Abstract
Bisphenol A (BPA) is one of the endocrine-disrupting chemicals that are ubiquitous in aquatic environments. Biodegradation is a major way to clean up the BPA pollution in sediments. However, information on the effective BPA biodegradation in anaerobic sediments is still lacking. The present study investigated the biodegradation potential of BPA in river sediment under nitrate- or sulfate-reducing conditions. After 120-day incubation, a high removal of BPA (93 or 89 %) was found in sediment microcosms (amended with 50 mg kg−1 BPA) under these two anaerobic conditions. Illumina MiSeq sequencing analysis indicated that Proteobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Gemmatimonadetes, and Actinobacteria were the major bacterial groups in BPA-degrading sediments. The shift in bacterial community structure could occur with BPA biodegradation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Mass consumption of bisphenol A (BPA)-containing products has resulted in a widespread contamination of BPA in various aquatic environments, such as municipal wastewater [20, 23], river water and sediment [11, 28, 29, 35], lake water and sediment [24], groundwater [23], and tap water [10, 23]. The ubiquity of BPA in aquatic environments has aroused increasing eco-environmental concerns, due to its estrogenic effect on both human beings and other organisms [8, 42]. Microbial degradation plays a major role in the attenuation of BPA in natural environments. So far, aerobic BPA biodegradation in bulk water and sediment has been well-documented [7, 13, 17, 18, 40]. Moreover, aerobic BPA degraders belonging to several different bacterial genera have been isolated from natural aquatic ecosystems, such as Pseudomonas [7, 17], Bacillus [7], Novosphingobium [31], Sphingomonas [26], and Streptomyces [19]. In addition, anaerobic degradation in sediment ecosystems can be of greater importance for the cleanup of BPA pollution because they are mainly anaerobic. However, it remains unclear whether or not a significant attenuation of BPA in natural environments can occur under anaerobic conditions. Several previous studies suggested that BPA biodegradation in surface water or sediment could be hampered under anaerobic conditions [7, 17, 32]. It is usually hard to isolate pollutant degraders from anaerobic ecosystems. To date, only an anaerobic BPA-degrading bacterium (Bacillus sp. GZ) has been isolated from the environment [21].
The abatement of contamination in the environment is carried out by the collaborative role of a whole microbial community, instead of the isolated degraders alone [12, 36, 40]. Knowledge of BPA-degrading sediment microbial community can aid in our better understanding of BPA biodegradation in aquatic environments [40]. Therefore, the present study was to investigate the biodegradation potential of BPA in river sediment under anaerobic conditions. The structure of bacterial community associated with BPA degradation and its shift was also characterized.
Materials and Methods
Microcosm Setup
The river sediment (0–10-cm depth) used for anaerobic biodegradation tests was collected from the Beiyun Canal (Beijing), which was polluted by the discharge from municipal wastewater treatment plant. The sediment was stored at 4 °C after being air-dried and homogenized. The sediment sample (pH 7.6) contained organic matter of 6.3 g kg−1. At the time of collection, the BPA level in river sediment was nearly 1.5 mg kg−1. The mineral salt medium (pH 7.0) used in the biodegradation experiments was prepared as previously described [6], including (g L−1): NH4Cl (2.7), MgCl2·6H2O (0.1), CaCl2 (0.08), FeCl2·4H2O (0.02), K2HPO4 (0.27), KH2PO4 (0.35), and resazurin (0.001). Each microcosm contained mineral salt medium (10 mL), sediment (3 g, dry weight), resazurin (1 mg L−1), and supplementary electron acceptor (NaNO3 or Na2SO4, 20 mmol L−1) in a 150-mL serum bottle. In this study, six different treatments in triplicate were carried out: (A) sediment +20 mmol L−1 NaNO3, (B) sterilized sediment +50 mg kg−1 BPA +20 mmol L−1 NaNO3, (C) sediment +50 mg kg−1 BPA +20 mmol L−1 NaNO3, (D) sediment +20 mmol L−1 Na2SO4, (E) sterilized sediment +50 mg kg−1 BPA +20 mmol L−1 Na2SO4, (F) sediment +50 mg kg−1 BPA +20 mmol L−1 Na2SO4, and (G) sediment +50 mg kg−1 BPA. The headspace in bottles was vacuumed for 10 s and then refilled with purified N2 gas three times, as previously described [34]. The bottles were sealed with rubber stoppers and aluminum seals. The autoclaved controls were obtained by repeated autoclaving at 120 °C (30 min, three successive days). All the sediment microcosms were incubated at 25 °C on a horizontal shaker (150 rpm) for 120 days. At days 0, 6, 30, 40, 57, 75, 98, and 120, bottles were sacrificed and 1 g of dried sediment samples was used for chemical analysis, while the remaining sediment was used for DNA extraction. The number of bottles for each treatment was determined according to our preliminary study.
