyses, we conclude that D-glucose is a preferred carbon source in S. peucetius var. caesius and that the sp7066 expression product, a putative non-PTS glucose permease, likely is a H+/symporter, localized to the membrane, and shows a strong specificity for D-glucose for inducing expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptomycetes plays a vital role in the environmental carbon cycle because they are capable of metabolizing complex compounds like chitin, cellulose, and lignin through a wide range of extracellular hydrolases [1]. This allows the availability of single carbon backbones for the growth and secondary metabolite production. In addition to extracellular enzymes, for many years, secondary metabolite production has been of great interest to industry and may be helpful in medicine [2]. The Streptomyces genus is one of the main players in the fermentative production of commercially used natural antibiotics [3, 4]. In their production, fermentation industries frequently use glucose as the main carbon source [4].
Despite the importance of glucose, not only for enzyme and antibiotic production, but also in morphogenesis and other regulatory mechanisms, little is known about its transport systems in the genus Streptomyces [5–8]. In the Streptomyces coelicolor genome, a glucose permease (GlcP) belonging to the major facilitator superfamily (MFS) of transporter and sharing 51 % identity with a glucose permease from Synechocystis sp. has been reported [9, 10]. This permease is encoded twice on the chromosome by the genes glcP1 and glcP2; the genes are situated at different loci. However, glucose is transported only by GlcP1 [11] and the sugar is intracellularly phosphorylated to glucose 6-phosphate by an ATP-glucose kinase (Glk). The biochemical characterization of heterologously expressed GlcP1 from S. coelicolor suggests that the permease constitutes a H+/symporter [11].
Streptomyces peucetius var. caesius is an actinomycete of industrial interest. It produces doxorubicin and other anthracyclines used for the treatment of different types of cancer [12]. A genome size of 8.7 Mb for S. peucetius ATCC 27952 has been reported [13]. In S. peucetius var. caesius, the GlcP ortholog was found to be sp7066. In a previous work, we demonstrated that glucose exerts carbon catabolite repression on anthracycline formation [7], but little is known about the glucose transporter or its regulation.
In this paper, we conducted an in silico analysis of the sequence of the S. peucetius var. caesius SP7066 predicted protein, in a first attempt to know its structure and cellular localization. Furthermore, the effect of various carbon sources on the expression of promoter GlcP through fusion with the GFP protein was examined.
Materials and Methods
Microorganisms and Culture Conditions
S. peucetius var. caesius and Escherichia coli strains are listed in Table 1. E. coli strains were cultivated in LB or Nutrient Agar supplemented (when required) with the appropriate antibiotics and incubated for 12 h at 37 °C. S. peucetius var. caesius was cultured in YM medium [14] or in nitrate-defined medium (NDM) [15] at 29 °C. When desired, NDM medium was supplemented with different carbon sources (100 mM). For growth, anthracycline formation, carbohydrate consumption, and sp7066 putative promoter expression, all liquid cultures were grown in baffled 250-mL Erlenmeyer flasks (Sigma-Aldrich, St. Louis, MO, USA) under continuous agitation (200 rpm). All microorganisms were maintained in 50 % glycerol at −70 °C.
Growth Determination
At desired intervals, 1-mL samples from the S. peucetius var. caesius cultures were washed with 0.75 % NaCl and read at 550 nm after homogenization.
DNA Isolation
Genomic DNA from S. peucetius var. caesius was isolated as previously described [16] from a pellet obtained by centrifugation (5000 × g, 10 min) of 50-mL cultures grown for 48 h in YM medium with 0.5 % glycine. For E. coli plasmid isolation, cultures were grown in LB media for 12 h and subjected to alkaline lysis [17].
Cloning of the GlcP Promoter from S. Peucetius var. Caesius
The putative GlcP promoter region was amplified by PCR from genomic DNA, using a set of specific primers: forward 5′-TGATCGTGATATCGCACCAGAAGCGGACCA-3′ and reverse: 3′-CGCAGCCGCCACCGGCATCTAGAGCCCGCT-5′. Restriction sites (underlined bold letters) were added for EcoRV and XbaI. The PCR product (350 bp) was ligated into the pGEM-T-easy vector (Promega, Madison, WI, USA), following the manufacturer’s instructions. Construction of the recombinant plasmid was verified by restriction mapping, PCR, and direct nucleotide sequencing of the PCR-amplified product. After sequencing, the resulting DNA fragments were analyzed by looking for consensus sequences of bacterial promoters using the Softberry program (http://www.softberry.com/).
