Abstract
Common occurrence of testate amoebae (TA) in the rhizosphere of mycorrhizal plants indicates existence of yet undocumented ecological interactions, involving three distinct groups of organisms: soil protists, mycorrhizal fungi, and their host plants. This tripartite relationship was to date investigated only to a limited extent, despite its probable importance for processes taking place in the mycorrhizosphere. In this study, we (1) explored spectra of different TA genera naturally associated with the rhizoplane of three autochthonous European Rhododendron species, (2) screened natural fungal colonization of the TA shells occupying the rhizoplane of selected rhododendrons, and (3) carried out two in vitro experiments addressing the question whether TA shells may serve as a nutrient source for ericoid mycorrhizal fungi (ErMF) and dark septate endophytes (DSE). Our field observations indicated that TA regularly associated with the rhizoplane of all screened rhododendrons and that ErMF and/or DSE associated with their roots possibly exploited the TA shells as a nutrient source. We were unable to detect any major differences among the TA spectra from the rhizoplanes with respect to the three Rhododendron species. The spectra were dominated by Diplochlamys, Centropyxis, Cyclopyxis, Euglypha, Trinema, and Assulina. Positive, neutral, and negative associations were found for various TA genera × Rhododendron species combinations. The highest fungal colonization was observed in Centropyxidae and Trigonopyxidae, reaching up to 45% of the shells in the case of Trigonopyxis. In the in vitro experiments, both ErMF Rhizoscyphus ericae and DSE Phialocephala fortinii regularly colonized TA shells, utilizing them as a source of nutrients. We hypothesize a complex relationship between ErMF–DSE and TA. If corroborated, it would represent an interesting nutrient loop in the mycorrhizosphere of ericaceous plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heterotrophic protists (protozoans) represent an important part of soil biota, playing a crucial role in decomposition of organic matter and nutrient–element cycling [2, 13, 19, 55, 58]. Testate amoebae (TA), also known as thecamoebae, testaceans, thecamoebians, or arcellaceans [42], are a polyphyletic group of aquatic or terrestrial heterotrophic protists, whose cytoplasm is enclosed within a discrete shell–testa, which normally consists of one chamber with a single aperture [46]. Most TA belong to one of two major groups: Arcellinida (Amoebozoa) and Euglyphida (Cercozoa) [42].
The diagnostic character of most TA is the architecture and composition of the shell. Several reports were published with respect to the latter character; for instance, Bartoš [12] mentioned that the TA shell was formed by a protein–chitinaceous matrix produced by the TA cytoplasm; Joyon and Charret [34] stated that the shell wall of Hyalosphenia papilio was composed of a mucoprotein; Moraczewski [43] reported that the shell wall of Arcella discoides contained several amino acids, similar to those found in the organic cement of agglutinate marine foraminifera [31]; Ogden and Hedley [46] distinguished proteinaceous, agglutinate, siliceous, and calcareous TA shells. The shells of most TA species are additionally strengthened with organic and/or inorganic particles (xenosomes), e.g., sand grains, diatoms etc. [12, 46].
TA occupy various habitats, including heathlands and peat bogs [42]. In these environments, TA represent an important part of the community of soil protists; for instance, Gilbert et al. [26] found that TA represented 48% of the total microbial biomass (excluding fungi) in a Sphagnum peatland, and Gilbert et al. [27] stated that TA contributed with nearly 14% to the total microbial biomass (including fungi) in a Sphagnum fallax–Carex rostrata fen. Hence, dead TA constitute a considerable pool of nutrients in such nutrient-impoverished habitats, however in an organic form, which is directly unavailable to plants.
The presence of soil protists in the rhizosphere significantly affects plant growth [14] and this effect can be mediated by interactions with mycorrhizal fungi [15]. Similar to protists, mycorrhizal fungi are considered as drivers of nutrient exchange in the rhizosphere, with fundamental effects on plant fitness [50, 51]. The important role of mycorrhizal fungi is pronounced in nutrient-poor habitats where they access organically bound nutrients and make them available to plants. Members of Ericaceae and ericoid mycorrhizal fungi (ErMF) are an excellent example of this phenomenon. Ericaceous plants typically inhabit soils poor in available nitrogen (N) and often dominate vegetation in heathlands and peat bogs [17, 49]. They regularly form ericoid mycorrhiza (ErM) with ErMF, which provide N supply to their hosts [56]. ErMF are able to utilize such complex substrates as peptides [3], proteins [4], chitin [37], fungal mycelium [38], or plant–mycorrhizal necromass [39]. Kerley and Read [39] showed that the typical ErMF Rhizoscyphus ericae (Read) Zhuang and Korf produced extracellular protease and chitinase when grown on mycorrhizal root biomass and used fungal chitin as a source of N, transporting it into host plant tissues. Analogously to plant–mycorrhizal necromass, R. ericae could use TA necromass, a cocktail of organic nutrients containing chitin and proteins, which is abundantly available in peatlands [26, 27], as a substrate for its growth and transport obtained N to host plants in exchange for photosynthetic carbon.
Interactions between soil protists and mycorrhizal fungi comprise dynamic and complex processes that may influence all partners entering the interaction: spore germination, root colonization, growth of fungal mycelium, or sporulation can be affected on the fungal side [23], production of biomass on the plant side [15], and composition of the community [32] on the protist side. Even though investigations of interactions between TA and soil fungi are scarce, they reveal consequences important for ecophysiology of the rhizosphere. Mycophagy is accepted as one of the TA’s nutrition strategies [19]. Couteaux [18] used malt extract to stimulate the growth of fungi in a nonsterile soil and screened the response of the TA community to this manipulation. The biomass of the fungal mycelium remained unaltered, but the incidence of the most abundant TA, Phryganella acropodia, increased with time. This led the author to hypothesize P. acropodia mycophagy, which was further supported by frequent observations of its shells attached to fungal hyphae. Gilbert et al. [28] reported that spores and mycelium of soil fungi represented 36% of the identified prey of the members of the TA complex Nebela tincta major–bohemica–collaris. This suggested that soil fungi, possibly including mycorrhizal, represented an important part of the diet for this species. Ingham and Massicotte [32] found that TA regularly inhabited the mycorrhizosphere of ectomycorrhizal (EcM) roots of five conifers and that their communities quantitatively and qualitatively differed depending on host plants and EcM fungi colonizing their roots. This indicated that the composition of an EcM community might determine the composition of a TA community in the mycorrhizosphere. However, mechanisms of this relationship were unknown. Ingham and Massicotte [32] suggested that mycorrhizal fungi could influence the protist community by controlling–altering the bacterial community in the rhizosphere.
