Abstract
Seagrasses, a small group of submerged marine macrophytes, were reported to lack mycorrhizae, i.e., the root-fungus symbioses most terrestrial plants use for nutrient uptake. On the other hand, several authors detected fungal endophytes in seagrass leaves, shoots, rhizomes, and roots, and an anatomically and morphologically unique dark septate endophytic (DSE) association has been recently described in the roots of the Mediterranean seagrass Posidonia oceanica. Nevertheless, the global diversity of seagrass mycobionts is not well understood, and it remains unclear what fungus forms the DSE association in P. oceanica roots. We isolated and determined P. oceanica root mycobionts from 11 localities in the northwest Mediterranean Sea with documented presence of the DSE association and compared our results with recent literature. The mycobiont communities were low in diversity (only three species), were dominated by a single yet unreported marine fungal species (ca. 90 % of the total 177 isolates), and lacked common terrestrial and freshwater root mycobionts. Our phylogenetic analysis suggests that the dominating species represents a new monotypic lineage within the recently described Aigialaceae family (Pleosporales, Ascomycota), probably representing a new genus. Most of its examined colonies developed from intracellular microsclerotia occupying host hypodermis and resembling microsclerotia of terrestrial DSE fungi. Biological significance of this hitherto overlooked seagrass root mycobiont remains obscure, but its presence across the NW Mediterranean Sea and apparent root intracellular lifestyle indicate an intriguing symbiotic relationship with the dominant Mediterranean seagrass. Our microscopic observations suggest that it may form the DSE association recently described in P. oceanica roots.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Seagrasses are a narrow ecological and taxonomical group of marine sessile vascular macrophytes well adapted to aquatic life that form extensive underwater meadows with significant ecosystem functions [1]. Seagrass meadows rank among the most productive aquatic ecosystems [2] and can store up to twice as much carbon as temperate and tropical forests [3]. Although the center of seagrass biodiversity lies in the Indomalaya ecozone [4], they occur in most coastal shallow areas, except for polar regions [5]. All seagrasses belong to the Alismatales and comprise some 12 genera with ca. 50–70 species [4, 6]. The Mediterranean Sea is home to four seagrass genera (Cymodocea, Halophila, Posidonia, and Zostera) encompassing four autochthonous (Cymodocea nodosa, Posidonia oceanica, Zostera marina, and Zostera noltii) and one alien invasive (Halophila stipulacea) species [7, 8]. Most of the Mediterranean sublittoral area occupied by seagrasses is dominated by the endemic P. oceanica (=Neptune seagrass). Its clonally reproducing meadows can spread up to 15 km while being hundreds to thousands years old [9]. However, many observations suggest that in certain areas, its populations are significantly declining due to a combination of mostly human-induced factors [7, 10, 11].
Seagrasses take up nutrients through their shoots and leaves, and hence, in comparison with terrestrial plants, significance of nutrient uptake through the roots is generally reduced [2, 12]. Congruently, seagrasses were reported to lack mycorrhizal symbioses [13], i.e., root-fungus symbioses utilized by most land plants to scavenge nutrients from recalcitrant substrates [14]. On the other hand, this lack might be as well due to comparably lower attention paid to seagrasses, especially in comparison with other plant guilds hosting mycorrhizal fungi, including freshwater aquatic plants and plants from saltmarshes and mangroves [15–17]. Nevertheless, similarly to terrestrial plants, seagrasses host endophytic fungi [18–22], although their ecophysiological function in the marine environment has not yet been understood. Fungal endophytes are commonly defined as mycobionts which live inside living plant tissues, lack localized interfaces or specialized hyphae for nutrient transfer, do not synchronize their development with plant development, and do not provide nutritional benefits to the plant [23]. The term “mycobiont” as used here is more general and may specifically refer to the situations where the ecophysiological function of the fungal symbiont remains unknown.