Fusion of the Putative sp7066 Promoter Region to egfp in pIJ8660
The plasmid pIJ8660, containing the reporter gene egfp (encoding the green fluorescence protein, GFP) [18], was isolated from E. coli DH5-α grown in LB media. Simultaneously, pPGlcPvector was extracted and both plasmids (pIJ8660 and pPGlcP) were separately digested with EcoRV and XbaI (Fermentas, Lafayette, CO, USA). Digested plasmids (2 ng pIJ8660 and 4 ng of the putative sp7066 promoter region) were ligated with 1 U T4 DNA ligase (Promega Madison, WI, USA) following the manufacturer’s instructions. DH5-α cells were electrotransformed (25 μE, 1.25 kV, 200 ʊ) with the fusion product, and cells allowed to grow in LB media with apramycin (50 μg/mL). The presence of the putative 310 bp promoter was verified by restriction analysis. The plasmid pIJ8660 containing the sp7066 putative promoter was named pUNAM7066.
E. coli Transformation and Intergeneric Conjugation
Electrocompetent E. coli ET12567 (methylation-defective) cells were electroporated with pUNAM7066, according to Sambrook [17]. Transformed cells (100 μL) containing pUNAM7066 were used to conjugate mycelial fragments of S. peucetius var. caesius [19, 20] and selected in nalidixic acid (NA) (25 μg/mL) with apramycin (50 μg/mL). Ten exconjugants were randomly selected and grown in YMG with apramycin (50 μg/mL) to verify GFP expression. As a negative control, an exconjugant containing the empty plasmid pIJ8660 was also obtained.
GFP Expression and Visualization
For GFP expression assays, the S. peucetius var. caesius exconjugants 7066 and 8660 were cultured for 36 h in NDM medium supplemented with 100 mM arabinose. At this time, 100 mM of the carbon source to be tested was added and the expression was determined every 15 min for a period of 2 h. For this purpose, the cells were collected, washed with 0.75 % NaCl, and lysed by sonication at 50 W (5 times for 30 s with 60-s gaps) in buffer PED (75 mM KH2PO4, pH 7, DTT 2 mM, EDTA 1 mM). After centrifugation (10,000 × g, 4 °C), 50 μL supernatant samples containing 20 μg protein were electrophoresed (7.5 % PAGE), using standard procedures [23].
After SDS-PAGE, the proteins were electrotransferred to PVDF membranes (Merck-Millipore, Germany) previously activated in pure methanol for 20 s, using an electrophoretic transfer cell (Mini Trans-Blot, Bio-Rad 170-3930, Hercules, CA, USA). The transfer was carried out at 60 V over 90 min in 1× transfer buffer, which contained (L-1) 14.4 g glycine, 3.0 g Tris, and 100-mL methanol in distilled water. The membrane (with transferred proteins) was incubated at room temperature for 1 h and 10 mL in blocking buffer (3 % (w/v) Difco skim milk, 0.05 % (v/v) Tween 20 (Sigma) in TBST. After incubation, the membrane was washed twice (1 min each) with buffered Tween (BT) (0.05 % (v/v) Tween 20 (Sigma) in PBS and incubated again for 2 h at room temperature with monoclonal anti-GFP 3E6 (Invitrogen, Carlsbad, CA, USA; dilution 1:1000). After incubation, the membrane was washed again twice (1 min each) with buffered TBST and the secondary antibody was added and incubated with gentle agitation for 1 h at the same temperature. Finally, after washing twice with buffered TBST (5–10 min each) and three times with TBST under gentle agitation for 5–10 min each, the membrane was assessed using the commercial substrate NBT/BCIP. The reaction procedure was stopped with distilled water and the protein was visualized.