Chemical and Molecular Analyses
The residual BPA in sediment was extracted and determined according to the literature [40]. Briefly, sediment was extracted twice with methanol (10 mL), using a 300-W ultrasonic processor. The mixture was filtered with a 0.22-μm syringe filter, and the BPA in filtrate was determined using a high-performance liquid chromatography by absorbance at 276 nm with the retention time of 5.8 min. The concentrations of nitrate and sulfate in liquid phase of microcosms were determined using ion chromatography (Dionex ICS-500, USA) equipped with an Iopac ASI4 analytical column, as previously described [34].
Genomic DNA of each sediment sample was extracted using the Powersoil DNA extraction kit (Mobio Laboratories). Sediment DNA were amplified using the primers 515F (5′-GTGCCAGCMGCCGCGG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′), targeting the V4 hypervariable regions of 16S rRNA genes [2, 22]. The amplicons from each triplicate individual treatment were mixed in equal amounts and then subject to Illumina MiSeq sequencing. The quality filtering of sequences was carried out according to the literature [1]. UPARSE pipeline was applied to assign operational taxonomic units (OTUs), and sequences with 97 % similarity were clustered into OTUs. The identity of the representative sequence for each OTU was assigned using the RDP classifier [33]. Shannon diversity index and rarefaction curve of each sediment sample were generated using the UPARSE pipeline [9]. The beta diversity analysis was carried out using UniFrac to compare bacterial community. Weighted unifrac with QIIME (http://qiime.org/index.html) was used for Weighted Pair Group Method with Arithmetic mean (WPGMA) Clustering. The sequences obtained from Illumina MiSeq sequencing analysis in this study were deposited in the NCBI short-read archive under accession number SRP045722.
Statistical Analysis
Analysis of variance (one-way analysis of variance) using the software SPSS 20 was applied to check for the quantitative differences in the residual BPA. P < 0.05 was considered to be statistically significant.
Results
BPA Biodegradation
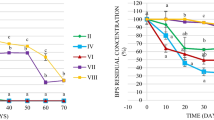
In this study, a negligible variation in the nitrate and sulfate concentrations was observed in the autoclaved controls (with treatments B and E). In contrast, nitrate loss was observed in the liquid phase of the sediment microcosm with treatment C (approximately 3 mmol L−1). A marked sulfate loss (nearly 2 mmol L−1) also occurred in the sediment microcosm with treatment F. These results confirmed the occurrence of nitrate or sulfate reduction in the non-sterilized sediment microcosms. Because of the hydrophobicity of BPA, the total amount of BPA in liquid phase of microcosms was negligible, compared with that in sediment during the incubation period (data not shown). At each sampling date (except day 0), the residual rate of BPA in the microcosm with treatment C was significantly lowered than that in the microcosm with treatment B (P < 0.05) (Fig. 1). In the microcosm with treatment C, the average residual rates of 65, 44, 29, and 7 % were observed at days 40, 75, 98, and 120, respectively. In contrast, an average residual rate of 80 % remained in the autoclaved control after 120-day incubation. These results indicated the occurrence of BPA biodegradation coupled with nitrate reduction. In addition, at each sampling date (except day 0), the residual rate of BPA in the microcosm with treatment F was significantly lowered than that in the microcosm with treatment E (P < 0.05). On day 120, the average residual rates of BPA in the microcosms with treatments F and E were 11 and 76 %, respectively. These results illustrated the occurrence of biological attenuation of BPA coupled with sulfate reduction. Moreover, an average residual rate of 85.7 % was still found in the microcosm with treatment G (without addition of nitrate or sulfate) after 120-day incubation. This suggested that a rapid anaerobic biodegradation of bisphenol A could not occur without external electron acceptors.