Despite its ubiquity and dominance in the Mediterranean Sea, the mycoflora of P. oceanica has been studied only by a few authors. The arguably oldest published information dates back to 1840s and reports two fungal species, Sphaeria biturbinata and Sphaeria posidoniae, in P. oceanica rhizomes [24]. More than a century later, Kohlmeyer [25] transferred these fungi in two new genera, Halotthia [with Halotthia posidoniae (Dur. et Mont.) Kohlm.] and Pontoporeia [with Pontoporeia biturbinata (Dur. et Mont.) Kohlm.]. Some 20 years later, Cuomo et al. [26] investigated the mycoflora of P. oceanica leaves, sheaths, rhizomes, and roots and found seven marine lignicolous fungi, namely H. posidoniae (occurred in 87 % of samples), Corollospora maritima Werdermann (70 %), Papulaspora halima Anastasiou (60 %), P. biturbinata (56 %), Lulworthia sp. (54 %), Phoma sp. (44 %), and Corollospora intermedia Schmidt (9 %). The authors pointed out that the detected diversity was comparably lower than in the intertidal grass Spartina alterniflora (29 species detected) and the mangrove Rhizopora mangle (31 species detected) as reported by Kohlmeyer and Kohlmeyer [27]. More recently, fungi associated with P. oceanica leaves, rhizomes, roots, and dead parts (matte) were studied by Panno et al. [28] who isolated 88 species in total, 14 of them being associated with the roots. Intriguingly, none of them belonged among those reported by Cuomo et al. [26], and most of them belonged to fast-growing sporulating species also known from terrestrial habitats, probably as a consequence of no surface sterilization of the seagrass organs prior to isolation. Another recent report is by Torta et al. [29] who however isolated only one fungus, contrary to the previous studies. Most recently, an anatomically and morphologically unique dark septate endophytic (DSE) association was discovered in the roots of P. oceanica at 11 localities in the northwest Mediterranean Sea [22], but the fungus or fungi forming this symbiosis have not been identified.
Terrestrial root endophytes may, among other possible functions, engage in nutrient uptake and transport to the host plant [30] and protect host roots against pathogen attacks [31]. On the other hand, nearly nothing is known about possible roles of endophytes in seagrass roots, and this is probably due to both the low number of seagrass mycobiont studies and their differing results. Therefore, we conducted a culture-dependent screening of P. oceanica endorhizal mycobionts at 11 localities in the NW Mediterranean Sea (the same as in [22]), using an isolation technique similar to those employed in the previous reports, but modified as follows: (1) In contrast to Panno et al. [28], we included a surface sterilization step to eliminate common saprobic contaminants, and (2) in contrast to Torta et al. [29], we prolonged the isolation period as some marine fungi are notoriously slow-growing [27]. We hypothesized that this approach coupled with the relatively large area under investigation would lead to detection of common P. oceanica endorhizal mycobionts, possibly comprising at least some of the previously reported fungi associated with P. oceanica roots. We also hoped that this approach could shed light on the fungus or fungi forming the recently discovered DSE association in P. oceanica roots [22].
Materials and Methods
Root Sampling
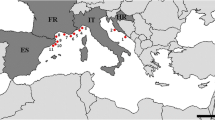
P. oceanica (L.) Delile roots were collected using free and scuba diving between June and September 2012 at 11 localities in the NW Mediterranean Sea in Croatia, Italy, France, and Spain (Fig. 1, Table 1; [22]). At each locality, roots of five different individuals (at least 3 m apart) were carefully excavated from the substrate, separated from the shoots, mixed to make a compound sample, and inserted into 50-ml plastic beakers filled with seawater. These were stored in the dark in a portable fridge and processed for isolation of mycobionts in the evening of the respective collection day.
Isolation of Posidonia oceanica Root Fungal Symbionts
Mycobionts were isolated from fresh P. oceanica roots using a portable flow box to prevent air contaminants. Healthy looking turgescent terminal roots (ca. 1–2 mm in diam.) were selected from bulk samples, surface-sterilized in 10 % SAVO (household bleach, Unilever ČR Ltd., Czech Republic; contains 4.5 % available chlorine) for 30 s, rinsed two times in sterile deionized water, cut into ca. 3–5-mm segments, and placed on agar media in four-compartment Petri dishes. We used potato dextrose agar (PDA; HiMedia Pvt. Ltd., India) and modified Melin-Norkrans agar (MMN) [32], both amended with Novobiocin sodium salt (50 mg/L; Sigma-Aldrich, Germany) to prevent growth of bacteria and NaCl (38 g/L) to adjust osmotic pressure [33]. Roots from two localities (#3 Cogoleto and #9 l′Escala) were additionally plated on PDA and MMN without NaCl. There were five dishes each with 16 root segments totaling 80 root segments per each medium/locality combination. The dishes were sealed with an air-permeable tape, kept at room temperature in the dark, and observed for hyphal growth each day during the first 2 weeks then once each week until 101 days. Obtained fungal colonies were conservatively grouped into morphotypes, and several representatives of each morphotype/medium/locality combination were transferred to new petri dishes with MMN + NaCl.
Microscopic Observations
Hyphal colonies emerging from the root segments were screened using an Olympus SZX12 stereomicroscope and a FEI Quanta 200 scanning electron microscope equipped with the Olympus ESEM mode. Hand semi-thin cross sections were made from several root segments with emerging dark mycelia to investigate their possible intraradical origin. The cross sections were screened with an Olympus BX60 microscope equipped with DIC. Photos were taken with an Olympus DP70 camera and adjusted for clarity in Adobe Photoshop (Adobe Systems, USA) and Paint.NET (Brewster, Jackson and contributors + Microsoft Corporation, USA) as needed. Figures were assembled in Adobe Photoshop and Paint.NET.