Although reports describing direct interactions between TA and mycorrhizal fungi are lacking, several interesting facts can be inferred from the literature focused on related topics. To summarize, (1) TA represent a quantitatively important part of soil biota in nutrient-poor peatland and forest soil habitats [26, 55]; (2) such habitats are often dominated by ericaceous plants, which regularly form ErM and/or DSE association [= an association with the dark septate endophytes (DSE)] [17, 35, 36, 49]; (3) in such habitats, ErMF and possibly also DSE help plants to access organic nutrients, namely N [41, 51]; (4) mycorrhizal fungi can access animal N [40]; (5) soil fungi represent an important food source for TA [28]. In addition, our previous observations indicate that (6) shells of probably dead TA are regularly associated with mycelium emerging from ericoid mycorrhizal roots (M. Vohník, unpublished data). These facts together indicate a possible complex relationship between mycorrhizal fungi and TA, which might also influence physiology and ecology of mycorrhizal plants.
In the present study, we investigated several aspects of this relationship, focusing mainly on phenomena related to the hypothesis that mycorrhizal fungi can use TA shells as a nutrient source. We describe the composition and similarity of the TA communities found in the rhizoplane of three native European Rhododendron species and quantify the number of TA shells which were colonized by fungal hyphae. Additionally, we report observations of the association between TA shells and fungal mycelium emerging from Rhododendron mycorrhizal roots. In two in vitro experiments, we address the question whether TA shells could serve as a source of nutrients and/or propagule carriers for ErMF R. ericae and DSE Phialocephala fortinii Wang and Wilcox.
Materials and Methods
Occurrence of TA in the Rhizoplane of Three Rhododendron Species
Three European Rhododendron species (Rhododendron hirsutum L., Rhododendron kotschyi Simk., and Rhododendron luteum Sweet) were screened for TA shells occurring in their rhizoplane. The roots of R. hirsutum were collected in June 2005 at Velika Planina plateau, Slovenia (~1,400 m a. s. l.); the roots of R. kotschyi were collected in September 2005 in the Carpathian Mts., Romania (1,724–2,505 m a. s. l.) and the roots of R. luteum were collected in September 2005 in a deciduous forest (tree dominants Fagus sylvatica L., Quercus petraea (Mattusch.) Liebl., Carpinus betulus L., and Castanea sativa Mill.) near the municipality of Boštanj, Slovenia (~220 m a. s. l.). For each species, four to seven individuals were screened. From each individual, three root samples (each containing approximately 15 cm of roots) were taken from the upper soil layer (depth 5–15 cm), inserted in plastic bags, and stored in a fridge (8°C) until screened.
Roots with adhering rhizospheric soil were gently washed under running tap water on a sieve (∅ 1 mm) to remove excessive substrate. Washed roots with adhering rhizoplanic substrate were divided into three parts. The first part was placed into 250-ml flasks with lactoglycerol and stored in the fridge for 1 month. After this period, the rhizoplanic material, which separated from the roots of R. hirsutum (n = 6), R. kotschyi (n = 7), and R. luteum (n = 4) due to gravity, was collected from the bottom of the flasks with plastic pipettes and was screened for TA shells. In each sample, at least 100 shells were collected, determined to the genus level according to Alekperov and Snegovaya [1], Balík [6–11], Bartoš [12], Deflandre [21, 22], Foissner and Korganova [25], Ogden and Hedley [46], and Patterson et al. [47], and percent frequency of each genera was counted. Qualitative and quantitative similarity of the TA spectra among all Rhododendron individuals was assessed by the cluster analysis using the Jaccard index [33, 52] and the Morisita index [44]. The affinity of the TA genera to the three Rhododendron species was assessed using Pearson’s χ 2 test (significant positive–negative association was considered if the absolute value of the normalized standard deviation between a pair of expected and observed frequencies was >3.5; otherwise, the association was considered neutral). BMDP 7.0 (Statistical Solutions; Saugus, USA) and PAST 1.4 [30] were used for statistical analyses.
Natural Colonization of TA Shells by Fungal Hyphae
The TA shells from separated rhizoplanic material were additionally screened for colonization by fungal hyphae. To accomplish this, one individual was randomly chosen per each Rhododendron species. At least 300 shells from its rhizoplane were collected, immersed in 0.05% trypan blue in lactoglycerol (lactic acid–glycerol–deionized water = 1:1:3) to stain fungal mycelium, and observed with an Olympus SZX12 inverted light microscope equipped with the Olympus Relief Contrast (maximum magnification ×400) and an Olympus BX60 light microscope equipped with DIC (maximum magnification ×1,000). A shell was considered as being colonized when fungal hyphae either grew on its surface or inside its lumen. Percentual frequency of the colonized shells was counted for each TA genera.
Association Between TA Shells and Rhododendron Roots
The remaining two parts of the roots were subjected to direct observation of associated TA shells using light microscopy and scanning electron microscopy (SEM). Prior to observation, one part was treated according to the methods commonly used for screening mycorrhizal colonization [16], i.e., autoclaved in 10% KOH for 20 min at 121°C, rinsed in 3% HCl, washed with running tap water, and autoclaved in 0.05% trypan blue in lactoglycerol for 20 min at 121°C and left overnight at room temperature (= the common treatment). With positive results, this treatment is commonly used in our laboratory to screen ErM and DSE association in field samples. The other part of the roots was directly immersed in a solution of 0.05% trypan blue in lactoglycerol and left overnight at room temperature (= the alternative treatment). Roots were destained by immersing in deionized water and screened for the presence of associated shells of TA and mycorrhizal structures using a dissecting microscope (magnification up to ×144). Because there were no TA shells associated with the roots in the common treatment, we further focused only on the roots from the alternative treatment. Here, mycorrhizal roots associated with TA shells via fungal mycelium were cut into 1-cm pieces and mounted onto nonpermanent slides for light microscopy or were directly examined using SEM. Slides were observed at high magnification (×400 or ×1,000) using the Olympus BX60 light microscope equipped with DIC. SEM photographs were taken in the Olympus ESEM™ mode at low temperatures (−6°C to −3°C) using a FEI Quanta 200 microscope. The Olympus ESEM™ mode enables screening of noncollapsed roots without previous treatment (e.g., coating or critical-point drying).