Percentage of sediment residual BPA in the microcosms with treatments B, C, E, F, and G. Treatment B, sterilized sediment + 50 mg kg−1 BPA + 20 mmol L−1 NaNO3; treatment C, sediment + 50 mg kg−1 BPA + 20 mmol L−1 NaNO3; treatment E, sterilized sediment + 50 mg kg−1 BPA + 20 mmol L−1 Na2SO4; treatment F sediment + 50 mg kg−1 BPA + 20 mmol L−1 Na2SO4; and treatment G, sediment + 50 mg kg−1 BPA. BPA values are the average of three independent experiments. Vertical bars indicate standard deviations
Bacterial Community Diversity and Composition
In this study, Illumina MiSeq sequencing analysis was applied to depict sediment bacterial communities in different microcosms. The obtained valid reads were normalized to 10,600 for the studied sediment samples in the microcosms with treatments A and C. Using a 3 % sequence dissimilarity cutoff, the number of OTUs in these sediment bacterial communities varied from 885 to 1115 (Table 1a). At days 75 and 120, the OTU number of the microcosm sample with treatment C was higher than that with treatment A. The appearance of a plateau in the rarefaction curve for each sediment sample indicated that the sediment community diversity had been well investigated at the sequencing depth used in this study (Fig. S1a). The Shannon index values of the sediment samples with treatments A and C ranged between 7.26 and 7.98. At days 75 and 120, the microcosm sample with treatment C had a lower Shannon diversity than that with treatment A. The number of normalized sequences was 9800 for the sediment samples in the microcosms with treatments D and F, composed of 891–1213 OTUs (Table 1b). On day 75, the OTU number of the microcosm sample with treatment F was lower than that with treatment D, while sample F120 had a higher OTU number than sample D120. Moreover, Fig. S1b also showed that the community diversities of these sediment samples had been well-captured, with the Shannon index value of 6.81–7.98. At days 75 and 120, the microcosm sample with treatment F had a higher bacterial diversity than that with treatment D.
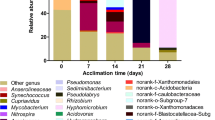
Nine frequently detected bacterial phyla were identified in the microcosm samples with treatments A, C, D and F, including Proteobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Gemmatimonadetes, Actinobacteria, Planctomycetes, Verrucomicrobia, and Acidobacteria (Fig. 2a, b). For the microcosm samples with treatments A and C, Proteobacteria (accounting for 31.7–47.3 %) was the largest phylum group in all samples, mainly consisting of Betaproteobacteria and Gammaproteobacteria. A large variation in the proportion of proteobacterial classes was found among these sediment samples (Fig. 3a). For the microcosm sample with treatment A, the relative abundance of Betaproteobacteria decreased from 21.2 % (day 0) to 8.4 % (day 75), followed by a slight rise to 9.7 % (day 120). A large decrease in the betaproteobacterial proportion was also found in the microcosm with treatment C after 75-day incubation, while a large increase occurred in subsequent incubation. In contrast, only a slight decrease in the betaproteobacterial proportion was observed in the microcosm with either treatment A or C. The relative abundance of Bacteroidetes showed a large increase in the microcosm with treatment A after 75-day incubation, followed by a slight decline in subsequent incubation. Compared with day 75, the proportion of Bacteroidetes showed a remarkable drop on day 120 in the microcosm with treatment C. Moreover, a slight variation in the proportions of Chloroflexi and Actinobacteria occurred in the microcosm with either treatment A or C, and Firmicutes and Gemmatimonadetes showed a slight increase during incubation. In addition, based on OTU level, the result of WPGMA clustering illustrated that sample AC0 was distantly separated from other sediment samples (Fig. 4a), which suggested a profound shift in the structures of sediment bacterial communities in both of the two microcosms during the incubation period. Sample C120 were distantly separated from sample C75 and more distantly from the cluster of samples A75 and A120. This suggested that BPA biodegradation coupled with nitrate-reducing condition had a strong impact on sediment bacterial community structure.