Determination of Total Protein and Residual Carbohydrate
For protein determination, samples were processed as reported previously [21], and assayed by the Bradford method (Bio-rad, Hercules, CA, USA), using bovine serum albumin as the standard. Remaining glucose in the culture medium was determined enzymatically, as described previously [7]. Arabinose was assayed by the phenol-sulfuric acid method [22].
Computer Analyses
Genome sequences of S. peucetius var. caesius were subjected to BLAST at the website of the National Center for Biotechnology Information at the National Institutes of Health, Bethesda, MD, USA. (www.ncbi.nlm.nih.gov). Alignments were corroborated with Clustal W. Additional bioinformatics predictions (e.g., secondary structures, promoter sequence) were performed using the corresponding algorithms available at www.softberry.com.
Results
In silico Analysis of Streptomyces Non-PTS Glucose Permease SP7066
Genome mining of microbial genome databases allowed the detection of various sp7066 orthologs. In fact, sp7066 exhibited 85 % identity to orthologs of Streptomyces griseus, S. coelicolor (GlcP1, SCO5578), S. lividans, and S. scabies, with over 99–100 % of the genome length covered [10, 24]. Based on the data available in our laboratory and BLAST analysis of the nucleotide sequence of the SCO7153 sugar transporter gene from S. coelicolor A3(2) (Gene ID: 1102591), and the genome sequence of S. peucetius, there was only one glcP gene copy in S. peucetius. Therefore, a glcP2 orthologue was not detected in this microorganism.
In agreement with MFS topology, an analysis of hydrophobicity and cellular location of the putative SP7066 amino acid sequence, performed with the support of TopPred (www.sbc.su.se), HMMTOP (www.enzim.hu/hmmtop/index), and ProtComp version 8 (www.softberry.com) programs, predicted the membrane to be the localization site of SP7066 and revealed a topology of six plus six transmembrane segments, interrupted by a large cytoplasmic loop (Fig. 1).
Cloning of the Putative sp7066 Promoter
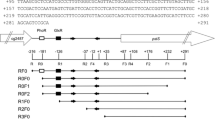
Once the sp7066 expression product was deemed related to the non-PTS glucose permease (GlcP), the gene was explored upstream of the initiation site (-10 and -35 consensus sequences) for the σ70 promoter. For this purpose, the BPROM (bacterial sigma70 promoter recognition program) was used, with has about 80 % accuracy and specificity for improved gene and operon prediction. This algorithm is best used in regions immediately upstream from the ORF start for improved gene and operon prediction in bacteria. Although the program do not consider the high GC content, from this search, the sequences 5′-AGGAATAGT-3′ for -10 and 5′-TTGACT-3′ for -35 were predicted to be sections of the putative promoter region (Fig. 2).
Analysis of the putative sp7066 promoter sequence. The -10 and -35 consensus regions are indicated (black down-pointing triangle) and highlighted with big bold letters. Additionally, the probably site for the transcription initiation site (black up-pointing triangle) and its reading direction (upward arrow with tip rightwards) are also indicated. Underlined sequences show the restriction sites for EcoRV and XbaI. The ribosome binding site is shown in bold letters, and the start codon is indicated by letters over a black arrow
The whole region was amplified by PCR, using S. peucetius var. caesius genomic DNA as template. The resulting product contained 216 bp upstream of the initiation site and 135 bp downstream of this site (Fig. 2). As shown in the figure, the upstream fragment included the putative -10 and -35 regions, sp7066 initiation site as well as the ribosomal binding site (RBS).
Fusion of the GlcP Promoter to the egfp Reporter Gene and its Transformation into S. peucetius var. caesius
The PCR product was subcloned into pGEM-T-easy vector, and after being released, it was fused to the reporter egfp gene. The new plasmid (pIJ8660 + sp7066 promoter) was named pUNAM7066. Several clones were verified using XbaI and EcoRV, releasing a 310 bp fragment, corresponding to the expected promoter size (Fig. 3).
Through intergeneric conjugation, pUNAM7066 was transferred to S. peucetius var. caesius. As a negative control, S. peucetius var. caesius was conjugated with pIJ8660 (empty vector) to verify that GFP responded to the sp7066 gene promoter.
