Abstract
A novel, strictly anaerobic, moderately thermophilic, endospore-forming, sulfate-reducing bacterium, designated TGB60-1T, was isolated from a hydrothermal sediment vent collected from the Tofua Arc in the Tonga Trench. The strain was characterized phenotypically and phylogenetically. The isolated strain was observed to be Gram-positive, with slightly curved rod-shaped cells and a polar flagellum. Strain TGB60-1T was found to grow anaerobically at 37–60 °C (optimum, 50 °C), at pH 6.0–8.5 (optimum, pH 7.0) and with 1.0–4.0 % (w/v) NaCl (optimum, 3.0 %). The electron acceptors utilised were determined to be sulfate, sulfite, and thiosulfate. Strain TGB60-1T was found to utilise pyruvate and H2 as electron donors. Strain TGB60-1T was determined to be related to representatives of the genus Desulfotomaculum and the closest relatives within this genus were identified as Desulfotomaculum halophilum SEBR 3139T, Desulfotomaculum alkaliphilum S1T and Desulfotomaculum peckii LINDBHT1T (92.7, 92.1, and 91.8 % 16S rRNA gene sequence similarity, respectively). The major fatty acids (>20 %) were identified as C16:0 and C18:1 ω7c. The G+C content of the genomic DNA of this novel bacterium was determined to be 53.9 mol%. Based on this polyphasic taxonomic study, strain TGB60-1T is considered to represent a novel species in the genus Desulfotomaculum, for which the name Desulfotomaculum tongense sp. nov. is proposed. The type strain of D. tongense is strain TGB60-1T (= KTCT 4534T = JCM 18733T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Desulfotomaculum was proposed by Campbell and Postgate (1965) to describe Gram-positive and obligately anaerobic sulfate-reducing bacteria that form heat-resistant endospores. Many Desulfotomaculum species have been isolated from a wide range of environments, including hot springs/geothermal groundwaters (Zeikus et al. 1983; Daumas et al. 1988; Nazina et al. 1988; Love et al. 1993; Henry et al. 1994; Liu et al. 1997; Goorissen et al. 2003; Kaksonen et al. 2006; Haouari et al. 2008), fresh water (Elsgaard et al. 1994; Kuever et al. 1999), cold marine sediments (Isaksen et al. 1994; Vandieken et al. 2006), oilfields (Rosnes et al. 1991; Beeder et al. 1995; Rees et al. 1995; Nilsen et al. 1996; Tardy-Jacquenod et al. 1996; Nazina et al. 2005), compost/manure (Fardeau et al. 1995; Pikuta et al. 2000; Krishnamurthi et al. 2013) and anaerobic bioreactors (Min and Zinder 1990; Tasaki et al. 1991; Weijma et al. 2000; Plugge et al. 2002; Parshina et al. 2005; Kaksonen et al. 2008; Jabari et al. 2013). Species in the genus Desulfotomaculum are grouped in clusters from Ia to If, according to Stackebrandt and Goebel (1997) and Kuever et al. (1999). Two species, Desulfotomaculum halophilum (Tardy-Jacquenod et al. 1998) and Desulfotomaculum alkaliphilum (Pikuta et al. 2000), are extremophiles affiliated to the cluster If, i.e., a halophile and an alkaliphile, respectively.
In the present study, we characterized a moderately thermophilic sulfate-reducing bacterial strain from a hydrothermal vent sediment collected from the Tofua Arc in the Tonga Trench, which was designated TGB60-1T and determined that it belongs to cluster If in the genus Desulfotomaculum. This study reports the physiological and biochemical characteristics of strain TGB60-1T.