Comparison of the quantitative contribution of the sequences affiliated with different phyla to the total number of sequences from the microcosm samples under nitrate- (a) and sulfate-reducing (b) conditions. The rare species with relative abundance less than 0.5 % are included as others. Uppercase letters refer to treatment, and digits indicate sampling date. Sample AC0 represents the composite sample from the microcosms with treatments A and C on day 0, while sample DF0 represents the composite sample from the microcosms with treatments D and F on day 0. Treatment A, sediment + 20 mmol L−1 NaNO3; treatment C, sediment + 50 mg kg−1 BPA + 20 mmol L−1 NaNO3; treatment D, sediment + 20 mmol L−1 Na2SO4; and treatment F, sediment + 50 mg kg−1 BPA + 20 mmol L−1 Na2SO4
Comparison of the quantitative contribution of the sequences affiliated with different proteobacterial classes to the total number of sequences from the microcosm samples under nitrate- (a) and sulfate-reducing (b) conditions. Uppercase letters refer to treatment, and digits indicate sampling date. Sample AC0 represents the composite sample from the microcosms with treatments A and C on day 0, while sample DF0 represents the composite sample from the microcosms with treatments D and F on day 0. Treatment A, sediment + 20 mmol L−1 NaNO3; treatment C, sediment + 50 mg kg−1 BPA + 20 mmol L−1 NaNO3; treatment D, sediment + 20 mmol L−1 Na2SO4; and treatment F, sediment + 50 mg kg−1 BPA + 20 mmol L−1 Na2SO4
WPGMA clustering of microcosm sediment samples under nitrate- (a) and sulfate-reducing (b) conditions based on OTU level. Uppercase letters refer to treatment, and digits indicate sampling date. Sample AC0 represents the composite sample from the microcosms with treatments A and C on day 0, while sample DF0 represents the composite sample from the microcosms with treatments D and F on day 0. Treatment A, sediment + 20 mmol L−1 NaNO3; treatment C, sediment + 50 mg kg−1 BPA + 20 mmol L−1 NaNO3; treatment D, sediment + 20 mmol L−1 Na2SO4; and treatment F, sediment + 50 mg kg−1 BPA + 20 mmol L−1 Na2SO4
A large variation in the proportions of bacterial phyla was found in the microcosms with treatments D and F (Fig. 2b). A remarkable decrease in the proteobacterial proportion was found in either of the two anaerobic microcosms. At days 75 and 120, the proportion of Gammaproteobacteria in all the microcosm samples was higher than those of the other proteobacterial classes (Fig. 3b). Bacteroidetes was the largest phylum group in samples D75 and D120, while it became much less abundant in samples F75 and F120. Firmicutes showed a slight increase in the microcosm with treatment D, while it experienced a remarkable increase in the microcosm with treatment F. After 120-day incubation, the proportion of Chloroflexi showed a slight increase in either of the two anaerobic microcosms, while the proportion of Actinobacteria decreased. Moreover, WPGMA clustering indicated that samples DF0, F75, and F120 were grouped together, while they were distantly separated from the cluster of samples D75 and D120 (Fig. 4b). This result showed that sediment bacterial community structure greatly changed in the BPA unamended microcosm with incubation time, while BPA biodegradation coupled with sulfate reduction had a relatively weak impact on sediment bacterial community structure.
There were 23 frequently detected genera in the microcosm samples with treatments A and C (Table 2a). Ochrobactrum, Thiobacillus, Pseudoxanthomonas, and Thermomonas were the predominant genus groups in all of the microcosm samples.
The evident variation in bacterial community structures among different sediment samples was also found at genus level. In addition, a total of 22 frequently detected genera were found in the microcosm samples with treatments D and F, with an evident variation in genus members and their proportions (Table 2b). Ochrobactrum, Thiobacillus, Acinetobacter, Pseudoxanthomonas, and Thermomonas showed the predominance in all of the samples.
Discussion
BPA-degrading microorganisms ubiquitously exist in aquatic ecosystems, and BPA can be rapidly biodegraded in river waters [13, 17, 18]. Moreover, several previous studies also showed the rapid BPA biodegradation in river sediment. Chang et al. reported that the residual BPA rate in river sediment (amended with 250 mg kg−1) was 3.5–21.9 % after 5 days of aerobic incubation [7]. Our previous study found that a nearly complete removal of BPA (more than 95 %) occurred in aerobic river sediment microcosms (amended with 180 or 450 mg kg−1) after 3–4 days of incubation [40]. Two previous studies showed the occurrence of effective BPA biodegradation during anaerobic sewage sludge digestion [27], and by activated sludge under anaerobic conditions [37], while no BPA loss was found in sediment ecosystems under anaerobic conditions even after a long incubation (140 or 162 days) [7, 32]. However, in this study, after 120-day incubation, a nearly complete removal of BPA (93 or 89 %) occurred in the river sediment microcosms under either nitrate- or sulfate-reducing conditions. The present study provided the first evidence for BPA biodegradation in river sediment under anaerobic conditions. However, anaerobic dissipation of bisphenol A without nitrate or sulfate amendment was frustrated. In addition, there was no time lag in the anaerobic BPA degradation under either nitrate- or sulfate-reducing conditions. In this study, the river sediment for anaerobic biodegradation tests was collected from a polluted river receiving the discharge from municipal wastewater treatment plant. Therefore, the sediment autochthonous microbial community had been acclimatized to BPA pollution, and a number of BPA-degrading microorganisms could present (see below). This might account for the quick attenuation of BPA under nitrate- or sulfate-reducing conditions.
