Abstract
Constructed wetlands (CWs) constitute an interesting alternative option to conventional systems for wastewater treatment. This technology is based on the utilization of the concerted activity of microorganisms for the removal of contaminants. Consequently, knowledge on the microbial assemblages dwelling CWs and the different environmental factors which can alter their activities is crucial for understanding their performance. In the last decades, the use of molecular techniques to characterize these communities and more recently, application of –omics tools, have broaden our view of microbial diversity and function in wastewater microbiology. In this manuscript, a review of the current knowledge on microbial diversity in CWs is offered, placing particular emphasis on the different molecular studies carried out in this field. The effect of environmental conditions, such as plant species, hydraulic design, water depth, organic carbon, temperature and substrate type on prokaryotic communities has been carefully revised, and the different studies highlight the importance of these factors in carbon, nitrogen and sulfur cycles. Overall, the novel –omics open a new horizon to study the diversity and ecophysiology of microbial assemblages and their interactions in CWs, particularly for those microorganisms belonging to the rare biosphere not detectable with conventional molecular techniques.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Constructed wetlands (CWs) are engineered systems formed by lagoons, shallow ponds or channels planted with aquatic vegetation (macrophytes), which constitute a decentralized solution to treat wastewater in small communities and dwellings within a controlled environment. They remove pollutants using natural processes such as volatilization, sedimentation, sorption, photodegradation, plant uptake, transpiration flux, accretion and chemical procedures [35], as well as microbial-mediated transformations [35, 62]. They are extensively used in many countries all over the world, such as Australia, Canada, Korea, Sweden, Taiwan, Thailand, the UK and the USA among others [69] due to their efficiency and low cost in terms of installation, maintenance and operation. Several types of constructed wetlands can be distinguished depending on the criteria like hydrology (free water surface flow and subsurface flow), type of macrophytes (free floating, emerged, submerged) and flow path (vertical or horizontal) [68]. Originally, CWs were used to treat domestic wastewaters, but now their applications comprise industrial and agricultural wastewaters, as well as landfill leachate, mine drainage or stormwater runoff [68]. Besides, the array of pollutants removed by them is large and include not only soluble labile forms of organic carbon but also hydrocarbons like toluene [44, 45, 47], herbicides [20], pesticides [46, 70], antimicrobial agents as triclosan [78] or pharmaceutical residues like ibuprofen [40] among others. Furthermore, removal of pathogenic microorganisms has also been a primary target for CWs [48, 67]. Recently, an emerging technology has arisen combining the use of microbial fuel cells (MFCs) and CWs, since redox conditions necessary for MFCs can be found naturally in CWs [18]; the combination of these two biological systems is aimed to produce electrical power while improving the wastewater treatment capacity. Also, other combined wetland-bioelectrochemical systems offer the simultaneous production of electrical current and H2O2 for disinfection of wastewater [5].
Many studies have highlighted the importance of microorganisms in CWs, since different processes and reactions are based on their concerted activity [1, 23, 28, 65]. For example, they constitute key participants in nitrogen and metal removal, as well as in sulfide oxidation, being dynamic players in carbon, nitrogen and sulfur cycling [23]. Thus, active populations of biofilms can be found associated to plant’s roots or to the filter bed material, and they are assumed to be responsible for the degradation performance of CWs [60]. Hence, the rhizosphere of CWs is considered to be the most important compartment where many organic compounds are transformed by microorganisms, due to the release of oxygen and organic exudates by the plant. As a consequence, knowledge of the diversity of the communities involved in these biological processes and their link to certain environmental factors is crucial in order to understand the functioning of these systems.
Several decades ago, before the advent of cultivation-independent techniques, it was arduous to ascertain the role of the key drivers of wastewater treatment processes, but nowadays, the scenario has overturned with the development of molecular methods, which circumvent cultivation and its inherent selectivity. Moreover, the current –omics era has extended our understanding on the diversity of these complex systems. In this manuscript, a review of the most recent findings concerning the diversity of prokaryotic microorganisms in CWs is presented, highlighting their influence in some relevant microbial processes and their correlation with environmental variables.
Molecular Approaches to Study Diversity in the –omics Era
In the last decades, a great number of studies dealing with molecular characterization of diversity in constructed wetlands have been published. The first culture independent approaches employed fingerprinting techniques such as denaturing gradient gel electrophoresis (DGGE) or terminal fragment length polymorphism (TRFLP) of PCR-amplified 16S ribosomal RNA (rRNA) gene fragments (Fig. 1), which allowed to obtain an overall pattern of the microbial community from different samples [8, 11, 12, 19, 31, 60]. DGGE further allows the assessment of population diversity by subsequent sequencing of bands. These methodologies are still currently used in a number of studies for assessing diversity in CWs [2, 20, 48, 54], although they present a bias associated to PCR amplification and primer sets (Table 1) and, overall, do produce a limited number of reads (bands or peaks) compared to tag sequencing of 16S rRNA, which prevent the former from being the future molecular methods in modern microbial ecology laboratories.
However, despite the disadvantages resulting from its use, PCR is still utilized to assess the distribution of certain functional genes and shed light on the diversity of main players in certain microbial processes, i.e. the nitrogen cycle [39]. By contrast, fluorescent in situ hybridization (FISH) with specific probes overcomes the problem of PCR bias, although the technique is limited in practice by the number of probes that are presently known (Table 1).