In addition to native Rhododendrons, roots of four rooted stem cuttings (size approximately 10 cm) of Rhododendron cv. Azurro cultivated 4 months in a growth chamber (16–8 h day–night, average temperature 20°C, light intensity 150 μmol m−2 s−1) in a nonsterile peat-based substrate (peat–zeolite–perlite = 1:1:1) were treated in the same manner and screened for association between TA shells and their roots.
Experiment 1—Colonization of TA Shells by P. fortinii and R. ericae
Cyclopyxis and Trigonopyxis shells were extracted with fine forceps from a water suspension of a peat-based substrate, collected from the rhizosphere of Vaccinium myrtillus L. in Modrava, Šumava National Park, Czech Republic in September 2005. These two TA genera were used because of their abundance in the suspension and shell size suitable for manipulation. Moreover, we found that Cyclopyxis and especially Trigonopyxis shells were frequently naturally colonized by fungal hyphae in the rhizoplanic samples from the Rhododendron roots examined in this study (see “Results”), which indicated their suitability for inoculation experiments. Grains of serially washed quartz sand (Provodínské písky Inc., Czech Republic; fraction <1 mm) were used as a negative control and were treated in the same manner as the TA shells.
Extracted shells–sand grains were transferred onto moistened PRAGOPOR 6 nitrocellulose membranes (Ø adjusted to 1 cm, pore size 0.4 μm; Pragochema Ltd., Czech Republic), placed in glass Petri dishes, five shells–grains per membrane, and autoclaved for 20 min at 121°C. Autoclaved membranes with adhering shells–grains were aseptically placed into plastic Petri dishes with water agar (WA).
P. fortinii and R. ericae axenic cultures were precultivated in plastic Petri dishes on WA for 1 month at room temperature in the dark. Small pieces of their mycelium (a few mm3) were obtained from margins of their actively growing colonies and transferred approximately 3 mm from each membrane. P. fortinii was the strain PFO-F (GenBank EF446149), which is deposited in the Culture Collection of Fungi (Department of Botany, Faculty of Science, Charles University in Prague, Czech Republic) under the accession number CCF 3586. It corresponds to the P. fortinii cryptic species CSP7 (= P. fortinii s. s.) sensu Grünig et al. [29] and forms DSE association with ericaceous plants [59]. R. ericae was a culture derived from the strain UAMH 6735 (GenBank AJ319078), isolated by Pearson and Read [48]. It forms ErM with ericaceous plants [60, 61].
There were three treatments (P. fortinii inoculated, R. ericae inoculated, and noninoculated), each containing in total 60 autoclaved TA shells on 12 membranes in four Petri dishes (three membranes in one dish, each membrane with five TA shells) and 60 autoclaved sand grains organized in the same manner. The dishes were sealed and incubated at room temperature in the dark for 2 months. After this period, the shells–grains were extracted with forceps and divided into three parts for each variant, each part containing 20 shells–grains on four membranes. The first part was screened for colonization with P. fortinii or R. ericae mycelium using light and/or scanning electron microscopy. The sand grains were observed by SEM only.
Experiment 2—TA Shells as a Source of Nutrients for P. fortinii and R. ericae
The second part of the shells–grains was aseptically transferred onto new autoclaved moistened nitrocellulose membranes, placed on WA in plastic Petri dishes (∅ 9 cm), sealed, and incubated at room temperature in the dark for 2 months. The third part of the shells–grains was transferred onto new autoclaved moistened nitrocellulose membranes, placed on serially washed quartz sand (the same provenience as the grains) in glass Petri dishes (∅ 5 cm), sealed, and incubated at room temperature in the dark for 2 months. The mycelium emerging from the shells–grains was observed and documented periodically each week. After 2 months, the shells were screened using light and scanning electron microscopy. The grains were observed by SEM only.
Results
Occurrence of TA in the Rhizoplane of Three Rhododendron Species
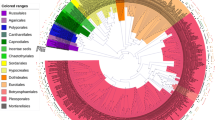
In total, we found 13 genera of TA to be associated with the rhizoplane of R. hirsutum, R. luteum, and R. kotschyi. They were (alphabetically): Arcella, Assulina, Centropyxis, Corythion, Cyclopyxis, Diplochlamys, Euglypha, Heleopera, Nebela, Pseudodifflugia, Tracheleuglypha, Trigonopyxis, and Trinema. Shells of Pseudodifflugia were found only in the R. hirsutum samples; all other TA genera were found in the rhizoplane of all three Rhododendron species (Table 1). On average, we were unable to identify to genus level, approximately 5–6% of the collected TA shells. The average value of the Jaccard index for all Rhododendron individuals was 0.777 (minimum 0.583 for R. hirsutum #1 vs. R. luteum #4 and #5; maximum 1 for R. hirsutum #1 vs. R. kotschyi #3, R. hirsutum #3 vs. R. luteum #3, R. kotschyi #4 vs. R. luteum #7, and R. luteum #4 vs. R. luteum #5). The average value of the Morisita index was 0.697 (minimum 0.276 for R. hirsutum #4 vs. R. luteum #4; maximum 0.969 for R. hirsutum #1 vs. R. luteum #3). The cluster analysis separated the observed TA spectra into two main clades, where different individuals of the three Rhododendron species were intermingled (Fig. 1). According to the Pearson’s χ 2 test, Diplochlamys was positively and Corythion, Nebela, and Tracheleuglypha negatively associated with the rhizoplane of R. hirsutum; Corythion, Tracheleuglypha, and Trigonopyxis positively and Trinema negatively associated with the rhizoplane of R. luteum; and Trinema and Nebela positively and Tracheleuglypha negatively associated with the rhizoplane of R. kotschyi (Table 1).
Cluster analysis of the spectra of the TA genera found in the rhizoplane of different individuals of the three Rhododendron genera. The UPGMA algorithm and the Morisita’s index were used to create the tree. For details, see “Materials and Methods”
Natural Colonization of TA Shells by Fungal Hyphae
The highest natural fungal colonization was found among shells of relatively scarce Trigonopyxis (approximately 45%, 42%, and 44% of the shells in the rhizoplane of R. hirsutum, R. luteum, and R. kotschyi, respectively), followed by Cyclopyxis (34%, 32%, and 36%) and Centropyxis (34%, 29%, and 32%). The shells of some TA genera differed in the fungal colonization with respect to Rhododendron species, whereas the colonization of others was similar in all Rhododendron rhizoplanes (Table 1).