To date, information on the phylogenetic composition of aerobic BPA-degrading sediment microbial community is still very limited. Chang et al. suggested that Pseudomonas may be the dominant bacteria in the degradation process of BPA in river sediment [5]. Our previous study indicated that Proteobacteria (mainly Gammaproteobacteria and Alphaproteobacteria) predominated in BPA-degrading sediment microcosm and further illustrated a shift in bacterial community structure and a loss of bacterial diversity with aerobic BPA biodegradation [40]. High-throughput sequencing, especially using the recently developed Illumina MiSeq platform, can yield detailed information on microbial community with greater throughput and less cost [2]. It has been successfully applied to detect microbial community involved in pollutant-biodegrading process [14, 22, 30]. In this study, Illumina MiSeq sequencing analysis was also applied to characterize BPA-degrading sediment bacterial communities. Under nitrate- and sulfate-reducing conditions, the BPA-degrading sediment bacterial communities were mainly composed of Proteobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Gemmatimonadetes, and Actinobacteria. However, the proportions of these major phylum groups varied with BPA biodegradation. Moreover, the results of WPGMA clustering also illustrated a shift in bacterial community structure with anaerobic BPA biodegradation.
Based on clone library analysis, our previous study found that microorganisms from several bacterial genera were remarkably enriched with aerobic BPA biodegradation and they might be involved in BPA attenuation in river sediment [40]. In this study, the relative abundance of several bacterial genera (Candidatus Solibacter, Bacillus, Clostridium, Sedimentibacter, Desulfotomaculum, Desulfurispora, and Acinetobacter) obtained a remarkable increase after 120-day incubation under sulfate-reducing condition. Members of genus Bacillus are known for their ability to degrade BPA under both aerobic and anaerobic conditions [16, 21, 24]. Clostridium species have been linked to the anaerobic biodegradation of toluene [39], and phenanthrene and pyrene [4]. Acinetobacter species can transform pyrene [41] and phenol [25]. Moreover, members of genera Desulfotomaculum and Desulfurispora are known as sulfate-reducing bacteria [3, 15, 38]. Therefore, the roles of these microorganisms in biodegrading aromatic compounds or sulfate reduction suggested that they might be involved in biodegradation of BPA in river sediment. The presence of a variety of potential BPA-degrading organisms might account for the quick attenuation of BPA under anaerobic conditions. However, no bacterial species from any known genus was found to be enriched with BPA biodegradation under nitrate-reducing condition. This suggested that there might be novel microorganisms associated with BPA biodegradation under nitrate-reducing condition.
Conclusions
A high level of BPA could be biodegraded in sediment microcosm under both nitrate- and sulfate-reducing conditions. Bacterial community structure could vary with BPA biodegradation. A variety of bacterial species might have links with sulfate-reducing BPA degradation in river sediment. However, further study is necessary in order to elucidate the links between sediment bacterial community and BPA biodegradation.