Currently, the rise of the –omics era has changed the present perception of diversity in wastewater treatment systems at an inconceivable scale [24]. The development of high-throughput sequencing (HTS) methods, like Illumina or 454-pyrosequencing among others, has proved increasingly valuable because they are able to produce for the first time millions of sequence reads both easy and inexpensive, while allowing large scale analysis of microbial assemblages and contributing to a better overview of the functioning of populations. Thus, metagenomic characterization of microbial communities (DNA sequencing of all genes) in CWs can provide information about the phylogenetic diversity and the genomic potential of these systems without bias, avoiding isolation of microorganisms and PCR (Table 1). On the other hand, metatranscriptomics (RNA sequencing) allows the analysis of gene expression, while metaproteomics results in the analysis of proteins, the processes in which they are involved and the reconstruction of the subsequent metabolic routes (Fig. 1).
The number of studies involving the different –omics in CWs is still scarce. In 2014, Bai et al. [7] used metagenomic analysis to explore the composition of the microbial community in the rhizosphere soil of a constructed wetland; by means of function annotation of metagenomic data, they were able to identify several biodegradation pathways associated with 14 xenobiotic compounds, as well as different genes involved in the nitrogen cycle and related to nitrogen fixation, nitrification and denitrification, along with genes involved in Fe and Mn transformations. At the same time, they could characterize the most abundant groups in this system, providing new insights in microbial mediated bioremediation of contaminants in CWs. Also, a metagenomic approach using 454-pyrosequencing was provided by Zhong et al. [79], who studied the composition of the bacterial community in horizontal subsurface flow CWs with and without front aeration, and shed light to the genera of microorganisms that could be involved in the nitrogen cycle concerning processes like nitrification, autotrophic and heterotrophic denitrification, or anaerobic ammonium oxidation; they found as well different dominant families of bacteria depending on the CW and related to sulfur cycle.
On the other hand, a metaproteomic approach was used by Lünsmann et al. [45] to investigate the microbial processes in the rhizosphere of a CW treating toluene-contaminated water, observing a stable aerobic toluene turnover by Burkholderiales during day and night, and an upregulation of polyhydroxyalkanoate synthesis during the day. Functional assignment of the proteins showed that main functions were cell envelope biogenesis, lipid and amino acid metabolism, as well as proteins belonging to the translation apparatus. In general, their study presented information about the microbial transformations occurring in the rhizosphere of CWs.
However, although the progress of–omics becomes unstoppable, culture-dependent methods should not be mistreated. Isolates are a source for laboratory models in physiological studies and are relevant to determine the role of newly discovered genes and functions through the abovementioned molecular methods.
Microbial Diversity in Constructed Wetlands
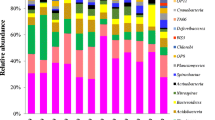
Fingerprinting methods based on 16S rRNA gene analysis such as DGGE or TRFLP have been used in the last decades to characterize the prokaryotic dynamics and structure in CWs (Table 2). More recently, these methods, which provide a rough estimate of the dominant bacterial composition, have been combined with HTS techniques, so that a more detailed picture of the communities which develop in these systems was obtained (Table 2). On the other hand, other authors have used only HTS to determine diversity through the analysis of the 16S rRNA gene or through a metagenomic approach (Table 3). Tables 2 and 3 include the most remarkable phylogenetic groups found in CWs using these techniques and their relative abundances when available.
In virtually all these studies, independently of the origin of influent waters, there is a permanent dominance of the phylum Proteobacteria, including members of the classes Alpha-, Beta-, Gamma-, Delta- or Epsilonproteobacteria, which are in different proportions depending on the conditions. This phylum is present in different types of samples from CWs, such as soil or sediment [4, 41, 78], rhizosphere [7, 44], manure influent [32], lagoon water [20, 32], inlet and outlet water [1] or biofilms from substrate particles [28, 79]. Within this group, the proteobacterial ammonia oxidizers (AOB), which include the Betaproteobacteria Nitrosomonas and Nitrosospira, and the Gammaproteobacteria Nitrosococcus (except Nitrosococcus mobilis, a betaproteobacterium), can carry out aerobic or anaerobic ammonia oxidation [57], while nitrite oxidizing bacteria (NOB) perform the conversion of nitrite to nitrate and include members of the genera Nitrobacter (Alphaproteobacteria) and Nitrococcus (Gammaproteobacteria) among others. Ammonium removal by nitrification is a well-documented process in different types of CWs [23], where sequence DGGE analyses have revealed the presence of two dominant AOB lineages corresponding in fact to Nitrosomonas europaea/Nitrosococcus mobilis and Nitrosospira [31, 63, 64]. Recent metagenomic analyses confirmed the presence of a complete set of nitrification genes (amo, hao) in both water and soil samples of a CW treating urban water associated to Nitrosomonas eutropha [7], suggesting that this bacterium might be responsible for nitrification activity in the rhizosphere soil. Some genera of Proteobacteria are also responsible for denitrification in CWs, a process spread in diverse phylogenetic groups, which involves the transformation of nitrate or nitrite into gaseous products (N2 and N2O). They are mainly facultative anaerobic chemoheterotrophic bacteria such as Pseudomonas, Aeromonas or Vibrio among others, although some AOB like Nitrosomonas sp. can also denitrify when grown under oxygen limitation [9].