Molecular Identification of Posidonia oceanica Root Fungal Symbionts
DNA was extracted from representatives of all morphotypes (Table 2) using Extract-N-Amp Plant Kits (Sigma-Aldrich, Germany) following manufacturer’s instructions. SSU (18S) ribosomal DNA (rDNA) was amplified using the NS1 + NS24 primer pair [34], ITS1-5.8S-ITS2 rDNA using the ITS1F + ITS4 primer pair [34, 35] and LSU (28S) rDNA using the LR0R + LR5 primer pair [36]. For PCR parameters and gel electrophoresis, see [37]. PCR products were purified and sequenced by Macrogen Europe Laboratory (Macrogen Inc., The Netherlands/South Korea) using the NS1, NS4, NS5 and NS24, ITS1F, and LR0R primers.
The obtained ITS rDNA sequences were screened in Finch TV v1.4.0 (geospiza.com/finchtv) for possible machine errors and edited when needed. They were subsequently aligned in BioEdit v7.0.5.3 using ClustalW [38], and the alignment was used as a matrix for NJ analysis in TOPALi v2.5 (topali.org). The threshold limit for grouping of sequences was set at 97 %. Sequences within separate groups were further aligned to screen their heterogeneity, and the most divergent were subjected to BLAST searches (megablast/blastn algorithms) in GenBank [39], and their taxonomic position was further checked with Blast Tree View (NJ, max. seq. difference 0.75). This was only sufficient for identification of the least abundant sequence group at the species level. The same approach with LSU sequences enabled identification of the second most frequent group at the genus level. The remaining group of the most frequent dark septate mycelia was subjected to the following phylogenetic analysis.
Taxon selection was patterned on reduced LSU-SSU dataset of Liu et al. [40] (TreeBASE matrix no. 14874). Sequence alignments were obtained using MAFFT 6 (http://mafft.cbrc.jp/alignment/software/) [41]. The dataset consisted of 57 sequences, 1894 positions (463 variable and 353 parsimony-informative sites). Maximum likelihood analyses were performed using PhyML 3.0 [42], and bootstrap support was obtained using 500 replicates. Evolutionary model (TN93+G+I) was determined for all datasets using MEGA 5.2.1 [43].
Results
From the total 880 surface-sterilized root segments, we obtained 177 relatively slow-growing fungal colonies. Contrary to endophyte isolations from roots of terrestrial plants [44], there were no contaminations by bacteria or fast-growing sporulating fungi. First hyphal colonies started to emerge from the surface-sterilized roots after ca. 4 weeks, and several dark brown to black colonies emerged even after 12 weeks of incubation. The best medium for mycobiont isolation was PDA with NaCl (141 isolates) followed by MMN with NaCl (32 isolates).
The 177 fungal isolates were conservatively grouped into three morphotypes (black, yellow, and ochre) with 160, 16, and 1 isolate, respectively. These morphotypes differed not only in colony color and morphology but also in the isolation time and growth rate, the black morphotype emerging significantly later and growing slower than yellow and ochre. Isolates of the most frequent black morphotype produced thick slow-growing melanized hyphae on the root surface and formed either aerial colonies on the MMN medium (Fig. 2a) or compact colonies on the PDA medium (Fig. 2b–d). The surface of the compact colonies was covered by terminally swollen hyphae resembling capitate cystidia (Fig. 2e; cf. [37]). The microscopic screening of the hand sections showed that most of the colonies emerging from root segments actually developed from intracellular melanized microsclerotia occurring in the P. oceanica root hypodermis (Fig. 2f). Isolates of the most frequent black morphotype were obtained from all but two localities (#8 Anse de Paulilles and #10 Tamariu) and dominated P. oceanica root mycobiont communities at five localities (Table 2). The second most frequent yellow morphotype grew faster and produced non-melanized hyaline to yellow hyphae forming compact colonies which were partly submerged in the cultivation medium. The yellow isolates were obtained at seven localities and were the only isolates obtained at one locality (#8 Anse de Paulilles), although in a low number (Table 2). In total, only one ochre isolate was obtained at a single locality (#10 Tamariu; Table 2). The great majority of black isolates were obtained on PDA with NaCl (134 isolates); on the other hand, there seemed to be no medium preferences in the case of the yellow isolates.