Expression of egfp Gene Under the Control of the S. peucetius var. caesius GlcP Promoter
Colonies showing the best growth in liquid medium were selected for a rapid GFP expression assay. Two of three selected colonies grown for 8 h in NDM media with 100 mM glucose exhibited an intense band at ~30 kDa which corresponded to the GFP size (data not shown).
The S. peucetius var. caesius 7066 exconjugant containing pIJ7066 (with GlcP promoter fused to the egfp reporter) and its control strain containing pIJ8660 (without insert) were grown in NDM supplemented with 100 mM glucose and growth, carbohydrate consumption, and egfp expression were compared. As shown in Fig. 4a and b, both strains grew well in NDM and used glucose efficiently. Under these conditions, sp7066 expression was detected from 2 to 72-h fermentation in the 7066 exconjugant (Fig. 4c) and no expression was detected in the strain containing pIJ8660 (Fig. 4d). With regard to the good growth shown in glucose, the 7066 strain grew relatively well in 100 mM arabinose but this carbohydrate was not well used as more than 50 % remained in the culture medium at 48-h fermentation (Fig. 4a and b). As expected, under these conditions, expression of EGFP was not detected.
a Growth kinetics and b carbohydrate remaining in cultures of S. peucetius var. caesius EII strain containing pUNAM7066 (white circle, white square) and the control strain transformed with pIJ8660 (white triangle), grown in NDM supplemented with 100 mM glucose (white circle, white triangle) or arabinose (white square). (c, d and e): immunoblots showing egfp expression of EII and control strain grown in glucose and EII grown in arabinose, respectively
The 7066 exconjugant was exposed to pulses of different carbohydrates at 100 mM concentration and the egfp expression analyzed by Western blot. As shown in Fig. 4, in the presence of glucose or 2-deoxyglucose (2-DOG), sp7066 was detected at 15 min incubation and the signal remained for at least 2 h. Although the glucose analog 2-DOG lacks an -OH group at the C-2 position, it was able to elicit sp7066 expression. However, no signal was detected in the presence of fructose or in the absence of these carbohydrates, suggesting glucose as the sp7066 inducer.
In some streptomycetes, a sugar phosphotransferase transport system (PTS) and its activation by fructose and N-acetylglucosamine (NacGlc) have been described [25, 26]. In S. coelicolor, GlcNAc is transported via the phosphoenolpyruvate-dependent PTS to form N-acetylglucosamine-6-phosphate. This compound has been proposed as a preferred carbon and nitrogen source for S. coelicolor [27]. Thus, to explore the possible interference of NacGlc on the glucose induction of sp7066 expression, pulses of this carbohydrate were directed in the presence of two NacGlc concentrations. As shown in Fig. 4d and e, gfp was expressed by glucose pulses independently of the NacGlc concentration being used.
Discussion
In S. coelicolor genome, more than 50 carbohydrate permeases have been identified by bioinformatic tools. Among them, 14 belong to the ABC family, four to the phosphotransferase system (PTS), two copies are from a putative transporter of the major facilitator superfamily (MFS), one member belongs to the superfamily of sodium-dependent solute transporters (SSS), and one is a facilitator of the major intrinsic proteins (MIP) [10].
In the genus Streptomyces, glucose utilization depends on two key proteins, a glucose permease GlcP and two glucose kinases [28, 29]. S. coelicolor has two genes to transport glucose (glcp1 and glcp2), but only glcp1 (sco5578) seems to be functional [28]. However, there is little information about this process.
In the present study, sp7066 was identified as the putative region encoding GlcP in S. peucetius var. caesius, exhibiting 85 % identity to orthologs of Streptomyces griseus, S. coelicolor (sco5578), S. lividans, and S. scabies [10, 24]. Contrary to S. coelicolor, only one glcp gene copy was found in S. peucetius. Additionally, hydrophobicity and cellular location analysis of the putative SP7066 amino acid sequence predicted the membrane as the location site of SP7066, with a topology of six plus six transmembrane segments, interrupted by a large cytoplasmic loop characteristic of the MFS transport family proteins [30].