Moreover, some pathogenic bacteria belong to the phylum Proteobacteria, and actually, proteobacterial indicator microorganisms (Enterobacteriaceae) are routinely used to investigate microbial removal in CWs [66]. In fact, CWs offer a combination of mechanisms suitable to remove pathogenic microorganisms [48]. Proteobacteria are also involved in sulfur cycling, an important process in CWs due to the considerable amounts of sulfur present in wastewater. The Deltaproteobacteria sulfate reducing bacteria (SRB) can oxidize organic substrates anaerobically and utilize sulfate as a terminal electron acceptor in CWs, although some works have showed that they can persist in oxic conditions [10, 15, 29, 59]. Different genera, such as Desulfobacter, Desulfovibrio, Desulfobulbus and Desulfobacterium have been detected in CWs by means of culture dependent and independent techniques [36, 43, 56]. Interestingly, SRB metabolism is closely related to precipitation of metal sulfides and they are considered of great interest in the bioremediation of metal-polluted environments [37]. In fact, different microbial processes can affect the mobility, toxicity and bioavailability of metals, particularly activities like biosorption, precipitation, redox tranformation, methylation, or microbe-plant interactions [37].
Another group usually abundant in CWs is the phylum Bacteroidetes, formed by Gram-negative chemoheterotrophic bacteria widely distributed in different environments, such as soil, sediments, seawater, aside from the guts and skin of animals. It is known by its ability to degrade complex organic matter, and it is suggested to be strongly involved in the degradation of aromatic compounds used in the post-tanning process [17], or in denitrification processes [2] from different CWs. A list of some genera of Bacteroidetes found in CWs is shown in Table 4.
Other prokaryotes, including Firmicutes, Actinobacteria, Acidobacteria, Planctomycetes, Chloroflexi, Sinergistetes, Deferribacteres, Nitrospirae, Cyanobacteria, Verrumicrobia or Archaea, can be found in different types of CWs (Tables 2 and 3), but usually their contribution is low, in most cases being part of the rare biosphere, that is, those species that are represented by a low number of individuals [52, 53]. Also, in these surveys, a great proportion of sequences can be hardly affliliated to a specific group, and remain as “uncultured” in the databases, highlighting the fact that further efforts to characterize these microorganisms are essential in order to explore the diversity and functional abilities of microbial communities in CWs.
Correlating Microbial Communities With Environmental Parameters in CWs
There is a large set of environmental conditions that can affect microbial physiology and community structure in CWs, and different publications have dealt with this issue in the last years. Microbial communities can be altered with different environmental factors, such as plant species, hydraulic design, availability of organic matter, temperature, dissolved oxygen (DO) or substrate type among others [6, 27, 58, 66, 73]. As a consequence, investigation of fluctuations in microbial populations can shed light to the response of a CW to variations in operational and environmental settings.
Several studies have highlighted the importance of vegetation in shaping microbial communities in CWs both directly through the comparison of populations in planted and unplanted CWs by molecular methods [6, 58, 60, 66] or indirectly by reporting loss of performance in nitrogen processing for unplanted wetlands [33]. The wetland rhizosphere seems to play an important role in the establishment of oxic-anoxic interfaces where aerobic and anaerobic microorganisms can operate in close vicinity, enhancing elemental cycling and therefore stimulating microbial activity [25, 50]. Plants can provide carbon compounds through the excretion of exudates and can release oxygen via the aerenchyma system; oxygen is consumed during the night and redox diurnal fluctuations are established, allowing the coexistence of oxic and anoxic conditions and probably contributing to niche differentiation in the rhizosphere [50]. Other effects of vegetation in CWs include the role of plants in the control of hydraulic efficiency due to modification of internal flow patterns, the influence on water temperature and energy balance, the decrease of reaeration due to wind blocking and shading or the provision of surfaces (litter and stems) to which microbes attach [33].
Gagnon et al. [25] showed that microbial density and activity increased when plants were present, and these values presented differences within plant species, being higher values associated with Phalaris arundinacea compared with Typha angustifolia and Phragmites australis. The effect of plant species in microbial profiles was further investigated by Sidrach-Cardona et al. [58], who studied how the presence of T. angustifolia or P. australis could influence the microbial composition of the rhizoplane, gravel biofilm and interstitial water in CWs; they concluded that plants exerted an effect on all the microbial communities examined within the mesocosms, confirming their influence in interstitial water and gravel-associated bacteria far beyond their roots. In the case of Phragmites, differences between environments were higher than in Typha. Faulwetter et al. [22], by means of quantitative PCR and DGGE, examined also effluent and biofilms associated to gravel and roots and observed a clear influence of plants on abundance and diversity of SRB and AOB populations. On the other hand, Arroyo et al. [6] obtained evidence of a most effective metal removal of P. australis compared to Typha latifolia, highlighting that those CWs planted with P. australis presented the highest diversity and richness, measured with PCR-amplified 16S rRNA clone libraries. Concerning the nitrogen cycle, Kadlec [33] reported a reduced ability of unvegetated CWs to process nitrogen compared to planted ones; nitrification and denitrification rates decreased probably due to a reduction in the surface area for microbe attachment. Differences between planted and unplanted CWs by means of quantitative PCR and HTS were also observed by Wang et al. [71], who concluded that both microbial community structure and abundance were positively affected by plants, being bacteria more abundant in the rhizoplane than in sand.
However, other works showed the lack of a strong vegetation effect on the structure of microbial communities [3, 8, 30], and the study of specific groups of organisms like metanotrophic bacteria by quantitative PCR revealed no significant differences in the quantities of Type I or Type II metanotrophic communities between planted and unplanted wetlands [16]. Sidrach-Cardona et al. [58] suggested that inadequate redox conditions for root development could have conducted to the absence of plant effects on microbial communities, a fact that is in accordance with Ravit et al.’s [55] proposals, which point out to less evident effects of plants on microbial communities in anthropogenically disturbed wetlands. In fact, Ahn et al. [3] indicated that the absence of results could be explained by the low growth of plants and sampling anomalies in their CWs. In any case, it is not clear the effect of plants in these systems, making evident that little is known about the mechanisms performing the shape of microbial communities and their functioning in different compartments of CWs.