Association Between TA Shells and Rhododendron Roots
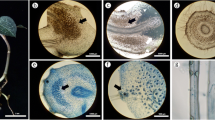
We documented the association between TA shells and roots via the mycelium of putative mycorrhizal fungi in all samples of all three Rhododendron species and Rhododendron cv. Azurro in the alternative treatment. We estimated that there was at least one TA shell associated with the mycorrhizal root by means of the fungal mycelium per 5 cm of the total root length. We found no associated TA shells in the samples from the common treatment. In some cases, TA shells appeared to be only loosely attached to the root surface by means of the fungal mycelium (Fig. 2a); in other cases, however, the association between TA shells and the root appeared to be very intimate (Fig. 2b). TA shells were frequently embedded in the fungal mycelium, often being partially decomposed (Fig. 2c), which substantially hampered their identification. Some of the objects with the TA shells’ appearance were filled with darkly pigmented thick-walled cells (Fig. 2d), which resembled the colonization pattern of the DSE P. fortinii (see below). All screened ericaceous plants developed ErM colonization and also possessed DSE association.
a A disintegrated unidentified TA shell loosely associated with the mycorrhizal root of Rhododendron cv. Azurro via fungal hyphae emerging from the root (arrow). SEM; bar = 10 μm. b A TA shell (cf. Centropyxis minuta) associated with the mycorrhizal root of Rhododendron cv. Azurro by means of the fungal hyphae. SEM; bar = 50μm. The detail shows the hyphae connecting the shell with the mycorrhizal root (arrows). SEM; bar = 25 μm. c Partially decomposed unidentified TA shell, surrounded by fungal hyphae (arrows). SEM; bar = 50 μm. d An object from the root surface of Rhododendron cv. Azurro, which resembles a TA shell colonized by fungal hyphae. The object is associated with a dark septate hypha (arrow) and filled with a septate multicellular structure, which shows similarity to microsclerotia formed by DSE fungi either in TA shells (Fig. 3a) or rhizodermal cells of higher plants (Fig. 3e). DIC; bar = 50 μm. e A T. arcula shell colonized by the mycelium of R. ericae, incubated on WA for 6 weeks. SEM; bar = 50 μm. f A T. arcula shell colonized by the mycelium of P. fortinii, incubated on WA for 6 weeks. Contrary to R. ericae, there is only loose net of hyphae covering the shell’s surface and the P. fortinii hyphae enter the shell’s lumen via its aperture (arrow). SEM; bar = 50 μm
Experiment 1—Colonization of TA Shells by P. fortinii and R. ericae
Both Cyclopyxis and Trigonopyxis shells were colonized by the mycelium of both ErMF R. ericae and DSE P. fortinii. Commonly, R. ericae mycelium almost completely covered the surface of the colonized shells (Fig. 2e). P. fortinii was slower in colonization and its mycelium usually did not cover the whole surface of the shells (Fig. 2f). P. fortinii hyphae often entered the shell via its aperture (Fig. 2f). The intracellular colonization of such shells consisted of short, thick, darkly colored, and thick-cell-walled hyphae, which usually occupied the whole lumen of the shell (Fig. 3a). This was connected with notable color change—the shells colonized intracellularly by P. fortinii were dark brown to black, which contrasted with the yellowish to light brownish color of the shells, which were colonized only superficially. Such intracellular colonization resembled microsclerotia, formed by DSE in the roots of higher plants (Fig. 3e). The surface of the sand grains was only poorly colonized by either single hyphae or a very loose weft of the mycelium of P. fortinii or R. ericae.
a A detail of a T. arcula shell intracellularly colonized by the mycelium of P. fortinii. The colonization pattern, i.e., dark, short, and thick cells forming dense coiled hyphae, resembles microsclerotia formed by DSE fungi in the rhizodermal cells of higher plants (e). SEM; bar = 1 μm. The detail shows general appearance of the colonized shell. DIC; bar = 10 μm. b A T. arcula shell as a propagule carrier and a nutrient source for the P. fortinii mycelium. The preinoculated shell was cultivated on a nitrocellulose membrane placed on water agar (WA) for 2 months. The mycelium emerges in all directions from the shell’s lumen and completely disintegrates its structure and shape. SEM; bar = 100 μm. The detail shows a sand grain, which served as a negative control. Nearly no mycelium developed from the preinoculated grain. SEM; bar = 100 μm. c A T. arcula shell as a propagule carrier and a nutrient source for the R. ericae mycelium. The preinoculated shell was cultivated on a nitrocellulose membrane placed on WA for 2 months. The shell is completely covered by the R. ericae mycelium, which partly disintegrates its structure. SEM; bar = 50 μm. d A T. arcula shell as a propagule carrier and a nutrient source for the P. fortinii mycelium. The preinoculated shell (arrow) was cultivated on a nitrocellulose membrane placed on moistened, serially washed quartz sand for 2 months. The mycelium radiates from the shell across the membrane. A dissecting microscope; bar = 250 μm. The detail shows the same situation using SEM. Bar = 100 μm. e Microsclerotia formed by P. fortinii in the rhizodermal cells of V. myrtillus L. in an aseptic culture. DIC; bar = 25 μm. f A T. arcula shell collected from the rhizosphere of V. myrtillus from the field. Note dark septate hyphae attached to its surface (arrows). DIC; bar = 50 μm
Experiment 2—TA Shells as a Source of Nutrients for P. fortinii and R. ericae
After transferring the P. fortinii- and R. ericae-precolonized shells–grains onto new membranes and during their 2-month cultivation on WA, both fungi were able to utilize the TA shells as a source of nutrients. This was indicated by vigorous growth of their hyphae, which radiated from the precolonized shells and lack of such growth from the precolonized sand grains. Vigorous growth was notable especially for the hyphae of P. fortinii, which expanded from the dark-colored precolonized shells in all directions, covering the whole surface of the shells and completely disorganizing their shape with progressing time (Fig. 3b). Also, the mycelium of R. ericae completely covered the surface of the precolonized shells and partially disorganized their shape with progressing time (Fig. 3c).
When the membranes with the precolonized shells–grains were transferred into the dishes with serially washed quartz sand, a similar situation occurred: the precolonized shells gave rise to new abundant P. fortinii or R. ericae mycelium (Fig. 3d), which was not the case of the precolonized sand grains. The dark-brown- to black-colored shells, which were intracellularly colonized by P. fortinii as described above, gave rise to more abundant mycelium than the yellow- to light-brown-colored shells, which lacked significant intracellular colonization.