Materials and methods
Bacterial strain and culture conditions
Sediment samples were collected during April 2011 from a hydrothermal vent in the Tofua Arc of the Tonga Trench (S24°27′54″, W176°47′55″) at a water depth of 400 m using a remotely operated vehicle onboard the Korean research icebreaker ARAON. The samples were stored anaerobically at 4 °C until their cultivation and analysis. The sediment samples were inoculated into anaerobic artificial seawater (ASW) medium and incubated at 50 °C for 2 weeks. The ASW medium contained the following (l−1): 25 g NaCl, 4.0 g MgCl2·6H2O, 4.3 g MgSO4, 0.33 g KCl, 0.25 g NH4Cl, 0.2 g KH2PO4, 0.1 g CaCl2·2H2O, 2 g sodium pyruvate, and 1 mg resazurin. The media were flushed under O2-free N2 gas for 30 min. Next, 10 ml of the medium was distributed into 22 ml serum vials (Wheaton) under a stream of O2-free N2 gas and the medium was adjusted to pH 6.9–7.2 with 1 M NaHCO3. Dithiothreitol (0.3 g l−1) was added as a reducing reagent. The vials were sealed immediately with butyl rubber stoppers (Bellco). The medium was autoclaved at 120 °C for 15 min, before 0.01 ml vitamin solution (Wolin et al. 1963), 0.01 ml trace element SL-10 solution (Widdel and Pfennig 1981) and 0.01 ml tungsten–selenite solution (Widdel and Bak 1992) were supplemented to each tube. The cultures were transferred and sampled using syringes. To isolate individual colonies, the samples were enriched at 50 °C then spread and cultivated on solidified ASW medium with 1.5 % (w/v) agarose in an anaerobic chamber (Coy). The gas conditions in the anaerobic chamber were 90 % N2, 5 % CO2, and 5 % H2. Regular long rod-shaped cells were observed in the enrichment cultures after 2 weeks of incubation. To obtain pure isolates, the enriched cultures were spread onto ASW plates in the anaerobic chamber. Colonies of an isolated strain designated as TGB60-1T were transferred successively onto fresh solid media. The colonies were suspended in 5 % (w/v) dimethyl sulfoxide and stored at −80 °C in a cryogenic freezer.
Physiological, morphological, and biochemical characteristics
Bacterial growth was determined by quantifying sulfide production from sulfate in the growth medium (Trüper and Schlegel 1964) and by measuring the optical density at 600 nm. The cells were tested using a Gram-staining kit (Difco), according to the manufacturer’s instructions, and confirmed with the KOH test (Ryu 1938). The cell morphology was examined using light microscopy (Eclipse 80i; Nikon) and thin-section transmission electron micrography (55 VP; Carl Zeiss); the presence of flagella was determined by transmission electron microscopy (EM-109; Carl Zeiss) after negative staining with 1 % (w/v) phosphotungstic acid. A motility test was performed using semi-solid agar (0.25 %, w/v). Spore formation was determined microscopically using phase-contrast microscopy (Eclipse 80i) and growth was tested after heat treatment at 80 °C for 12 h. The optimal conditions for growth were determined using ASW medium with 0–16 % (w/v) NaCl (at intervals of 1 %) and at 25, 30, 37, 40, 50, 60, and 70 °C. To analyze the growth in different pH conditions, the medium was adjusted at pH 5.0–10.0 (at intervals of 0.5 pH units) with sterile 1 N HCl or NaOH under O2-free N2 gas and calibrated at 25 °C using a pH meter. For the growth test with yeast extract or tryptone, ASW medium was supplemented with 0.01 % (w/v) yeast extract or tryptone instead of the vitamin solution. To analyze the utilization of different carbon sources, pyruvate was replaced by each of the following substrates in the ASW medium (adjusted to pH 7.0 at 25 °C): H2/CO2 (80:20, 170 kPa), 10 mM pyruvate, acetate, lactate, malate, formate, ethanol, butanol, fumarate, succinate, propionate, glycerol, n-butyrate, isobutyrate, methanol, 2-propanol, lysine and methionine, 5 mM valerate, acetone, glucose, fructose, alanine, cysteine, glutamate, aspartate, glycine betaine, glycine, phenol and benzoate, 2 mM crotonate, octanoate, nonanoate, tartrate, 2-oxoglutarate, thioglycolate, thioacetamide and nicotinate, 2.5 mM palmitate, 0.5 mM catechol and 0.25 mM indole. The utilization of carbon sources was determined by measuring the bacterial growth (OD600) or the production of hydrogen sulfide. To test the capacity for using different electron acceptors, MgSO4 was replaced with MgCl2·6H2O in the ASW medium. The utilization of various electron acceptors was tested in the presence of pyruvate as the carbon and energy source. The electron acceptors were added as autoclaved or filter-sterilized stock solutions. The final concentrations of the electron acceptors were 20 mM Na2SO4, 2 mM Na2SO3, 10 mM Na2S2O3, 10 g (l−1) elemental sulfur (S0), 20 mM iron(III) citrate and 10 mM KNO3. The reduction of electron acceptors was determined by measuring the bacterial growth and by measuring changes in the electron acceptors: hydrogen sulfide production from Na2SO4, Na2SO3, Na2S2O3, and S0; ferrous iron production from iron(III) citrate; and nitrate removal. Hydrogen sulfide, ferrous iron and nitrate were analyzed as described by Trüper and Schlegel (1964), Lovley and Phillips (1986), and Benson (2002), respectively. End products of pyruvate oxidation with sulfate were analyzed using HPLC for the liquid medium and gas chromatography for the gases released after 2 days of incubation at 50 °C. Desulfoviridin and c-type cytochromes in the crude bacterial extract were analyzed as described by Postgate (1959).