Morphology of the pleosporalean mycobiont (Pleosporales sp. MV-2012, the black morphotype) isolated from Posidonia oceanica roots. a Aerial hyphae emerging from a surface-sterilized root plated on modified Melin-Norkrans agar (MMN) with sodium chloride (NaCl, 38 g/L). Stereomicroscopy, bar = 500 μm. b, c Typical structures formed by the pleosporalean mycobiont when growing out from segments of Posidonia oceanica roots incubated in the dark on potato dextrose agar (PDA) with NaCl. Scanning electron microscopy (SEM), bars = 1 mm. d Compact black colonies formed on a surface-sterilized root segment plated on PDA with NaCl, identical in nature with those depicted at b and c using SEM. Stereomicroscopy, bar = 500 μm. e A detail of some superficial hyphae with terminal swellings developed on the surface of the compact black colonies (as in d). Light microscopy with DIC, bar = 20 μm. f A cross section of a root segment with aerial hyphae originating from intracellular microsclerotia (arrows). MMN with NaCl, light microscopy with DIC, bar = 20 μm
DNA was successfully isolated and amplified from all examined isolates, and gel electrophoresis yielded a single band in all tested samples. All sequences were of sufficient quality and length. Three separate groups were delimited within our sequence dataset. Using BLAST searches, two sequence groups could be identified at the species/genus level: The ochre isolate P07 was identified as Fuscoporia torulosa (Basidiomycetes, Hymenochaetaceae) and the yellow isolates P01–P04, P12, P13, P21, and P32 as Lulworthiales sp. MV-2012 (Ascomycetes) (Tables 2 and 3). The dominant conspecific black isolates (Pleosporales sp. MV-2012) showed the closest similarity to members of the Aigialaceae family (Ascomycetes, Pleosporales) such us Aigialus grandis (SSU: 98 % to AF441172), Rimora mangrovei (LSU: 93 % to GU479798), or Fissuroma aggregata (ITS: 87 % to JN846718) (Tables 2 and 3). In the phylogenetic analysis based on SSU-LSU rDNA, they formed a sister lineage to the genus Aigialus and together with Ascocratera manglicola and Rimora mangrovei formed a strongly supported clade within Aigialaceae (Fig. 3) [40]. The respective sequences were deposited in GenBank under accession numbers KC145421-4, KC145426-32, and KC412711-21 (ITS rDNA), KC736937-46 (LSU rDNA), and KJ210571 (SSU rDNA).
A PhyML tree based on a combined dataset of SSU (894 bp) and LSU (919 bp) rDNA sequences of the pleosporalean mycobiont (Pleosporales sp. MV-2014, the black morphotype) from Posidonia oceanica roots. Bootstrap support values greater than 50 % are given above the nodes. Thickened branches represent 100 % bootstrap values. The clade of the pleosporalean mycobiont dominating the cultivable fungal communities in P. oceanica roots is highlighted in green color. The tree was rooted with Myriangium hispanicum (Myriangiales) and Dothidea insculpta (Dothideales). For details on the sequences used, see “Materials and Methods”
Discussion
Only a few studies focused on root mycobionts of seagrasses in general and P. oceanica in particular. Here, we detected only three cultivable fungal species associated with P. oceanica surface-sterilized roots. Moreover, one of them, F. torulosa, was represented by only a single isolate. This number is therefore comparable to Cuomo et al. [26] and Torta et al. [29] who detected only one species and comparably lower than Panno et al. [28] who detected 14 fungal species associated with the seagrass roots. In contrast to Cuomo et al. [26], we did not detect C. maritima, and in agreement with Torta et al. [29], we detected Lulworthiales sp. MV-2012 with nearly identical ITS rDNA sequences to their “Lulwoana sp.”. In contrast to our hypothesis, we did not detect any of the species reported by Panno et al. [28]. On the other hand, Panno et al. [28] did not detect any species reported by Cuomo et al. [26] and concluded that this might be due to seasonality of sampling as they took their samples in spring while Cuomo and colleagues in autumn. We argue with this conclusion and point out that a more likely explanation is the method used by Panno and colleagues for isolation of P. oceanica mycobionts: they used no surface sterilization but only serial washing and blended the samples with sterile seawater. Such an approach leads to preferential isolation of fast-growing surface-dwelling saprobes in contrast to true endorhizal mycobionts [45]. This is especially true in the aquatic environment where submerged solid substrates entrap substantial numbers of fungal propagules which may be present in dormant or minimally active state [46]. Congruently, the spectra of root-associated fungi reported by Panno and colleagues were dominated by well-known saprobes, including species of Acremonium, Penicillium, etc., which cannot be interpreted as true P. oceanica endophytes. The low effectiveness of serial washing as a surface sterilization technique in seagrass research is further discussed in Newell and Fell [47]. In agreement with our hypothesis, it thus seems that the sterilization technique used in this study eliminated all surface-dwelling fungi and revealed true endorhizal mycobionts of P. oceanica. On the other hand, Shoemaker and Wyllie-Echeverria [48] obtained an assemblage of 36 fungi, mostly dominated by saprobes/parasites probably well-adapted to the marine environment, such as Aspergillus, Cylindrocarpon, Penicilium, and Trichoderma, from rhizomes of three temperate Pacific seagrasses even after surface sterilization with sodium hypochlorite.