A second important attribute in CWs is hydraulic design, since different operation and configuration strategies can lead to variations in performance due to changes in redox conditions. Conventional CWs are categorized according to water flow in free water (FW) surface and subsurface flow (SSF) wetlands. FW systems have a better exchange of oxygen than SSF CWs and therefore, have a higher redox and a higher advantage in nitrate removal, while SSF wetlands might provide a more anoxic environment and might have more surface area for microbial attachment and for growth of denitrifying bacteria [42]. Other innovative designs have emerged in the last years, such as circular-flow corridor CWs, towery hybrid CWs, baffled subsurface-flow CWs, or microbial fuel cells CWs [72], which try to intensify performance for wastewater treatment, for instance enhancing nitrogen removal.
Several works have examined the influence of flow on community composition. Thus, Lin et al. [42] studied nitrate removal in CWs with the same size but diverse flow patterns (FW surface vs SSF) and concluded that, in general, there was no significant difference between both types of CWs. In contrast, Arroyo et al. [6] observed that, besides plant presence, the type of flow seemed to be the main design parameter to increase efficiency to remove zinc and arsenic, being the Proteobacteria phylum, characterized by 16S rRNA gene amplification and cloning, the most abundant group under all conditions. Zinc and arsenic removal was better in FW flow CWs, although in the case of arsenic, those CWs planted with P. australis offered higher rates; in the case of zinc, no interaction between vegetation and flow was observed. Sidrach-Cardona et al. [58] also demonstrated that hydraulic configuration, together with the type of vegetation, was important in shaping microbial communities in FW and SSF CWs. However, the choice of the type of treatment wetland is not clear, and apart from the nature of contaminant to be removed, aspects like size, cost, operability, health issues and ancillary benefits should be considered [34].
In addition to flow, another key design factor for CWs is water depth, which in SSF systems has been normally set at 0.60 m to maximize growth and plant effects [14]. Water depth influences the oxygen transfer coefficient from atmosphere to water, and governs the water volume in contact with macrophytes, leading to a compartmentalization of metabolic processes within the wetland, but although it is an important feature to consider, it has not been included in performance models as a variable. Truu et al. [65] concluded that depth gradient was the most important spatial pattern to determine microbial community structure in a horizontal SSF CW treating domestic wastewater. Besides, in the study from Morató et al. [48], it was shown that shallower horizontal SSF CWs (0.27 m) had higher redox values and consequently more oxidized conditions; accordingly, microbial communities were more related to aerobic microorganisms. Also, shallow wetlands combined with fine gravel granulometry were more effective when removing pathogens such as total coliforms and Escherichia coli, as well as Clostridium spores, a result partially explained by the fact that a larger volume of water is contacting macrophyte roots in this type of CWs. On the other hand, cluster analysis of the DGGE patterns separated samples according to water depth, showing that it was an important feature to consider for microbial community structure.
Bacterial populations can also be affected by environmental variables like organic carbon availability. The carbon source usually comes from wastewater, soil, or products excreted by plant roots. It has been considered that organic carbon can promote denitrification, but in those waters with high nitrate or carbon deficient content, such as waters from agricultural drainage or from secondary treated nitrified effluents, denitrification can be greatly sustained by root exudates [73, 77]. The alternative of adding external organic sources to enhance denitrification revealed that planted wetlands were more efficient in nitrate removal than unplanted ones after organic matter supply, although the loss of the added carbon by other microbial processes increased costs and limited its utilization in CWs [73]. Chang et al. [13] demonstrated that organic matter especially affected the index of fatty acids in CWs, used as biomarkers to characterize microbial community structure. On the other hand, Wu et al. [73] also reported that total organic carbon had a high impact on some denitrifiers. Nevertheless, other environmental factors, such as dissolved oxygen, can be crucial in microbial processes like nitrogen transformation, since nitrifying bacteria need oxygen, and it is recognized that nitrification is the first limiting step for nitrogen removal. Thus, artificial aeration can be an alternative to enhance nitrogen removal by rising AOB and NOB, while increasing the wetland performance [21]. Furthermore, other factors, such as phosphorus loading, appear to be critical in the treatment efficiency of CWs; high phosphorus loading seems to have a recognizable impact by decreasing and altering the microbial diversity associated to soil [3], and consequently, microbial communities may be used as potential indicators of phosphorus dynamics in CWs.
CW performance and their associated microbial communities are also sensitive to climate and seasonal changes. During winter, low temperatures have a negative impact in plant activity and microbial growth. Wu et al. [73] made an interesting revision about the different wetland configurations reported to intensify pollutant removal under low temperature conditions. Other works, however, highlighted the seasonal effect on microbial composition. Thus, Morató et al. [48] observed a reduction of E. coli in summer, while heterotrophic bacteria decreased in winter, and Paranychianakis et al. [51] described a seasonal shift in the composition of denitrifiers. Beyond the plant influence, Faulwetter et al. [22] described as well a seasonal effect on abundance and richness of SRB and AOB, while Wang et al. [71] concluded in their results that removal efficiencies of contaminants and microbial community structure differed in winter. DGGE analysis of samples from different lab-scale horizontal SSF CWs revealed also that seasonal variations had an effect on the diversity and composition of microbial populations [76]. Other studies reported seasonal changes in microbial communities with indirect methods, such as Song et al. [61], who used BIOLOG-GN plates to describe the metabolic features of airborne microbial communities in CWs and found different degrees of carbon compound utilization between seasons.