Discussion
To date, the relationship between soil protists and fungi has not been frequently investigated. One direction of the research was focused on investigations of communities of protists in the rhizosphere. With this respect, we are aware of two papers relevant to our study: Ingham and Massicote [32] and recently published Sutton and Wilkinson [57].
Ingham and Massicote [32] explored ectomycorrhizospheres of Pinus ponderosa, Pseudotsuga menziesii, Picea sitchensis, Tsuga heterophylla, and Abies grandis and identified six types of protist communities, which always included ciliates, TA, naked amoebae, and flagellates but differed qualitatively and quantitatively with respect to different host plants and different symbiotic fungi. Protists generally tended to be more abundant and diverse in roots colonized by fungi from the EcM genera Rhizopogon than in roots symbiotic with EcM fungus Thelephora terrestris or DSE fungi from the Mycelium radicis atrovirens complex (formed predominantly by P. fortinii cryptic species) or in nonmycorrhizal roots. TA were relatively poor in species number and abundance, especially when compared with flagellates, and included Nebela, Euglypha, Valkanovia, Difflugia, Trinema, Corythionopsis, and Arcella, the first genera being the most abundant across all combinations of host species and mycorrhizal fungi.
Contrary to Ingham and Massicote [32], we found no clear differences in the TA spectra among the rhizoplanes of the three Rhododendron species. Additionally, these frequencies differed qualitatively and quantitatively from those found in the coniferous ectomycorrhizospheres. This discrepancy might be caused by several factors because designs of both studies varied in many ways. We focused on environmental samples, whereas the results of Ingham and Massicote were derived from a greenhouse inoculation experiment. We examined ErM- and DSE-colonized hair roots of mature ericaceous plants from a single genus, whereas the other study targeted EcM roots of young coniferous seedlings from several genera. Not least, ericaceous plants associate with unique spectrum of fungi, which does not include EcM fungi from the Rhizopogon genera. Moreover, in contrast to EcM fungi, ErMF or DSE do not coat root tips with a compact fungal sheath; it is very likely that, in our study, TA had direct access to the root surface and its products whereas, in the other study, TA interacted mostly with the fungal mantle. On one hand, these facts seriously hamper comparison of the results; on the other, they emphasize the need for further screening of preferably environmental root samples of plants from various ecosystems and with different mycotrophy, to make future comparisons possible.
Recently, Sutton and Wilkinson [57] published an interesting study elucidating the effects of an invasive shrub Rhododendron ponticum L. on TA communities in woodland soils in NW England. They investigated six urban broadleaved woods in the Liverpool area, comparing sites with presence–absence of the R. ponticum cover, and found that its presence apparently modified the TA community living in the soil (the Rhododendron-affected soils had a smaller number of TA taxa along with a smaller total number of individual TA than the non-Rhododendron soils).
Sutton and Wilkinson [57] claimed that their study provided “circumstantial evidence of a role for Rhododendron leaf chemistry in affecting soil living microbes.” In other words, Rhododendron leaf litter, which was the only (or at least the most frequent) ground cover in the studied sites (D. M. Wilkinson, personal communication), was supposed to be the major force driving composition of the TA communities below the R. ponticum shrubs. It is difficult to estimate such an effect in our study; unlike R. ponticum, both R. hirsutum and R. kotschyi are small leaved and form relatively low shrubs surrounded by grass vegetation. From the three Rhododendron species examined in this study, R. luteum was the most similar to R. ponticum, both by its size and habitat preference. However, at the study locality in Slovenia, its leaves were not a dominant part of leaf litter. Thus, we suppose that, in our study, the effect of the Rhododendron leaf litter on TA communities was relatively marginal. However, various effects of Rhododendron leaf litter on surrounding vegetation and its mycorrhizae are expected [although not always evident in field: see Nilsen et al. [45] and references therein] and Sutton and Wilkinson [57] showed its relevance also for TA ecology.
The soils from below the R. ponticum shrubs in Sutton and Wilkinson [57] contained the following TA genera (total numbers of individuals in presence–absence of R. ponticum across all six studied sites in brackets): Hyalosphenia (106/76), Trinema (84/103), Corythion (66/68), Centropyxis (58/76), Euglypha (38/112), Trigonopyxis (18/1), Difflugia (7/23), and Nebela (7/7); all except Hyalosphenia and Difflugia were detected also in our study. The authors found that two TA species, Corythion dubium and Trigonopyxis arcula, were positively associated with the Rhododendron-affected soils. Our results show that Corythion and Trigonopyxis positively associated with the rhizoplane of R. luteum (not R. hirsutum or R. kotschyi), the relatively most similar species to R. ponticum, as already stated. It is of interest that, already two decades before the studies of Sutton and Wilkinson [57] and ours, Foissner [24] suggested that Trigonopyxis and Corythion were positively associated with mor soils, a common soil type under rhododendrons [20].
Our root samples originated from different Rhododendron species and were collected in different environments. Then, why were the average frequencies of different TA genera relatively similar? If we omit the effect of Rhododendron leaf litter [57], the answer could be in the conclusion that similar protist communities could be on roots of different host plants but with the same mycorrhizal fungi [32]. We have not identified the respective fungi colonizing the screened roots; on the other hand, it is assumed that ErM and DSE association are formed by only limited number of symbiotic fungi and that most ErM and DSE associations are formed by R. ericae and P. fortinii, respectively [56]. As a consequence, relatively uniform spectrum of ErM–DSE fungi could attract a relatively uniform spectrum of TA. Alternatively, and possibly more accurately, the similarity of the TA spectra in the Rhododendron rhizoplanes could be the result of general uniformity of ericaceous roots [49] and environments they inhabit [17]. Apparently, further investigations are needed on the relative importance of microenvironment versus host plant and root-inhabiting fungi identity as a determinant of TA community structure.