At most of the localities, the P. oceanica root cultivable mycobiont communities were dominated by a pleosporalean fungal species hitherto not reported in seagrasses, the Pleosporales sp. MV-2012. This was surprising especially when taking into account the results of Torta et al. [29] who used a very similar isolation protocol and indeed detected the “Lulwoana sp.”, probably conspecific with our Lulworthiales sp. MV-2012, i.e., the second most frequent mycobiont isolated in this study. There are two probable explanations for the lack of the pleosporalean mycobiont within the fungal spectra reported by Torta and colleagues. First, the fungus was simply absent or present in very low frequencies which prevented its detection by the isolation method used; this may be true also for the two of our localities not yielding the pleosporalean mycobiont. Second, the cultivation period utilized by Torta and colleagues, i.e., 4 weeks, was too short for successful isolation of the slow-growing pleosporalean mycobiont which new colonies appeared even after 12 weeks of incubation. This could be resolved by re-visiting the investigated localities and prolonging the cultivation period. Obviously, the pleosporalean mycobiont could be targeted also by culture-independent molecular methods, e.g., amplification of its DNA using specific primers.
The phylogenetic analysis placed the Pleosporales sp. MV-2012 in the vicinity of Aigialus ssp., Ascocratera manglicola, and Rimora mangrovei within Aigialaceae. The recently established Aigialaceae family currently encompasses five genera: Aigialus, Ascocratera, and Rimora represent marine intertidal fungi colonizing mangrove bark and wood in various tropical geographic locations while Fissuroma and Neoastrosphaeriella colonize dead bamboo and palm tissues and are known from Australia, Japan, Philippines, and Thailand [40, 49]. This study thus extends the know range of Aigialaceae habitats for the roots of the Mediterranean endemic seagrass P. oceanica. Whether the second most frequent mycobiont of P. oceanica roots isolated in this study, the Lulworthiales sp. MV-2012, has any closer relationship with the Lulworthia sp. detected in fibrous remains of old P. oceanica leaf sheaths by Cuomo et al. [26] will probably remain unresolved because at present, direct comparison of the respective sequences is impossible. The least frequent species in this study, F. torulosa, is mainly known as a basidiomycetous wood decay saprobe causing white pocket rot in dead and living hardwood trees in Europe [50]. This seems to be the first report on the occurrence of this species in the marine environment. However, due its low isolation frequency, significance of this finding remains unknown.
Our microscopic observations suggested that the pleosporalean mycobiont developed its in vitro colonies from melanized intracellular microsclerotia occurring in the seagrass root hypodermis which were morphologically identical to those reported by Vohník and colleagues [22]. Since the pleosporalean mycobiont also produced thick, septate, a melanized hyphae, it can be ranked among DSE, a miscellaneous group of ubiquitous endophytes colonizing roots of most terrestrial plants [51–53]. Pleosporalean fungi are often reported as DSE in roots of plants from arid and semi-arid regions [54, 55]. On the other hand, although the Pleosporales comprise many freshwater and marine, mostly saprobic, genera [56], to our knowledge, they have not yet been reported as DSE in roots of seagrasses. Intriguingly, Torta et al. [29] claimed that the “Lulwoana sp.” (Lulworthiales) detected by them in P. oceanica roots belonged to DSE despite that no evidence was given that it produced dark septate hyphae or melanized intracellular microsclerotia. Under our cultivation conditions, isolates of the Lulworthiales sp. MV-2012 did not produce menalized hyphae typical for DSE. Torta et al. [29] also provided no evidence that their “Lulwoana sp.” had any connection to the reported intracellular microsclerotia, except for being isolated from some of the collected P. oceanica roots. The identity of microsclerotia reported by Torta and colleagues could be resolved by techniques targeting single cells, i.e., laser capture microdissection, followed by PCR, possibly employing specific primers targeted at the “Lulwoana sp.” DNA.
Based on the data available at present, relatively little can be speculated about the biological significance of the dominant pleosporalean mycobiont. Some DSE can form structures morphologically resembling hyphal interfaces produced by mycorrhizal fungi for communication and nutrient exchange with their host plant [52, 57, 58], and the DSE colonization pattern in P. oceanica roots reported by Vohník et al. [22] comprises dense parenchymatous nets/hyphal sheaths on the root surface which may morphologically resemble hyphal mantles produced by some ectomycorrhizal fungi. On one hand, based on the microscopic observations, it seems plausible that this DSE association is formed by the pleosporalean mycobiont discovered in this study. On the other hand, it remains to be investigated whether this mycobiont engages in nutrient uptake and transport to the host plant in exchange for carbohydrates as reported for other DSE [30]. Other possible functions may include protection of host roots against pathogen attacks [31] or modifications of the distribution of rhizosphere-associated organisms [59, 60] as suggested for other fungi possessing melanized hyphae. Moreover, P. oceanica forms characteristic peat-like sediment (matte) which can be several meters thick and which is exceptionally resistant to decay [1]. Such stocks of organic nutrients are usually inaccessible for plants without the aid of symbiotic bacteria or fungi, and it may be that the pleosporalean mycobiont plays a role during the decomposition of matte.