References
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624
Cha IT, Roh S, Kim SJ, Hong HJ, Lee HW, Lim WT, Rhee SK (2013) Desulfotomaculum tongense sp nov., a moderately thermophilic sulfate-reducing bacterium isolated from a hydrothermal vent sediment collected from the Tofua Arc in the Tonga Trench. Antonie Van Leeuwenhoek 104:1185–1192
Chang BV, Chang IT, Yuan SY (2008) Anaerobic degradation of phenanthrene and pyrene in mangrove sediment. Bull Environ Contam Toxicol 80:145–149
Chang BV, Liu JH, Liao CS (2014) Aerobic degradation of bisphenol-A and its derivatives in river sediment. Environ Technol 35:416–424
Chang BV, Lu ZJ, Yuan SY (2009) Anaerobic degradation of nonylphenol in subtropical mangrove sediments. J Hazard Mater 165:162–167
Chang BV, Yuan SY, Chiou CC (2011) Biodegradation of bisphenol-A in river sediment. J Environ Sci Health Part A-Toxic/Hazard Subst Environ Eng 46:931–937
Chouhan S, Yadav SK, Prakash J, Swati, Singh SP (2014) Effect of Bisphenol A on human health and its degradation by microorganisms: a review. Ann Microbiol 2014(64):13–21
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Esteban S, Gorga M, Gonzalez-Alonso S, Petrovic M, Barcelo D, Valcarcel Y (2014) Monitoring endocrine disrupting compounds and estrogenic activity in tap water from Central Spain. Environ Sci Pollut Res 21:9297–9310
Esteban S, Gorga M, Petrovic M, Gonzalez-Alonso S, Barcelo D, Valcarcel Y (2014) Analysis and occurrence of endocrine-disrupting compounds and estrogenic activity in the surface waters of Central Spain. Sci Total Environ 466:939–951
Fuentes S, Mendez V, Aguila P, Seeger M (2014) Bioremediation of petroleum hydrocarbons: catabolic genes, microbial communities, and applications. Appl Microbiol Biotechnol 98:4781–4794
Ike M, Jin CS, Fujita M (2000) Biodegradation of bisphenol A in the aquatic environment. Water Sci Technol 42:31–39
Ju F, Zhang T (2014) Novel microbial populations in ambient and mesophilic biogas-producing and phenol-degrading consortia unraveled by high-throughput sequencing. Microb Ecol 68:235–246
Kaksonen AH, Spring S, Schumann P, Kroppenstedt RM, Puhakka JA (2007) Desulfurispora thermophila gen. nov., sp nov., a thermophilic, spore-forming sulfate-reducer isolated from a sulfidogenic fluidized-bed reactor. Int J Syst Evol Microbiol 57:1089–1094
Kamaraj M, Sivaraj R, Venckatesh R (2014) Biodegradation of Bisphenol A by the tolerant bacterial species isolated from coastal regions of Chennai, Tamil Nadu, India. Int Biodeterior Biodegrad 93:216–222
Kang JH, Kondo F (2002) Bisphenol A degradation by bacteria isolated from river water. Arch Environ Contam Toxicol 43:265–269
Kang JH, Kondo F (2005) BPA degradation in river water is different from that in seawater. Chemosphere 60:1288–1292
Kang JH, Ri N, Kondo F (2004) Streptomyces sp strain isolated from river water has high bisphenol A degradability. Lett Appl Microbiol 39:178–180
Kotowska U, Kapelewska J, Sturgulewska J (2014) Determination of phenols and pharmaceuticals in municipal wastewaters from Polish treatment plants by ultrasound-assisted emulsification-microextraction followed by GC-MS. Environ Sci Pollut Res 21:660–673
Li GY, Zu L, Wong PK, Hui XP, Lu Y, Xiong JK, An TC (2012) Biodegradation and detoxification of bisphenol A with one newly-isolated strain Bacillus sp GZB: kinetics, mechanism and estrogenic transition. Bioresour Technol 114:224–230
Liao XB, Chen C, Zhang JX, Dai Y, Zhang XJ, Xie SG (2014) Operational performance, biomass and microbial community structure: impacts of backwashing on drinking water biofilter. Environ Sci Pollut Res. doi:10.1007/s11356-014-3393-7
Luo YL, Guo WS, Ngo HH, Nghiem LD, Hai FI, Zhang J, Liang S, Wang XCC (2014) A review on the occurrence of micropollutants in the aquatic environment and their fate and removal during wastewater treatment. Sci Total Environ 473:619–641
Matsumura Y, Hosokawa C, Sasaki-Mori M, Akahira A, Fukunaga K, Ikeuchi T, Oshiman KI, Tsuchido T (2009) Isolation and characterization of novel bisphenol- A-degrading bacteria from soils. Biocontrol Sci 14:161–169