The other direction of the protists vs. soil fungi research examined trophic relationships between both groups of organisms. Couteaux [18] investigated the relationship between TA community and soil fungi by addition of malt extract (ME) into experimental microcosms. She concluded that one of the TA species monitored in her study, P. acropodia, was mycophagic, i.e., consumed mycelium of soil fungi, growth of which was stimulated by introduction of ME. Gilbert et al. [28] drew additional attention to the nutritional importance of fungal mycelium for soil protists by showing that it could represent a considerable part (up to 36%) of the diet of some TA species. In contrast, we had focused on the trophic relationship from the fungal perspective and showed that TA shells collected from environmental root samples were regularly colonized by fungal mycelium, and the colonization in some genera reached considerable frequency. Moreover, we found that relatively large Trigonopyxis (but not smaller Corythion) shells, which were positively associated with the rhizoplane of R. luteum (this study) and the rhizosphere of relatively similar R. ponticum [57], had the highest colonization by fungal hyphae. Such possible preferential association (and fungal colonization) might give a chance for evolution of more complex relationships between TA and its “host” plants and/or their symbiotic fungi. On the other hand, Trigonopyxis maintained high colonization level also in the rhizoplanes of the two remaining Rhododendron species, and we did not find a tendency of the other highly colonized TA genera (Centropyxis, Cyclopyxis) to positively associate with any Rhododendron rhizoplane. A species-level taxonomic resolution might clear this discrepancy; Sutton and Wilkinson [57] found that Corythion dubium but not Corythion pulchellum and Corythion–Trinema spp. were positively associated with the Rhododendron-affected soils. Similar situation might be with different species of Centropyxis and Cyclopyxis.
The discussed findings point at frequent interactions between TA and soil fungi. From the fungal point of view, interactions with TA may have two different modes of functioning: passive and active. In the former mode, fungal hyphae (dead or alive) are only mechanically glued to alive TA shells and are de facto organic xenosomes. Slowly moving TA could serve as carriers for vital fungal propagules, which, attached to the surface, wait for their chance to rapidly colonize the shell after its death. In the latter mode, there is an indication of active fungal growth towards–from the shell and we can expect that the fungus immediately explores the shell (or its parts) as a nutrient source. From the TA (or rather the TA researcher) point of view, frequent and genera-specific fungal colonization might hamper proper assessment of TA community composition. The identified Trigonopyxis shells were represented relatively poorly in our rhizospheric samples, which could reflect either their genuine scarcity or the possibility that they were decomposed or deformed by fungi more frequently–rapidly than other genera. This might be true especially for TA with larger shells (Centropyxis, Cyclopyxis, Trigonopyxis, etc.) since these appear to be colonized more frequently than genera with smaller shells (see “Results”). Indeed, it would make more sense for the fungi to preferentially infect larger shells, as these will presumably contain more resources for the fungi to use. On the other hand, it might be that larger colonized shells are more likely to be recognized as such. Clearly, future investigations of TA communities from mycorrhizospheres should consider the fact that fungal colonization could rapidly and totally alter shape of TA shells.
Nutrients obtained by soil fungi from TA shells could be transported directly to the host plant in exchange for photosynthetic carbon if the fungi were mycorrhizal and associated with roots. We showed that direct connections between presumably dead TA and ericaceous roots by means of symbiotic fungi, which is a prerequisite for the hypothetical nutrient flow, were relatively common in the ericaceous rhizoplane. The magnitude of this nutrient exchange remains to be determined. However, the relatively high incidence of such connections and high fungal colonization of some TA shells in the mycorrhizosphere indicate that the “TA–root symbiotic fungi–host plants” association is not an exceptional curiosity. Subsequently, a question arose: Why had this association not been reported earlier? We see two probable reasons: (1) the common treatment (see “Materials and Methods”) prevents observation of fragile TA shells and their more fragile association with the fungal mycelium emerging from the root and (2) colonization by ErMF and DSE notably changes shape of the colonized shells. To date, TA species are identified mostly using the morphology of their shells [42, 46]; very likely, those colonized by soil fungi, especially in later stages of decomposition, would not be recognized as TA at all (cf. Fig. 2c and Fig. 3b, c).
The proposed nutrient flow between TA and plants by means of mycorrhizal fungi has two steps: nutrient uptake by the fungus followed by transport to the plant. We had focused on the first step and demonstrated in vitro that TA shells represented an acceptable nutrient source for ErMF R. ericae and DSE P. fortinii. It could be argued that WA or the membranes (see “Materials and Methods”) served as an additional source of nutrients for the mycelium expanding from the shells. However, it was shown that the nitrocellulose membranes used in the experiments were resistant to microbial degradation [5] and the results from WA were repeated on serially washed quartz sand. Moreover, the differences between the growth of mycelium from the preinoculated shells and the sand grains indicated that both fungi did utilize the shells as a source of nutrients. Certainly, it would be interesting to determine which parts of the TA shell serve as the actual source of nutrients for the colonizing fungus. It could be a wide range of organic compounds forming the shell or even the cytoplasm of the amoeba. With this respect, it appeared that ErMF and DSE used different colonization strategies: while the ErMF readily colonized all available shells, suggesting utilization of the whole TA necromass, the DSE appeared to ignore empty shells, suggesting preference of the organic matter of the cell in the lumen to the organic substances forming the shell. Primarily triggered by organic compounds, P. fortinii later developed multicellular structures consisting of short, thick-walled hyphae inside the lumen of some shells. Such structures notably resembled intracellular microsclerotia formed by DSE in roots of higher plants [36]. Thus, a dead TA shell could represent a feasible shelter for reproductive structures of DSE, similar to root cells for microsclerotia or cavities of dead seeds for spores of arbuscular mycorrhizal fungi [53, 54].
Our observations and findings together with the published papers on related topics (see “Introduction”) bring support to the existence of the second step: transport of nutrients derived from TA shells to host plants. However, its corroboration will depend on a direct demonstration, probably with the help of isotopically labeled compounds. Likewise, it remains to be established whether such highly probable interaction has any significant effect on the host plant. With this respect, we might parallel our deductions to a nutrient cycle demonstrated by Klironomos and Hart [40], who investigated a tripartite relationship between Pinus strobus, an EcM fungus Laccaria bicolor, and a springtail Folsomia candida in a microcosmos experiment. The authors reported that P. strobus was able to derive up to 25% of its N from either dead or live soil-dwelling arthropods via its fungal partner. Interestingly, L. bicolor was able to immobilize the animals before infecting them—do ErMF or DSE have similar capabilities towards TA? As the authors concluded, “EcM plants could indirectly depredate soil arthropods for a significant source of nitrogen through their fungal partners”—could ErM plants derive a part of their nitrogen from TA in a similar way? According to Klironomos and Hart, their results revealed “a nitrogen cycle of far greater flexibility and efficiency than was previously assumed, where the fungal partner uses animal-origin nitrogen to ‘barter’ for the carbon from the host tree.” However, it remained unclear whether the observed phenomenon was widespread or functional under natural conditions. In contrast, our observations and results suggest that the interaction between TA and root-symbiotic fungi is both widespread and functional under natural conditions. This might have profound impacts on all partners entering this interaction, as mentioned throughout the discussion. Hopefully, our report will encourage further investigations to elucidate ecological significance of the proposed “TA–mycorrhizal fungi–host plants” bidirectional nutrient loop.