To conclude, we found that cultivable fungal assemblages colonizing healthy P. oceanica roots across the NW Mediterranean Sea were composed of only three species and were dominated by a single pleosporalean mycobiont. Such a narrow spectrum of root mycobionts across a wide distribution area markedly contrasts with available data on root mycobiont communities from terrestrial ecosystems. The pleosporalean mycobiont has not been previously reported in seagrasses or any other host/ecosystem and probably represents a member of a hitherto undescribed genus in the Aigialaceae family. It apparently forms melanized intracellular microsclerotia in P. oceanica roots and dark septate hyphae in culture and therefore possesses the main characteristics of the ubiquitous terrestrial DSE. This work thus extends the known range of pleosporalean DSE for the marine environment. Our microscopic observations support the view that this fungus forms the recently described DSE association in P. oceanica roots [22]. Regular occurrence of the pleosporalean mycobiont in P. oceanica roots across the NW Mediterranean Sea and its apparent intracellular lifestyle indicate its non-random symbiotic relationship with the dominant Mediterranean seagrass which certainly deserves further investigation.
References
Hemminga MA, Duarte CM (2000) Seagrass ecology. Cambridge University Press, Cambridge
Hemminga MA, Harrison PG, van Lent F (1991) The balance of nutrient losses and gains in seagrass meadows. Mar Ecol Prog Ser 71:85–96
Fourqurean JW, Duarte CM, Kennedy H, Marba N, Holmer M et al (2012) Seagrass ecosystems as a globally significant carbon stock. Nat Geosci 5:505–509
Green EP, Short FT (2003) World atlas of seagrasses. University of California Press, Berkeley
den Hartog C (1970) Seagrasses of the world. North-Holland Publishing, Amsterdam
den Hartog C, Kuo J (2007) Taxonomy and biogeography of seagrasses. In: Larkum AWD, Orth RJ, Duarte CM (eds) Seagrasses: Biology, Ecology and Conservation. Springer, Dordrecht
Borum J, Duarte CM, Krause-Jensen D, Greve TM (2004) European seagrasses: an introduction to monitoring and management. A publication by the EU project Monitoring and Managing of European Seagrasses EVK3-CT-2000-00044. ISBN: 87-89143-21-3
Gambi MC, Barbieri F, Bianchi CN (2009) New record of the alien seagrass Halophila stipulacea (Hydrocharitaceae) in the western Mediterranean: a further clue to changing Mediterranean Sea biogeography. Mar Biodivers Rec 2:1–7
Arnaud-Haond S, Duarte CM, Diaz-Almela E, Marbá N, Sintes T et al (2012) Implications of extreme life span in clonal organisms: millenary clones in meadows of the threatened seagrass Posidonia oceanica. PLoS One 7:e30454. doi:10.1371/journal.pone.0030454
Peirano A, Bianchi NC (1995) Decline of the seagrass Posidonia oceanica in response to environmental disturbance: a simulation-like approach off Liguria (NW Mediterranean Sea). Proceedings of the 3rd European Marine Biological Symposium, Southampton
Leriche A, Pasqualini V, Boudouresque C-F, Bernard G, Bonhomme P et al (2006) Spatial, temporal and structural variations of a Posidonia oceanica seagrass meadow facing human activities. Aquat Bot 84:287–293
Stapel J, Aarts TL, van Duynhoven BHM, de Groot JD, van den Hoogen PHW, Hemminga MA (1996) Nutrient uptake by leaves and roots of the seagrass Thalassia hemprichii in the Spermonde Archipelago, Indonesia. Mar Ecol Prog Ser 134:195–206
Nielsen SL, Thingstrup I, Wigand C (1999) Apparent lack of vesicular-arbuscular mycorrhiza (VAM) in the seagrasses Zostera marina L. and Thalassia testudinum Banks ex Konig. Aquat Bot 63:261–266
Smith SE, Read DJ (2008) Mycorrhizal symbiosis. Academic Press, London
Kothamasi D, Kothamasi S, Bhattacharyya A, Kuhad RC, Babu CR (2006) Arbuscular mycorrhizae and phosphate solubilising bacteria of the rhizosphere of the mangrove ecosystem of Great Nicobar Island, India. Biol Fertil Soils 42:358–361
Welsh AK, Burke DJ, Hamerlynck EP, Hahn D (2010) Seasonal analyses of arbuscular mycorrhizae, nitrogen-fixing bacteria and growth performance of the salt marsh grass Spartina patens. Plant Soil 330:251–266
Kohout P, Sýkorová Z, Čtvrtlíková M, Rydlová J, Suda J et al (2012) Surprising spectra of root-associated fungi in submerged aquatic plants. FEMS Microbiol Ecol 80:216–235