Determination of the16S rRNA gene sequence and phylogenetic analysis
The 16S rRNA gene sequence of isolate TGB60-1T was determined for the phylogenetic analysis. The genomic DNA was extracted from isolate TGB60-1T using a commercial DNA extraction kit (Solgent). The 16S rRNA gene was amplified from the genomic DNA using the universal bacterial primers, 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT-3′) (Lane 1991). The purified PCR products were sequenced by Cosmo Genetech Co. Ltd using 27f, 338f, 786r, and 1492r primers. The 16S rRNA gene sequence was assembled using SeqMan (DNAStar). The 16S rRNA gene sequences of related taxa were obtained from the GenBank database and used for the phylogenetic analysis. Sequence alignments were performed using SILVA (http://www.arb-silva.de/aligner) and considered the secondary structure of the rRNA gene (Pruesse et al. 2007). Gaps were edited using the BioEdit program (Hall 1999). The evolutionary distances were calculated using Kimura’s two-parameter model (Kimura 1983). The phylogenetic trees were constructed with the neighbor-joining (NJ) (Saitou and Nei 1987), minimum-evolution (ME) (Nei et al. 1998), and maximum-likelihood (ML) (Felsenstein 1981) methods using the MEGA 5 program (Tamura et al. 2011) with 1,000 randomly selected bootstrap replicates. The nucleotide similarity values of the 16S rRNA gene sequences were calculated using EzTaxon (http://www.eztaxon-e.ezibiocloud.net/) (Kim et al. 2012).
Analysis of cellular fatty acids and determination of the DNA G+C content
To analyze the cellular fatty acids, strain TGB60-1T was cultivated for 3 days in ASW medium at 50 °C and pH 7.0. The cellular fatty acids were saponified, methylated, and extracted according to the Sherlock Microbial Identification System protocol (MIDI 2001). The fatty acids were analyzed by gas chromatography (Hewlett Packard 6890) and identified using the Microbial Identification software package (Sasser 1990). The DNA G+C content of strain TGB60-1T was determined using a fluorometric technique based on real-time PCR (Gonzàlez and Saiz-Jimenez 2002).