References
Alekperov I, Snegovaya N (2000) The fauna of testate amoebae (Rhizopoda, Testacea) in freshwater basins of Apsheron peninsula. Protistology 1:135–147
Aoki Y, Hoshino M, Matsubara T (2007) Silica and testate amoebae in a soil under pine-oak forest. Geoderma 142:29–35
Bajwa R, Read DJ (1985) The biology of mycorrhiza in the Ericaceae: IX. Peptides as nitrogen sources for mycorrhizal and non-mycorrhizal plants. New Phytol 101:459–467
Bajwa R, Abuarghub S, Read DJ (1985) The biology of mycorrhiza in the Ericaceae. X. The utilization of proteins and the production of proteolytic enzymes by mycorrhizal plants. New Phytol 101:469–486
Baláž M, Vosátka M (2001) A novel inserted membrane technique for studies of mycorrhizal extraradical mycelium. Mycorrhiza 11:291–296
Balík V (1990) Zur Kenntnis der Bodentestaceen (Rhizopoda: Testacea) Südböhmens. Act Mus Boh Meridion in Č. Budějovice 30:1–12 [in Czech with German summary]
Balík V (1992a) Krytenky (Rhizopoda, Testacea) z několika lokalit z chráněné krajinné oblasti Beskydy (Severní Morava, ČSFR). Čas Slez Muz Opava 41:31–40 [in Czech with English summary]
Balík V (1992b) Testate amoebae fauna (Protozoa, Rhizopoda, Testacea) from the marsh habitats in the Krkonoše Mountains National Park (Czechoslovakia). Opera Corcontica 29:139–154 [in Czech with English summary]
Balík V (1992c) Die Testaceen (Rhizopoda, Testacea) aus dem Naturschutzgebiet Trojmezná hora im Böhmerwald (Tschechoslowakei). Act Mus Boh Meridion in Č. Budějovice 32:69–78 [in Czech with German summary]
Balík V (1994) Testaceenfauna (Protozoa, Rhizopoda, Testacea) aus dem Naturschutzgebiet Žofínský prales (Novohradské hory Gebirge) in Südböhmen (Tschechische Republik). Act Mus Boh Meridion in Č. Budějovice 34:55–67 [in Czech with German summary]
Balík V (1997) Fauna krytenek (Protozoa, Testacea) západní části Tatranského Národního Parku (Slovenská Republika). Štúdie o Tatranskom Národnom Parku 2:103–122 [in Czech]
Bartoš E (1954) Koreňonožce radu Testacea. Vydavaťelstvo Slovenskej Akadémie Vied, Bratislava, pp 1–185 [in Slovak]
Bonkowski M (2004) Protozoa and plant growth: the microbial loop in soil revisited. New Phytol 162:617–631
Bonkowski M, Cheng W, Griffiths BS, Alphei J, Scheu S (2000) Microbial–faunal interactions in the rhizosphere and effects on plant growth. Eur J Soil Biol 36:135–147
Bonkowski M, Jentschke G, Scheu S (2001) Contrasting effects of microbial partners in the rhizosphere: interactions between Norway Spruce seedlings (Picea abies Karst.), mycorrhiza (Paxillus involutus (Batsch) Fr.) and naked amoebae (protozoa). Appl Soil Ecol 18:193–204
Brundrett M, Bougher N, Dell B, Grove T, Malajczuk N (1996) Working with mycorrhizas in forestry and agriculture. ACIAR Monograph 32. ACIAR, Canberra, p 374
Cairney JWG, Meharg AA (2003) Ericoid mycorrhiza: a partnership that exploits harsh edaphic conditions. Eur J Soil Sci 54:735–740
Coûteaux M-M (1985) Relationships between testate amoebae and fungi in humus microcosms. Soil Biol Biochem 17:339–345
Coûteaux M-M, Darbyshire JF (1998) Functional diversity amongst soil protozoa. Appl Soil Ecol 10:229–237
Cross JR (1975) Rhododendron ponticum L. J Ecol 63:345–364
Deflandre G (1929) Le genre Centropyxis Stein. Archiv für Protistenkunde 67:322–375 [in French]
Deflandre G (1936) Etude monographique sur le genre Nebela Leidy. Annales de Protistologie 5:201–286 [in French]
Fitter AH, Garbaye J (1994) Interactions between mycorrhizal fungi and other soil organisms. Plant Soil 159:123–132
Foissner W (1987) Soil protozoa: fundamental problems, ecological significance, adaptations in Ciliates and Testaceans, bioindicators, and guide to the literature. Prog Protistol 2:69–212
Foissner W, Korganova GA (2000) The Centropyxis aerophila complex (Protozoa: Testacea). Acta Protozool 39:257–273
Gilbert D, Amblard C, Bourdier G, Francez A-J (1998a) The microbial loop at the surface of a peatland: structure, function, and impact of nutrient input. Microb Ecol 35:83–93
Gilbert D, Amblard C, Bourdier G, Francez A-J (1998b) Short-term effect of nitrogen enrichment on the microbial communities of a peatland. Hydrobiologia 373–374:111–119
Gilbert D, Mitchell EAD, Amblard C, Bourdier G, Francez A-J (2003) Population dynamics and food preferences of the testate amoeba Nebela tincta major-bohemica-collaris complex (Protozoa) in a Sphagnum peatland. Acta Protozool 42:99–104
Grünig CR, McDonald BA, Sieber TN, Rogers SO, Holdenrieder O (2004) Evidence for subdivision of the root-endophyte Phialocephala fortinii into cryptic species and recombination within species. Fungal Genet Biol 41:676–687
Hammer R, Harper DAT, Ryan PD (2001) PAST: Palaeontological Statistics software package for education and data analysis. Palaeontol Electronica 4:9
Hedley RH (1963) Cement and iron in the arenaceous foraminifera. Micropaleontology 9:433–441
Ingham ER, Massicotte HB (1994) Protozoan communities around conifer roots colonized by ectomycorrhizal fungi. Mycorrhiza 5:53–61
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270 [in French]
Joyon L, Charret R (1962) Sur l’ultrastructure du Thécamoebien Hyalosphenia papilo (Leidy). C R Acad Sci 255:2661–2663 [in French]
Jumpponen A (2001) Dark septate endophytes—are they mycorrhizal? Mycorrhiza 11:207–211