Wilson WL (1998) Isolation of endophytes from seagrasses from Bermuda. MSc. thesis, University of New Brunswick. National Library of Canada, Ottawa
Devarajan PT, Suryanarayanan TS, Geetha V (2002) Endophytic fungi associated with the tropical seagrass Halophila ovalis (Hydrocharitaceae). Indian J Mar Sci 31:73–74
Sakayaroj J, Preedanon S, Supaphon O, Jones EBG, Phongpaichit S (2010) Phylogenetic diversity of endophyte assemblages associated with the tropical seagrass Enhalus acoroides in Thailand. Fungal Divers 42:27–45
Mata JL, Cebrián J (2013) Fungal endophytes of the seagrasses Halodule wrightii and Thalassia testudinum in the north central Gulf of Mexico. Bot Mar 56:541–545
Vohník M, Borovec O, Župan I, Vondrášek D, Petrtýl M, Sudová R (2015) Anatomically and morphologically unique dark septate endophytic association in the roots of the Mediterranean endemic seagrass Posidonia oceanica. Mycorrhiza. doi:10.1007/s00572-015-0642-7
Brundrett M (2006) Understanding the roles of multifunctional mycorrhizal and endophytic fungi. In: Schulz B, Boyle C, Sieber TN (eds) Microbial root endophytes. Springer, Berlin
Durieu de Maisonneuve C, Montagne JFC (1869) Pyrenomycetes Fr. In: Bory de Saint-Vincent J, Durieu de Maisonneuve C (eds) Exploration Scientifique de l´Algérie, Botanique
Kohlmeyer J (1963) Zwei neue Ascomyceten-Gattungen auf Posidonia-Rhizomen. Nova Hedwigia 6:5–13
Cuomo V, Vanzanella F, Fresi E, Cinelli F, Mazzella L (1985) Fungal flora of Posidonia oceanica and its ecological significance. Trans Brit Mycol Soc 84:35–40
Kohlmeyer J, Kohlmeyer E (1979) Marine mycology—the higher fungi. Academic Press, New York
Panno L, Bruno M, Voyron S, Anastasi A, Gnavi G et al (2013) Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanica. N Biotechnol 30:685–694
Torta L, Lo Piccolo S, Piazza G, Burruano S, Colombo P et al (2015) Lulwoana sp., a dark septate endophyte in roots of Posidonia oceanica (L.) Delile seagrass. Plant Biol 17:505–511
Usuki F, Narisawa K (2007) A mutualistic symbiosis between a dark septate endophytic fungus, Heteroconium chaetospira, and a nonmycorrhizal plant, Chinese cabbage. Mycologia 99:175–184
Tellenbach C, Sieber TN (2012) Do colonization by dark septate endophytes and elevated temperature affect pathogenicity of oomycetes? FEMS Microbiol Ecol 82:157–168
Marx DH (1969) Influence of ectotrophic mycorrhizal fungi on resistance of pine roots to pathogenic infections I. Antagonism of mycorrhizal fungi to root pathogenic fungi and soil bacteria. Phytopathology 59:153–163
Caye G, Bulard C, Meinesz A, Loques F (1992) Dominant role of seawater osmotic pressure on germination in Cymodocea nodosa. Aquat Bot 42:187–193
White TJ, Bruns TD, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis N, Gelfand D, Sninsky J, White T (eds) PCR—protocols and applications—a laboratory manual. Academic Press, New York
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for Basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246
Vohník M, Sadowsky JJ, Kohout P, Lhotáková Z, Nestby R, Kolařík M (2012) Novel root-fungus symbiosis in Ericaceae: sheathed ericoid mycorrhiza formed by a hitherto undescribed basidiomycete with affinities to Trechisporales. PLoS One 7:e39524. doi:10.1371/journal.pone.0039524
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Sym Ser 41:95–98
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Liu JK, Phookamsak R, Jones EBG, Zhang Y, Ko-Ko TW et al (2011) Astrosphaeriella is polyphyletic, with species in Fissuroma gen. nov., and Neoastrosphaeriella gen. nov. Fungal Divers 51:135–154
Katoh K, Toh H (2008) Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinform 9:212. doi:10.1186/1471-2105-9-212
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W et al (2010) New algorithms and methods to estimate Maximum likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Tamura K, Peterson D, Peterson N, Stecher G, Nei M et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Vohník M, Mrnka L, Lukešová T, Bruzone MC, Kouhout P, Fehrer J (2013) The cultivable endophytic community of Norway spruce ectomycorrhizas from microhabitats lacking ericaceous hosts is dominated by ericoid mycorrhizal Meliniomyces variabilis. Fungal Ecol 6:281–292