Ojha A, Mishra AK, Vashisht AK (2013) Isolation of phenol degrading bacteria from industrial waste water and their growth kinetic assay. J Pure Appl Microbiol 7:683–690
Sakai K, Yamanaka H, Moriyoshi K, Ohmoto T, Ohe T (2007) Biodegradation of bisphenol A and related compounds by Sphingomonas sp strain BP-7 isolated from seawater. Biosci Biotechnol Biochem 71:51–57
Samaras VG, Stasinakis AS, Thomaidis NS, Mamais D, Lekkas TD (2014) Fate of selected emerging micropollutants during mesophilic, thermophilic and temperature co-phased anaerobic digestion of sewage sludge. Bioresour Technol 162:365–372
Selvaraj KK, Shanmugam G, Sampath S, Larsson DGJ, Ramaswamy BR (2014) GC-MS determination of bisphenol A and alkylphenol ethoxylates in river water from India and their ecotoxicological risk assessment. Ecotoxicol Environ Safe 99:13–20
Shi JH, Liu XW, Chen QC, Zhang H (2014) Spatial and seasonal distributions of estrogens and bisphenol A in the Yangtze River Estuary and the adjacent East China Sea. Chemosphere 111:336–343
Tan B, Dong XL, Sensen CW, Foght J (2013) Metagenomic analysis of an anaerobic alkane-degrading microbial culture: potential hydrocarbon-activating pathways and inferred roles of community members. Genome 56:599–611
Toyama T, Kainuma Y, Kikuchi S, Mori K (2012) Biodegradation of bisphenol A and 4-alkylphenols by Novosphingobium sp strain TYA-1 and its potential for treatment of polluted water. Water Sci Technol 66:2202–2208
Voordeckers JW, Fennell DE, Jones K, Haggblom MM (2002) Anaerobic biotransformation of tetrabromobisphenol A, tetrachlorobisphenol A, and bisphenol A in estuarine sediments. Environ Sci Technol 36:696–701
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wang B, Huang B, Jin W, Wang Y, Zhao SM, Li FR, Hu P, Pan XJ (2012) Seasonal distribution, source investigation and vertical profile of phenolic endocrine disrupting compounds in Dianchi Lake, China. J Environ Monit 14:1275–1282
Wang B, Huang B, Jin W, Zhao SM, Li FR, Hu P, Pan XJ (2013) Occurrence, distribution, and sources of six phenolic endocrine disrupting chemicals in the 22 river estuaries around Dianchi Lake in China. Environ Sci Pollut Res 20:3185–3194
Wang Z, Yang Y, Sun W, Xie S, Liu Y (2014) Nonylphenol biodegradation in river sediment and associated shifts in community structures of bacteria and ammonia-oxidizing microorganisms. Ecotoxicol Environ Safe 106:1–5
Wang L, Zhao JM, Li YM (2014) Removal of bisphenol A and 4-n-nonylphenol coupled to nitrate reduction using acclimated activated sludge under anaerobic conditions. J Chem Technol Biotechnol 89:391–400
Watanabe M, Kojima H, Fukui M (2013) Desulfotomaculum intricatum sp nov., a sulfate reducer isolated from freshwater lake sediment. Int J Syst Evol Microbiol 63:3574–3578
Winderl C, Penning H, von Netzer F, Meckenstock RU, Lueders T (2010) DNA-SIP identifies sulfate-reducing Clostridia as important toluene degraders in tar-oil-contaminated aquifer sediment. ISME J 4:1314–1325
Yang YY, Wang Z, Xie SG (2014) Aerobic biodegradation of bisphenol A in river sediment and associated bacterial community change. Sci Total Environ 470–471:1184–1188
Yuan HY, Yao J, Masakorala K, Wang F, Cai MM, Yu C (2014) Isolation and characterization of a newly isolated pyrene-degrading Acinetobacter strain USTB-X. Environ Sci Pollut Res 21:2724–2732
Zhang WW, Yin K, Chen LX (2013) Bacteria-mediated bisphenol A degradation. Appl Microbiol Biotechnol 97:5681–5689
Acknowledgments
This work was financially supported by special fund of the State Key Joint Laboratory of Environment Simulation and Pollution Control (No. 14Y02ESPCP).
Author information
Authors and Affiliations
Corresponding author
Additional information
Yuyin Yang and Zhao Wang contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
(DOCX 94 kb)
Rights and permissions
About this article
Cite this article
Yang, Y., Wang, Z., He, T. et al. Sediment Bacterial Communities Associated with Anaerobic Biodegradation of Bisphenol A. Microb Ecol 70, 97–104 (2015). https://doi.org/10.1007/s00248-014-0551-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0551-x