When sp7066 was explored upstream of the initiation site for its σ70 promoter, the sequences 5′-AGGAATAGT-3′ for -10 and 5′-TTGACT-3′ for -35 were predicted as the putative promoter region. This predicted sequences fitted well with the promoter sequences reported for glcp in S. coelicolor [31]. Comparison of these sequences with other streptomycetes promoter sequences was not possible since among the 139 Streptomyces promoters compiled, only about 20 % showed conserved core promoter sequences, similar to those recognized by Escherichia coli σ70 [32]. This may reflect the complex regulatory network existing in Streptomyces, which is reflected by the degeneracy of reported promoter sequences [33, 34].
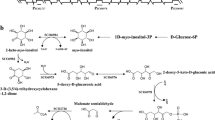
By fusing the GlcP promoter to the reporter egfp gene, it was found that induction of sp7066 responded to the presence of glucose and its non-metabolizable analog 2-DOG. This analog has been used widely in the selection of derepressed mutants with enhanced sugar catabolic enzyme production [21, 35]. A similar result with 2-DOG was reported for the chicken liver glucose transporter, Ch-Gt2 [36]. Other carbohydrates, including NacGlc and fructose, did not cause expression of sp7066, consistent with their transport by the PTS [25] (Fig. 5). On the contrary, it has been reported that GlcP from Zymomonas mobilis is able to transport fructose [37].
Effect of different carbon sources on the expression of sp7066 promoter. Immunoblots obtained from cultures exposed to the presence of a glucose, b 2-deoxyglucose, c D-fructose. d Glucose plus 10 and e glucose plus 100 mM N-acetylglucosamine. Carbohydrates were utilized at 100 mM concentration. Additionally, the cyclic structures of utilized carbohydrates are shown. Results are representative of two independent experiments
In conclusion, S. peucetius var. caesius showed a marked preference for D-glucose for growth. Glucose was incorporated by the sp7066 expression product (GlcP) and the transcription of this product was induced by D-glucose and its analog 2-deoxyglucose.
References
Hodgson, D. A. (2000). Advances in Microbial Physiology, 42, 47–238.
Paradkar, A., Trefzer, A., Chakraburtty, R., & Stassi, D. (2003). Critical Reviews in Biotechnology, 23, 1–27.
Watve, M. T. (2001). Archives of Microbiology, 176, 386–390.
Demain, A. L., & Sanchez, S. (2009). Journal of Antibiotics, 62, 5–16.
Pope, M. K., Green, B. D., & Westpheling, J. (1996). Molecular Microbiology, 19, 747–756.
van Wezel, G. P., White, J., Young, P., Postma, P. W., & Bibb, M. J. (1997). Molecular Microbiology, 23, 537–549.
Escalante, L., Ramos, I., Imriskova, I., Langley, E., & Sanchez, S. (1999). Applied Microbiology and Biotechnology, 52, 572–578.
Seo, J. W., Ohnishi, Y., Hirata, A., & Horinouchi, S. (2002). Journal of Bacteriology, 184, 91–103.
Zhang, C. C., Durand, M. C., Jeanjean, R., & Joset, F. (1989). Molecular Microbiology, 3, 1221–1229.
Bertram, R., Schlicht, M., Mahr, K., Nothaft, H., Saier, M. H., & Titgemeyer, F. (2004). Journal of Bacteriology, 186, 1362–1373.
van Wezel, G. P., Mahr, K., König, M., Traag, B. A., Pimentel-Schmitt, E. F., Willimek, A., & Titgemeyer, F. (2005). Molecular Microbiology, 55, 624–636.
Hutchinson, C. R., & Colombo, A. L. (1999). Journal of Industrial Microbiology & Biotechnology, 23, 647–652.
Parajuli, N., Basnet, D. B., Lee, H. C., Sohng, J. K., & Liou, K. (2004). Achives of Biochemistry and Biophysics, 425, 233–241.
Guzmán, S., Carmona, A., Escalante, L., Imriskova, I., López, R., Rodríguez-Sanoja, R., Ruiz, B., Servín-González, L., Sanchez, S. S., & Langley, E. (2005). Microbiology-SGM, 151, 1717–1723.