Substrate type can influence, too, the formation of microbial biofilms and the microbial assemblage structure in wetlands, as well as its system performance. In this case, molecular methods have also proved to be useful for the investigation of substrate effect on bacterial community composition. Thus, Vacca et al. [66], by using fingerprinting analyses, reported differences on rhizospheral microbial communities depending on filter material (expanded clay and sand). Likewise, Calheiros et al. [12], by numerical analysis of DGGE profiles, indicated that bacterial richness and community structure were affected by the type of substrate (different types of expanded clay aggregates and fine gravel) and the presence of T. latifolia. On the other hand, Guan et al. [27] utilized more recently a HTS technique (Illumina MiSeq) to investigate the influence of substrate type (sand, zeolite and gravel) on microbial communities, demonstrating a clear effect of soil material on the different bacterial groups detected. Some authors have used methodologies other than molecular techniques to investigate the link between microbial community structure and filter media, such as Li et al. [38], who compared the microbial community assemblages of eight types of substrate (zeolites, anthracite, shale, vermiculite, gravel, ceramic filter media, bio-ceramic and steel lag) by measuring the phospholipid fatty acid (PLFA) composition, concluding that PLFA profiles exhibited significant differences among the diverse materials. However, Gorra et al. [26] did not observe a clear effect of substrate (gravel, ceramic wastes, magnetite, zeolite and soil with marble sand) on AOB populations.
Future Directions
Introduction of –omics technologies in the study of CWs microbiology has certainly broadened our knowledge of the diversity in these systems at an unprecedented scale. However, the great amount of information obtained from these studies has not solved some questions about their functioning that still remain to be elucidated. For instance, which are the key species becoming crucial for the performance of a CW? A few metagenomic studies have provided insights about detailed species composition and potential functions in these systems (see Tables 2 and 3 and included references), but information is still scattered and scarce and does not usually provide knowledge of the core microbiome present in CWs. The core microbiome refers to the suite of microorganisms shared within similar environments, and determining its key components helps to understand how that particular system functions across complex microbial populations.
Although usually many authors try to link microbial composition and function, these two issues are not necessarily related, since in some cases, closely related microorganisms may have diferent metabolic features, while on the other hand, several physiological characteristics like the denitrification capacity are spread in different phylogenetic lineages. The different –omics can solve these obstacles by providing, through the combination of metagenomics and metatranscriptomics, the knowledge of microbial identification and gene expression under different conditions and time scales. Consequently, the future challenge to obtain a deeper understanding of CWs will be the integration of molecular tools with ecological theory, enhancing the development of mathematical models able to predict population structure and population functioning in variable environments, including their interactions and activities. In this sense, the use of model CWs can constitute a useful framework for formulating and testing hypotheses about optimization and design of wastewater treatment systems. As a remarkable example, in a context different from CWs but related to wastewater treatment, Nielsen et al. [49] developed a conceptual ecosystem model for describing microbial communities in enhanced biological phosphorus removal plants through the presentation of a core microbiome; the model offers knowledge of the dominant microorganisms present in these systems, with emphasis on key ecophysiological factors controlling their presence and activity and interspecies interactions. This kind of information lacks for CWs, and accordingly, more effort should be invested in this direction for the future management and design of these systems; some aspects commented above, such as the influence of plant presence, hydraulic design or seasonal variations on microbial communities could be contextualized and modelled to predict detailed metabolic patterns for relevant core species.
However, even though the different –omics offer an extraordinary potential to investigate structure-function relationships, they are not free of some limitations concerning mostly the interpretation of metagenomics and metatranscriptomics data, particularly due to the amazing number of sequences encoding hypothetical proteins generated using these tools, so that further efforts should be directed to improve their output.
Another question from ecological interest concerns all those microorganisms that are present in low numbers and were invisible until the development of HTS techniques. Are those microorganisms involved in the rare biosphere particularly active when favourable conditions appear and thus important for the system functioning? There are evidences from other environments that show that some members of the rare biosphere may actually become active under the appropriate conditions, although this is not always the case [53]. However, the rare biosphere is an unexplored field that has not been investigated in CWs, and the new sequencing technologies open the window to allow the experimental study of these rare bacteria.