Jumpponen A, Trappe JM (1998) Dark septate endophytes: a review of facultative biotrophic root-colonizing fungi. New Phytol 140:295–310
Kerley SJ, Read DJ (1995) The biology of mycorrhiza in the Ericaceae. XVIII. Chitin degradation by Hymenoscyphus ericae and transfer of chitin–nitrogen to the host plant. New Phytol 131:369–375
Kerley SJ, Read DJ (1997) The biology of mycorrhiza in the Ericaceae. XIX. Fungal mycelium as a nitrogen source for the ericoid mycorrhizal fungus Hymenoscyphus ericae and its host plant. New Phytol 136:691–701
Kerley SJ, Read DJ (1998) The biology of mycorrhiza in the Ericaceae. XX. Plant and mycorrhizal necromass as nitrogenous substrates for ericoid mycorrhizal fungus Hymenoscyphus ericae and its host. New Phytol 139:353–360
Klironomos JN, Hart MM (2001) Animal nitrogen swap for plant carbon. Nature 410:651–652
Mandyam K, Jumpponen A (2005) Seeking the elusive function of the root-colonising dark septate endophytic fungi. Stud Mycol 53:173–189
Mitchell EAD, Charman DJ, Warner BG (2008) Testate amoebae analysis in ecological and paleoecological studies of wetlands: past, present and future. Biodivers Conserv doi:10.1007/s10531-007-9221-3
Morcazewski J (1961) Testacea du littoral peu profound du lac Kisajno (Region des lacs de Mazurie). Polskie Archiwum Hydrobiol 9:175–194 [in Polish and French]
Morisita M (1959) Measuring the dispersion of individuals and analysis of the distributional patterns. Mem Fac Sci Kyushu Univ Ser E (Biol) 2:215–233
Nilsen ET, Walker JF, Miller OK, Semones SW, Lei TT, Clinton BD (1999) Inhibition of seedling survival under Rhododendron maximum (Ericaceae): could allelopathy be a cause? Am J Bot 86:1597–1605
Ogden CG, Hedley RH (1980) An atlas of freshwater testate amoebae. Oxford University Press, Oxford, p 222
Patterson RT, Dalby A, Kumar A, Henderson LA, Boudreau EA (2002) Arcellaceans (thecamoebians) as indicators of land-use change: settlement history of the Swan Lake area, Ontario as a case study. J Paleolimn 28:297–316
Pearson V, Read DJ (1973) The biology of mycorrhiza in the Ericaceae. I. The isolation of the endophyte and synthesis of mycorrhiza in aseptic culture. New Phytol 72:371–379
Read DJ (1996) The structure and function of the ericoid mycorrhizal root. Ann Bot 77:365–374
Read DJ, Perez-Moreno J (2003) Mycorrhizas and nutrient cycling in ecosystems—a journey towards relevance? New Phytol 157:475–492
Read DJ, Leake JR, Perez-Moreno J (2004) Mycorrhizal fungi as drivers of ecosystem processes in heathland and boreal forest biomes. Can J Bot 82:1243–1263
Real R, Vargas JM (1996) The probabilistic basis of Jaccard’s index of similarity. Syst Biol 45:380–385
Rydlová J, Vosátka M (2000) Sporulation of symbiotic arbuscular mycorrhizal fungi inside dead seeds of a non-host plant. Symbiosis 29:231–248
Rydlová J, Batkhuugyin E, Vosátka M (2004) Sporulation of four arbuscular mycorrhizal fungi isolates inside dead seed cavities and glass capillaries. Symbiosis 36:269–284
Schröter D, Brussaard L, De Deyn G, Poveda K, Brown VK, Berg MP, Wardle DA, Moore J, Wall DH (2004) Trophic interactions in a changing world: modelling aboveground–belowground interactions. Basic Appl Ecol 5:515–528
Smith SE, Read DJ (1997) Mycorrhizal Symbiosis, 2nd edn. Academic, London, pp 323–345
Sutton CA, Wilkinson DM (2007) The effects of Rhododendron on testate amoebae communities in woodland soils in North West England. Acta Protozool 46:333–338
Vargas R (1990) Avances en microbiologia de suelos: los protozoarios y su importancia en la mineralizacion del nitrogeno. Agronomía Costarricense 14(1):121–134 [in Spanish]
Vohník M, Lukančič S, Bahor E, Regvar M, Vosátka M, Vodnik D (2003) Inoculation of Rhododendron cv. Belle-Heller with two strains of Phialocephala fortinii in two different substrates. Folia Geobotanica 38:191–200
Vohník M, Albrechtová J, Vosátka M (2005) The inoculation with Oidiodendron maius and Phialocephala fortinii alters phosphorus and nitrogen uptake, foliar C:N ratio and root biomass distribution in Rhododendron cv. Azurro. Symbiosis 40:87–96
Vohník M, Fendrych M, Albrechtová J, Vosátka M (2007) Intracellular colonization of Rhododendron and Vaccinium roots by Cenococcum geophilum, Geomyces pannorum and Meliniomyces variabilis. Folia Microbiologica 52:407–414
Acknowledgments
This study was supported by the Academy of Sciences of the Czech Republic (research projects AV0Z60050516 and AV0Z50110509) and Ministry of Education, Youth, and Sports of the Czech Republic (research program LC06063). M. Vohník was financially supported by the Grant Agency of the Czech Republic (GACR 206/03/H137). The authors thank Dominik Vodnik, Simon Lukančič, and Matyáš Fendrych for help with collection of the environmental samples, Vladimír Balík and Alena Kubátová for useful information at the beginning of the work and Edward A. D. Mitchell, David M. Wilkinson, and anonymous reviewers for valuable comments on earlier versions of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vohník, M., Burdíková, Z., Albrechtová, J. et al. Testate Amoebae (Arcellinida and Euglyphida) vs. Ericoid Mycorrhizal and DSE Fungi: A Possible Novel Interaction in the Mycorrhizosphere of Ericaceous Plants?. Microb Ecol 57, 203–214 (2009). https://doi.org/10.1007/s00248-008-9402-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-008-9402-y