Results and discussion
The colonies of strain TGB60-1T were observed to be black and measured <1 mm in diameter. The cells of strain TGB60-1T were observed to be straight, 0.4–0.5 μm in diameter and 1.5–2.0 μm in length. The cells were found to be motile, possess a flagellum and stain Gram-positive (based on Gram-staining and the KOH test). The presence of a Gram-positive cell wall type was confirmed using thin-section transmission electron micrography (Fig. 1). The spores were observed to be located at the cell termini. Strain TGB60-1T was found to be able to grow with 1.0–4.0 % (w/v) NaCl (optimum, 3.0 %), but not in the absence of NaCl or at >5.0 % (w/v). Thus, NaCl is required for growth. Growth of strain TGB60-1T was observed at 37–60 °C (optimum, 50 °C) and at pH 6.0–8.5 (optimum, 7.0). Strain TGB60-1T was found to grow with yeast extract or tryptone instead of vitamins. Strain TGB60-1T is able to utilise pyruvate as a sole carbon and energy source and H2 is utilised autotrophically as an electron donor with CO2 in the presence of sulfate as the terminal electron acceptor. Strain TGB60-1T was determined to reduce sulfate, thiosulfate, and sulfite to sulfide in the presence of pyruvate as an electron donor and carbon source. However, elemental sulfur, nitrate, and iron (III) citrate were not reduced in the presence of pyruvate or H2 with CO2. Pyruvate was not fermented in the absence of sulfate. The end products of pyruvate oxidation with sulfate were identified as acetate and CO2. In the crude bacterial extract, desulfoviridin and c-type cytochromes were not detected. A comparison of the characteristics of strain TGB60-1T and closely related type strains in the genus Desulfotomaculum showed that the isolate could be distinguished from the reference strains (Table 1).
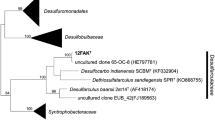
The 16S rRNA gene sequence of strain TGB60-1T determined is 1,447 bp in length (GenBank/EMBL/DDBJ accession number JX183068). The phylogenetic analysis based on the 16S rRNA gene sequences showed that strain TGB60-1T belongs to the family Peptococcaceae and the lineage Desulfotomaculum, which was supported by the high bootstrap values of the NJ, ME and ML phylogenetic trees (84, 85, and 84 %, respectively) (Fig. 2). Strain TGB60-1T is closely related to D. halophilum SEBR 3139T, D. alkaliphilum S1T and Desulfotomaculum peckii LINDBHT1T (92.7, 92.1, and 91.8 % 16S rRNA gene sequence similarity, respectively) in cluster If of the genus.
Phylogenetic tree based on the 16S rRNA gene sequences of strain TGB60-1T and closely related taxa, which were constructed using the neighbor-joining algorithm. The numbers on the nodes indicate the bootstrap values (>70 %) calculated using the neighbor-joining/minimum-evolution/maximum-likelihood methods, which are expressed as percentages of 1,000 replicates. Moorella glycerini JW-AS-Y6T and Calicoprobacter algeriensis TH7C1T were used as outgroups. Bar 0.01 accumulated change per nucleotide
The major fatty acids of strain TGB60-1T were identified as C16:0 (45.6 %) and C18:1 ω7c (22.8 %). The other fatty acids in strain TGB60-1T were identified as C18:1 ω9c (7.0 %), C16:1 ω7c (5.6 %), C16:0 DMA (dimethyl acetal) (4.2 %), iso-C18:0 (3.6 %), C17:0 (2.7 %), C17:0 cyclo (2.3 %), iso-C16:0 (2.1 %), C18:1 ω5c (2.1 %) and C16:0 ω9c (2.0 %). The DNA G+C content of the genomic DNA of strain TGB60-1T was determined to be 53.9 mol%, which is similar to that of D. halophilum SEBR 3139T (56.3 %) but different from those of the other closely related type strains (Tardy-Jacquenod et al. 1998; Pikuta et al. 2000; Jabari et al. 2013). Based on the phenotypic and genomic differences described here, strain TGB60-1T can be assigned to a novel species in the genus Desulfotomaculum, for which the name Desulfotomaculum tongense sp. nov. is proposed.