References
Abed RMM, Al-Kharusi S, Prigent S, Headley T (2014) Diversity, distribution and hydrocarbon biodegradation capabilities of microbial communities in oil-contaminated cyanobacterial mats from a constructed wetland. Plos One 9(12), e114570. doi:10.1371/journal.pone.0114570
Adrados B, Sánchez O, Arias CA, Bécares E, Garrido L, Mas J, Brix H, Morató J (2014) Microbial communities from different types of natural wastewater treatment systems: vertical and horizontal flow constructed wetlands and biofilters. Water Res 55:304–312
Ahn C, Gillevet PM, Sikaroodi M (2007) Molecular characterization of microbial communities in treatment microcosm wetlands as influenced by macrophytes and phosphorus loading. Ecol Indic 7(4):852–863
Ansola G, Arroyo P, Sáenz de Miera LE (2014) Characterisation of the soil bacterial community structure and composition of natural and constructed wetlands. Sci Total Environ 473–474:63–71
Arends JBA, Van Denhouwe S, Verstraete W, Boon N, Rabaey K (2014) Enhanced disinfection of wastewater by combining wetland treatment with bioelectrochemical H2O2 production. Bioresour Technol 155:352–358
Arroyo P, Ansola G, de Miera LES (2013) Effects of substrate, vegetation and flow on arsenic and zinc removal efficiency and microbial diversity in constructed wetlands. Ecol Eng 51:95–103
Bai Y, Liang J, Liu R, Hu C, Qu J (2014) Metagenomic analysis reveals microbial diversity and function in the rhizosphere soil of a constructed wetland. Environ Technol 35(20):2521–2527
Baptista JC, Davenport RJ, Donnelly T, Curtis TP (2008) The microbial diversity of laboratory-scale wetlands appears to be randomly assembled. Water Res 42:3182–3190
Bock E, Schmidt I, Stüven R, Zart D (1995) Nitrogen loss caused by denitrifying Nitrosomonas cells using ammonium or hydrogen as electron donors and nitrite as electron acceptor. Arch Microbiol 163(1):16–20
Brune A, Frenzel P, Cypionka H (2000) Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol Rev 24(5):691–710
Calheiros CSC, Duque AF, Moura A, Henriques IS, Correia A, Rangel AOSS, Castro PML (2009) Changes in the bacterial community structure in two-stage constructed wetlands with different plants for industrial wastewater treatment. Bioresour Technol 100(13):3228–3235
Calheiros CSC, Duque AF, Moura A, Henriques IS, Correia A, Rangel AOSS, Castro PML (2009) Substrate effect on bacterial communities from constructed wetlands planted with Typha latifolia treating industrial wastewater. Ecol Eng 35(5):744–753
Chang J-J, Wu S-Q, Liang K, Wu Z, Liang W (2015) Comparative study of microbial community structure in integrated vertical-flow constructed wetlands for treatment of domestic and nitrified wastewaters. Environ Sci Pollut Res 22(5):3518–3527
Cooper PF, Job GD, Green MB, Shutes RBE (1996) Reed beds and constructed wetlands for wastewater treatment. WRc publications, Medmenham
Cypionka H (2000) Oxygen respiration by Desulfovibrio species. Annu Rev Microbiol 54:827–848
DeJournett TD, Arnold WA, LaPara TM (2007) The characterization and quantification of methanotrophic bacterial populations in constructed wetland sediments using PCR targeting 16S rRNA gene fragments. Appl Soil Ecol 35(3):648–659
Desta AF, Assefa F, Leta S, Stomeo F, Wamalwa M, Njahira M, Appolinaire D (2014) Microbial community structure and diversity in an integrated system of anaerobic-aerobic reactors and a constructed wetland for the treatment of tannery wastewater in Modjo, Ethiopia. Plos One 9(12), e115576. doi:10.1371/journal.pone.0115576
Doherty L, Zhao Y, Zhao X, Hu Y, Hao X, Xu L, Liu R (2015) A review of a recently emerged technology: constructed wetland—microbial fuel cells. Water Res 85:38–45
Dong X, Reddy GB (2010) Soil bacterial communities in constructed wetlands treated with swine wastewater using PCR-DGGE technique. Bioresour Technol 101(4):1175–1182
Elsayed OF, Maillard E, Vuilleumier S, Imfeld G (2014) Bacterial communities in batch and continuous-flow wetlands treating the herbicide S-metolachlor. Sci Total Environ 499(1):327–335
Fan J, Zhang J, Guo W, Liang S, Wu H (2016) Enhanced long-term organics and nitrogen removal and associated microbial community in intermittently aerated subsurface flow constructed wetlands. Bioresour Technol 214:871–875
Faulwetter JL, Burr MD, Parker AE, Stein OR, Camper AK (2013) Influence of season and plant species on the abundance and diversity of sulfate reducing bacteria and ammonia oxidizing bacteria in constructed wetland microcosms. Microb Ecol 65(1):111–127
Faulwetter JL, Gagnon V, Sundberg C, Chazarenc F, Burr MD, Brisson J, Camper AK, Stein OR (2009) Microbial processes influencing performance of treatment wetlands: a review. Ecol Eng 35(6):987–1004
Ferrera I, Sánchez O (2016) Insights into microbial diversity in wastewater treatment systems: how far have we come? Biotechnol Adv 34(5):790–802
Gagnon V, Chazarenc F, Comeau Y, Brisson J (2007) Influence of macrophyte species on microbial density and activity in constructed wetlands. Water Sci Technol 56(3):249–254
Gorra R, Coci M, Ambrosoli R, Laanbroek HJ (2007) Effects of substratum on the diversity and stability of ammonia-oxidizing communities in a constructed wetland used for wastewater treatment. J Appl Microbiol 103(5):1442–1452
Guan W, Yin M, He T, Xie S (2015) Influence of substrate type on microbial community structure in vertical-flow constructed wetland treating polluted river water. Environ Sci Pollut Res 22:16202–16209