Dekleva, M. L., & Strohl, W. R. (1987). Canadian Journal of Microbiology, 33, 129–132.
Hopwood, D. A. (2006). Annual Review in Genetics, 40, 1–23.
Sambrook, J., Fritsch, E., & Maniatis, T. (2000). Molecular cloning: a laboratory manual (2nd ed.). Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
Sun, J., Kelemen, G. H., Fernandez, J. M., & Bibb, M. (1999). Microbiology, 145, 2221–2227.
Paranthaman, S., & Dharmalingam, K. (2003). Applied and Environmental Microbiology, 69, 84–91.
Blaesing, F., Agnes, M., Silke, V., Gunter, Z., Stefan, P., & Erich, L. (2005). Journal of Biotechnology, 120, 146–161.
Segura, D., González, R., Rodriguez, R., Sandoval, T., Escalante, L., & Sanchez, S. (1996). Asia Pacific Journal of Molecular Biology and Biotechnology, 4, 30–36.
Southgate, D. A. T. (1976). Applied Science Publishers. London: England.
Laemmli, U. K. (1970). Nature, 227, 680–685.
Zhang, Z., Schwartz, S., Wagner, L., & Miller, W. (2000). Journal of Computational Biology, 7, 203–214.
Nothaft, H., Dresel, D., Willimek, A., Mahr, K., Niederweis, M., & Titgemeyer, F. (2003). Journal of Bacteriology, 185, 7019–7023.
Wang, F., Xiao, X., Saito, A., & Schrempf, H. (2002). Molecular Genetics and Genomics, 268, 344–351.
Swiatek, M. A., Tenconi, E., Rigali, S., & van Wezel, G. P. (2012). Journal of Bacteriology, 194, 1136–1144.
van Wezel, G. P., Konig, M., Nothaft, H., Thomae, A. W., Bibb, M., & Titgemeyer, F. (2005). Journal of Molecular Microbiology and Biotechnology, 12, 67–74.
Ruiz-Villafán, B., Rodríguez-Sanoja, R., Aguilar-Osorio, G., Gosset, G., & Sanchez, S. (2014). Applied Microbiology and Biotechnology, 98, 6061–6071.
Pao, S. S., Paulsen, I. T., & Saier, M. H. (1998). Microbiology and Molecular Biology Reviews, 62, 1–34.
Pérez-Redondo, R., Santamarta, I., Bovenberg, R., Martín, J. F., & Liras, P. (2010). Microbiology, 156, 1527–1537.
Strohl, W. R. (1992). Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Research, 20, 961–974.
Kang, J. G., Hahn, M. Y., Ishihama, A., & Roe, J. H. (1997). Identification of sigma factors for growth phase-related promoter selectivity of RNA polymerases from Streptomyces coelicolor A3(2). Nucleic Acids Research, 25, 2566–2573.
Wang, W., Li, X., Wang, J., Xiang, S., Feng, X., & Yang, K. (2013). An engineered strong promoter for streptomycetes. Applied and Environmental Microbiology, 79(14), 4484–4492.
Xu, X.-J., Li, J.-H., ZHANG, D.-X., Chen, J., & Du, G.-H. (2009). Food Science, 30, 120–123.
Wang, F., Tsai, M., & Wang, C. (1994). Archives of Biochemistry and Biophysics, 310, 172–179.
Parker, C., Peekhaus, N., Zhang, X., & Conway, T. (1997). Applied and Environmental Microbiology, 63, 3519–3525.
Acknowledgments
We are indebted to Marco A. Ortíz for strain preservation studies. A. Romero was supported by a postgraduate fellowship from CONACyT, Mexico. Part of this work was supported by the grant IN208000 from PAPIIT, DGAPA, UNAM.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Romero, A., Ruiz, B., Sohng, J.K. et al. Functional Analysis of the GlcP Promoter in Streptomyces peucetius var. caesius . Appl Biochem Biotechnol 175, 3207–3217 (2015). https://doi.org/10.1007/s12010-015-1493-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1493-6