Description of Desulfotomaculum tongense sp. nov
Desulfotomaculum tongense (tong.en’se. N.L. neut. adj. tongense, of or belonging to Tonga). Cells are motile, Gram-positive and spore-forming rods (0.4–0.5 × 1.5–2.0 μm). Growth occurs at 37–60 °C (optimum, 50 °C), pH 6.0–8.5 (optimum, pH 7.0) and with 1.0–4.0 % NaCl (optimum, 3.0 %). Requires sodium ions for growth. Pyruvate and hydrogen are utilised with CO2 as electron donors in the presence of sulfate. Growth occurs with yeast extract or tryptone in the absence of vitamins. Cannot utilise acetate, lactate, malate, formate, ethanol, butanol, fumarate, succinate, propionate, glycerol, n-butyrate, isobutyrate, valerate, crotonate, octanoate, nonanoate, palmitate, tartrate, benzoate, 2-oxoglutarate, thioglycolate, thioacetamide, methanol, 2-propanol, acetone, glucose, fructose, catechol, alanine, lysine, methionine, cysteine, glutamate, aspartate, glycine betaine, glycine, indole, phenol or nicotinate in the presence of sulfate. Pyruvate is not fermented in the absence of sulfate. Sulfate, thiosulfate and sulfite are reduced, but not elemental sulfur, nitrate or iron (III) citrate. No desulfoviridin and c-type cytochromes are observed in crude bacterial extracts. The major fatty acids (>20 %) are C16:0 and C18:1 ω7c. The DNA G+C of the type strain content is 53.9 mol%.
The type strain, TGB60-1T (= KCTC 4534T = JCM 18733T), was isolated from a hydrothermal vent sediment collected from the Tofua Arc in the Tonga Trench. The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain TGB60-1T is JX183068.
References
Beeder J, Torsvik T, Lien T (1995) Thermodesulforhabdus norvegicus gen. nov., sp. nov., a novel thermophilic sulfate-reducing bacterium from oil field water. Arch Microbiol 164(5):331–336
Benson HJ (2002) Microbiological applications: a laboratory manual in general microbiology. McGraw-Hill, NY
Campbell LL, Postgate JR (1965) Classification of the spore-forming sulfate-reducing bacteria. Bacteriol Rev 29(3):359–363
Daumas S, Cord-Ruwisch R, Garcia JL (1988) Desulfotomaculum geothermicum sp. nov., a thermophilic, fatty acid-degrading, sulfate-reducing bacterium isolated with H2 from geothermal ground water. Antonie Van Leeuwenhoek 54(2):165–178
Elsgaard L, Prieur D, Mukwaya GM, Jorgensen BB (1994) Thermophilic sulfate reduction in hydrothermal sediment of lake tanganyika, East Africa. Appl Environ Microbiol 60(5):1473–1480
Fardeau ML, Ollivier B, Patel B-KC, Dwivedi P, Ragot M, Garcia JL (1995) Isolation and characterization of a thermophilic sulfate-reducing bacterium, Desulfotomaculum thermosapovorans sp. nov. Int J Syst Bacteriol 45:218–221
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17(6):368–376
Gonzàlez JM, Saiz-Jimenez C (2002) A fluorimetric method for the estimation of G+C mol% content in microorganisms by thermal denaturation temperature. Environ Microbiol 4:770–773
Goorissen HP, Boschker HT, Stams AJ, Hansen TA (2003) Isolation of thermophilic Desulfotomaculum strains with methanol and sulfite from solfataric mud pools, and characterization of Desulfotomaculum solfataricum sp. nov. Int J Syst Evol Microbiol 53(5):1223–1229
Hall TA (1999) BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Haouari O, Fardeau ML, Cayol JL, Casiot C, Elbaz-Poulichet F, Hamdi M, Joseph M, Ollivier B (2008) Desulfotomaculum hydrothermale sp. nov., a thermophilic sulfate-reducing bacterium isolated from a terrestrial Tunisian hot spring. Int J Syst Evol Microbiol 58(11):2529–2535
Henry EA, Devereux R, Maki JS, Gilmour CC, Woese CR, Mandelco L, Schauder R, Rensen CC, Mitchell R (1994) Characterization of a new thermophilic sulphate-reducing bacterium Thermodesulfovibrio yellowstonii, gen. nov. and sp. nov.: its phylogenetic relationship to Thermodesulfobacterium commune and their origins deep within the bacterial domain. Arch Microbiol 161:62–69