He T, Guan W, Luan Z, Xie S (2016) Spatiotemporal variation of bacterial and archaeal communities in a pilot-scale constructed wetland for surface water treatment. Appl Microbiol Biotechnol 100:1479–1488
Holmer M, Storkholm P (2001) Sulfate reduction and sulphur cycling in lake sediments: a review. Freshw Biol 46(4):431–451
Iasur-Kruh L, Hadar Y, Milstein D, Gasith A, Minz D (2010) Microbial population and activity in wetland microcosms constructed for improving treated municipal wastewater. Microb Ecol 59(4):700–709
Ibekwe AM, Grieve CM, Lyon SR (2003) Characterization of microbial communities and composition in constructed dairy wetland wastewater effluent. Appl Environ Microbiol 69(9):5060–5069
Ibekwe AM, Ma J, Murinda S, Reddy GB (2016) Bacterial community dynamics in surface flow constructed wetlands for the treatment of swine waste. Sci Total Environ 544:68–76
Kadlec RH (2008) The effects of wetland vegetation and morphology on nitrogen processing. Ecol Eng 33(2):126–141
Kadlec RH (2009) Comparison of free water and horizontal subsurface treatment wetlands. Ecol Eng 35(2):159–174
Kadlec RH, Wallace S (2009) Treatment wetlands. CRC press, Boca Raton
King JK, Harmon SM, Fu TT, Gladden JB (2002) Mercury removal, methylmercury formation, and sulfate-reducing bacteria in wetland mesocosms. Chemosphere 46(6):859–870
Kosolapov DB, Kuschk P, Vainshtein MB, Vatsourina AV, Wießner A, Kästner M, Müller RA (2004) Microbial processes of heavy metal removal from carbon-deficient effluents in constructed wetlands. Eng Life Sci 4(5):403–411
Li M, Zhou Q, Tao M, Wang Y, Jiang L, Wu Z (2010) Comparative study of microbial community structure in different filter media of constructed wetland. J Environ Sci 22(1):127–133
Li X, Zhang M, Liu F, Li Y, He Y, Zhang S, Wu J (2015) Abundance and distribution of microorganisms involved in denitrification in sediments of a Myriophyllum elatinoides purification system for treating swine wastewater. Environ Sci Pollut Res 22(22):17906–17916
Li Y, Wu B, Zhu G, Liu Y, Ng WJ, Appan A, Tan SK (2016) High-throughput pyrosequencing analysis of bacteria relevant to cometabolic and metabolic degradation of ibuprofen in horizontal subsurface flow constructed wetlands. Sci Total Environ 562:604–613
Ligi T, Oopkaup K, Truu M, Preem J-K, Nolvak H, Mitsch WJ, Mander Ü, Truu J (2014) Characterization of bacterial communities in soil and sediment of a created riverine wetland complex using high-throughput 16S rRNA amplicon sequencing. Ecol Eng 72:56–66
Lin Y-F, Jing S-R, Lee D-Y, Chang Y-F, Shih K-C (2008) Nitrate removal from groundwater using constructed wetlands under various hydraulic loading rates. Bioresour Technol 99(16):7504–7513
Lloyd JR, Klessa DA, Parry DL, Buck P, Brown NL (2004) Stimulation of microbial sulphate reduction in a constructed wetland: microbiological and geochemical analysis. Water Res 38(7):1822–1830
Lünsmann V, Kappelmeyer U, Benndorf R, Martinez-Lavanchy PM, Taubert A, Adrian L, Duarte M, Pieper DH, von Bergen M, Müller JA, Heipieper HJ, Jehmlich N (2016) In situ protein-SIP highlights Burkholderiaceae as key players degrading toluene by para ring hydroxylation in a constructed wetland model. Environ Microbiol 18(4):1176–1186
Lünsmann V, Kappelmeyer U, Taubert A, Nijenhuis I, von Bergen M, Heipieper HJ, Müller JA, Jehmlich N (2016) Aerobic toluene degraders in the rhizosphere of a constructed wetland model show diurnal polyhydroxyalkanoate metabolism. Appl Environ Microbiol 82(14):4126–4132
Lv T, Zhang Y, Carvalho PN, Zhang L, Button M, Arias CA, Weber KP, Brix H (2016) Microbial community metabolic function in constructed wetland mesocosms treating the pesticides imazalil and tebuconazole. Ecol Eng. doi:10.1016/j.ecoleng.2016.07.004
Martínez-Lavanchy PM, Chen Z, Lünsmann V, Marin-Cevada V, Vilchez-Vargas R, Pieper DH, Reiche N, Kappelmeyer U, Imparato V, Junca H, Nijenhuis I, Müller JA, Kuschk P, Heipieper HJ (2015) Microbial toluene removal in hypoxic model constructed wetlands occurs predominantly via the ring monooxygenation pathway. Appl Environ Microbiol 81(18):6241–6252
Morató J, Codony F, Sánchez O, Martín Pérez L, García J, Mas J (2014) Key design factors affecting microbial community composition and pathogenic organism removal in horizontal subsurface flow constructed wetlands. Sci Total Environ 481:81–89
Nielsen PH, Mielczarek AT, Kragelund C, Nielse JL, Saunders AM, Kong Y, Hansen AA, Vollertsen J (2012) A conceptual model system of microbial communities in enhanced biological phosphorus removal plants. Water Res 44:5070–5088
Nikolausz M, Kappelmeyer U, Széleky A, Rusznyák KM, Kästner M (2008) Diurnal redox fluctuation and microbial activity in the rhizosphere of wetland plants. Eur J Soil Biol 44(3):324–333
Paranychianakis NV, Tsiknia M, Kalogerakis N (2016) Pathways regulating the removal of nitrogen in planted and unplanted subsurface flow constructed wetlands. Water Res 102:321–329
Pedrós-Alió C (2007) Ecology. Dipping into the rare biosphere. Science 315(5809):192–193
Pedrós-Alió C (2012) The rare bacterial biosphere. Annu Rev Mar Sci 4:449–466
Ramond J-B, Welz PJ, Tuffin MI, Burton SG, Cowan DA (2013) Assessment of temporal and spatial evolution of bacterial communities in a biological sand filter mesocosm treating winery wastewater. J Appl Microbiol 115(1):91–101
Ravit B, Ehenfeld JG, Häggblom MM (2006) Effects of vegetation on root-associated microbial communities: a comparison of disturbed versus undisturbed estuarine sediments. Soil Biol Biochem 38:2359–2371