Sieber TN (2002) Fungal root endophytes. In: Waisel Y, Eshel A, Kafkafi (eds) Plant roots—the hidden half, 3rd edn. Marcel Dekker, New York
Park D (1974) Accumulation of fungi by cellulose exposed in a river. Trans Br Mycol Soc 63:437–447
Newell SY, Fell JW (1982) Surface sterilization and the active mycoflora of leaves of a seagrass. Bot Mar 25:339–346
Shoemaker G, Wyllie-Echeverria S (2013) Occurrence of rhizomal endophytes in three temperate northeastpacific seagrasses. Aquat Bot 111:71–73
Suetrong S, Schoch CL, Spatafora JW, Kohlmeyer J, Volkmann-Kohlmeyer B et al (2009) Molecular systematics of the marine Dothideomycetes. Stud Mycol 64:155–173
Tomšovský M, Jankovský L (2007) DNA sequence analysis of extraordinary fruiting specimens of Fuscoporia torulosa (Phellinus torulosus) on Pyrus spp. Czech Mycol 59:91–99
Addy HD, Piercey MM, Currah RS (2005) Microfungal endophytes in roots. Can J Bot 83:1–13
Vohník M, Albrechtová J (2011) The co-occurrence and morphological continuum between ericoid mycorrhiza and dark septate endophytes in roots of six European Rhododendron species. Folia Geobot 46:373–386
Bruzone MC, Fontenla SB, Vohník M (2015) Is the prominent ericoid mycorrhizal fungus Rhizoscyphus ericae absent in the Southern Hemisphere’s Ericaceae? A case study on the diversity of root mycobionts in Gaultheria spp. from northwest Patagonia, Argentina. Mycorrhiza 25:25–40
Porras-Alfaro A, Herrera J, Sinsabaugh RL, Odenbach KJ, Lowrey T, Natvig DO (2008) Novel root fungalconsortium associated with a dominant desert grass. Appl Environ Microbiol 74:1308–1315
Knapp DG, Kovács GM, Zajta E, Groenewald JZ, Crous PW (2015) Dark septate endophytic pleosporalean genera fromsemiarid areas. Persoonia 35:87–100
Zhang Y, Fournier J, Schoch CL, Crous PW, de Gruyter J et al (2009) Multi-locus phylogeny of Pleosporales: a taxonomic, ecological and evolutionary re-evaluation. Stud Mycol 64:85–102
Vohník M, Lukančič S, Bahor E, Regvar M, Vosátka M, Vodnik D (2003) Inoculation of Rhododendron cv. Belle-Heller with two strains of Phialocephala fortinii in two different substrates. Folia Geobot 38:191–200
Lukešová T, Kohout P, Větrovský T, Vohník M (2015) The potential of dark septate endophytes to form root symbioses with ectomycorrhizal and ericoid mycorrhizal middle European forest plants. PLoS One 10:e0124752. doi:10.1371/journal.pone.012475259
Vohník M, Burdíková Z, Albrechtová J, Vosátka M (2009) Testate amoebae (Arcellinida & Euglyphida) vs. ericoid mycorrhizal and DSE fungi: a possible novel interaction in the mycorrhizosphere of ericaceous plants? Microb Ecol 57:203–214
Vohník M, Burdíková Z, Vyhnal A, Koukol O (2011) Interactions between testate amoebae and saprotrophic microfungi in a Scots pine litter microcosm. Microb Ecol 61:660–668
Acknowledgments
Financial support was provided by the Grant Agency of Charles University in Prague (GAUK 68313/PrF/B-BIO). This study is a part of the long-term research projects of the Institute of Botany ASCR (RVO 67985939) and Faculty of Science, Charles University in Prague (MŠMT LO1417). Lukáš Kalous and Miloslav Petrtýl organized a workshop on marine biology in Borak, Croatia, which substantially stimulated our research at its beginning, Kateřina Rylková and David Vondrášek helped with collection of some root samples, Ivan Župan helped with obtaining a permit for sampling in Croatia (Croatian Ministry of Environmental and Nature Protection, Klasa UP/I-612-07/13-48/48, Urbroj 517-07-1-1-1-13-2), and Symbio-m Ltd. (Lanškroun, Czech Republic) provided a portable flow box for isolation of P. oceanica root mycobionts; all these contributions are greatly acknowledged. The authors also acknowledge inspiring comments of two anonymous reviewers which helped to improve earlier versions of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vohník, M., Borovec, O. & Kolařík, M. Communities of Cultivable Root Mycobionts of the Seagrass Posidonia oceanica in the Northwest Mediterranean Sea Are Dominated by a Hitherto Undescribed Pleosporalean Dark Septate Endophyte. Microb Ecol 71, 442–451 (2016). https://doi.org/10.1007/s00248-015-0640-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-015-0640-5