Russell RA, Holden PJ, Wilde KL, Neilan BA (2003) Demonstration of the use of Scenedesmus and Carteria biomass to drive bacterial sulfate reduction by Desulfovibrio alcoholovorans isolated from an artificial wetland. Hydrometallurgy 71(1–2):227–234
Schmidt I, Sliekers O, Schmid M, Bock E, Fuerst J, Gijs Kuenen J, Jetten MSM, Strous M (2003) New concepts of microbial treatment processes for the nitrogen removal in wastewater. FEMS Microbiol Rev 27(4):481–492
Sidrach-Cardona R, Sánchez O, Garrido L, Mas J, Bécares E (2015) Molecular characterization of microbial communities in constructed wetlands: the effect of plant species, organic matter and hydraulic design. In: Barret LM (ed) Wastewater treatment. Nova Science Publishers, Inc, New York, pp 45–67
Sigalevich P, Baev MV, Teske A, Cohen Y (2000) Sulfate reduction and possible aerobic metabolism of the sulfate-reducing bacterium Desulfovibrio oxyclinae in a chemostat coculture with Marinobacter sp. strain MB under exposure to increasing oxygen concentrations. Appl Environ Microbiol 66(11):5013–5018
Sleytr K, Tietz A, Langergraber G, Haberl R, Sessitsch A (2009) Diversity of abundant bacteria in subsurface vertical flow constructed wetlands. Ecol Eng 35(6):1021–1025
Song ZW, Wang L, Xu AL, Wu DD, Xia Y (2015) Carbon sources metabolic characteristics of airborne microbial communities in constructed wetlands. Huang Jing Ke Xue 36(2):415–420
Stottmeister U, Wiessner A, Kuschk P, Kappelmeyer MK, Bederski RA, Müller H, Moormann H (2003) Effects of plants and microorganisms in constructed wetlands for wastewater treatment. Biotechnol Adv 22(1–2):93–117
Sundberg C, Stendahl JSK, Tonderski K, Lindgren P-E (2007) Overland flow systems for treatment of landfill leachates—potential nitrification and structure of the ammonia-oxidising bacterial community during a growing season. Soil Biol Biochem 39(1):127–138
Tietz A, Hornek R, Langergraber G, Kreuzinger N, Haberl R (2007) Diversity of ammonia oxidizing bacteria in a vertical flow constructed wetland. Water Sci Technol 56(3):242–247
Truu M, Juhanson J, Truu J (2009) Microbial biomass, activity and community composition in constructed wetlands. Sci Total Environ 407(13):3958–3971
Vacca G, Wand H, Nikolausz M, Kuschk P, Kästner M (2005) Effect of plants and filter material on bacterial removal in pilot-scale constructed wetlands. Water Res 39(7):1361–1373
Vymazal J (2005) Removal of enteric bacteria in constructed treatment wetlands with emergent macrophytes: a review. J Environ Sci Health 40(6–7):1355–1367
Vymazal J (2011) Constructed wetlands for wastewater treatment: five decades of experience. Environ Sci Technol 45(1):61–69
Vymazal J (2013) Emergent plants in free water surface constructed wetlands: a review. Ecol Eng 61P:582–592
Vymazal J, Brezinova T (2015) The use of constructed wetlands for removal of pesticides from agricultural runoff and drainage: a review. Environ Int 75C:11–20
Wang Q, Xie H, Ngo HH, Guo W, Zhang J, Liu C, Liang S, Hu Z, Yang Z, Zhao C (2016) Microbial abundance and community in subsurface flow constructed wetland microcosms: role of plant presence. Environ Sci Pollut Res 23:4036–4045
Wu S, Kuschk P, Brix H, Vymazal J, Dong R (2014) Development of constructed wetlands in performance intensifications for wastewater treatment: a nitrogen and organic matter targeted review. Water Res 57C:40–55
Wu Y, Han R, Yang X, Fang X, Chen X, Yang D, Zhang R (2016) Correlating microbial community with physicochemical indices and structures of a full-scale integrated constructed wetland system. Appl Microbiol Biotechnol 100(15):6917–6926
Xu M, Liu W, Li C, Xiao C, Ding L, Xu K, Geng J, Ren H (2016) Evaluation of the treatment performance and microbial communities of a combined constructed wetland used to treat industrial park wastewater. Environ Sci Pollut Res 23(11):10990–11001
Yi X-H, Jing D-D, Wan J, Ma Y, Wang Y (2016) Temporal and spatial variations of contaminant removal, enzyme activities, and microbial community structure in a pilot horizontal subsurface flow constructed wetland purifying industrial runoff. Environ Sci Pollut Res 23(9):8565–8576
Yin J, Jiang L, Wen Y, Yao Z, Zhou Q (2009) Treatment of polluted landscape lake water and community analysis of ammonia-oxidizing bacteria in constructed wetland. J Environ Sci Health A Tox Hazard Subst Environ Eng 44(7):722–731
Zhai X, Piwpuan N, Arias CA, Headley T, Brix H (2013) Can root exudates from emergent wetland plants fuel denitrification in subsurface flow constructed wetland systems? Ecol Eng 61P:555–563
Zhao C, Xie H, Xu J, Xu X, Zhang J, Hu Z, Liu C, Liang S, Wang Q, Wang J (2015) Bacterial community variation and microbial mechanism of triclosan (TCS) removal by constructed wetlands with different types of plants. Sci Total Environ 505:633–639
Zhong F, Wu J, Dai Y, Yang L, Zhang Z, Cheng S, Zhang Q (2015) Bacterial community analysis by PCR-DGGE and 454-pyrosequencing of horizontal subsurface flow constructed wetlands with front aeration. Appl Microbiol Biotechnol 99(3):1499–1512
Acknowledgments
The author thanks Dr. Isabel Ferrera for her help. This work was supported by the Spanish grant CTM2015-70340-R from the Ministerio de Economía y Competitividad.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sánchez, O. Constructed Wetlands Revisited: Microbial Diversity in the –omics Era. Microb Ecol 73, 722–733 (2017). https://doi.org/10.1007/s00248-016-0881-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0881-y