Isaksen MF, Bak F, Jørgensen BB (1994) Thermophilic sulfate reducing bacteria in cold marine sediment. FEMS Microbiol Ecol 14:1–8
Jabari L, Gannoun H, Cayol JL, Hamdi M, Ollivier B, Fauque G, Fardeau ML (2013) Desulfotomaculum peckii sp. nov., a moderately thermophilic member of the genus Desulfotomaculum, isolated from an upflow anaerobic filter treating abattoir wastewaters. Int J Syst Evol Microbiol 63(6):2082–2087
Kaksonen AH, Spring S, Schumann P, Kroppenstedt RM, Puhakka JA (2006) Desulfotomaculum thermosubterraneum sp. nov., a thermophilic sulfate-reducer isolated from an underground mine located in a geothermally active area. Int J Syst Evol Microbiol 56(11):2603–2608
Kaksonen AH, Spring S, Schumann P, Kroppenstedt RM, Puhakka JA (2008) Desulfotomaculum alcoholivorax sp. nov., a moderately thermophilic, spore-forming, sulfate-reducer isolated from a fluidized-bed reactor treating acidic metal- and sulfate-containing wastewater. Int J Syst Evol Microbiol 58:833–838
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62(3):716–721
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University, Cambridge
Krishnamurthi S, Spring S, Kumar PA, Mayilraj S, Klenk HP, Suresh K (2013) Desulfotomaculum defluvii sp. nov., a sulfate-reducing bacterium isolated from the subsurface environment of a landfill. Int J Syst Evol Microbiol 63(6):2290–2295
Kuever J, Rainey FA, Hippe H (1999) Description of Desulfotomaculum sp. Groll as Desulfotomaculum gibsoniae sp. nov. Int J Syst Bacteriol 49(4):1801–1808
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–147
Liu Y, Karnauchow TM, Jarrell KF, Balkwill DL, Drake GR, Ringelberg D, Clarno R, Boone DR (1997) Description of two new thermophilic Desulfotomaculum spp., Desulfotomaculum putei sp. nov., from a deep terrestrial subsurface, and Desulfotomaculum luciae sp. nov., from a hot spring. Int J Syst Bacteriol 47:615–621
Love CA, Patel BKC, Nicholas PD, Stackebrandt E (1993) Desulfotomaculum australicum, sp. nov., a thermophilic sulfate-reducing bacterium isolated from the Great Artesian Basin of Australia. Syst Appl Microbiol 16:244–251
Lovley DR, Phillips EJP (1986) Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl Environ Microbiol 51:683–689
MIDI (2001) Sherlock microbial identification system. MIDI Inc, Newark, DE
Min H, Zinder SH (1990) Isolation and characterization of a thermophilic sulfate-reducing bacterium Desulfotomaculum thermoacetoxidans sp. nov. Arch Microbiol 153:399–404
Nazina TN, Ivanova AE, Kanchaveli LP, Rozanova EP (1988) A new spore-forming thermophilic methylotrophic sulfate-reducing bacterium, Desulfotomaculum kuznetsovii sp. nov. Microbiologia 57:823–827
Nazina TN, Rozanova EP, Beliakova EV, Lysenko AM, Poltaraus AB, Turova TP, Osipov GA, Beliaev SS (2005) Description of ‘Desulfotomaculum nigrificans subsp. salinus’ as a new species Desulfotomaculum salinum sp. nov. Microbiology (English translation of Microbiologiia) 74:567–574
Nei M, Kumar S, Takahashi K (1998) The optimization principle in phylogenetic analysis tends to give incorrect topologies when the number of nucleotides or amino acids used is small. Proc Natl Acad Sci USA 95(21):12390–12397
Nilsen RK, Torsvik T, Lien T (1996) Desulfotomaculum thermocisternum sp. nov., a sulfate reducer isolated from a hot North Sea oil reservoir. Int J Syst Bacteriol 46:397–402
Parshina SN, Sipma J, Nakashimada Y, Henstra AM, Smidt H, Lysenko AM, Lens PN, Lettinga G, Stams AJ (2005) Desulfotomaculum carboxydivorans sp. nov., a novel sulfate-reducing bacterium capable of growth at 100 % CO. Int J Syst Evol Microbiol 55(5):2159–2165
Pikuta E, Lysenko A, Suzina N, Osipov G, Kuznetsov B, Tourova T, Akimenko V, Laurinavichius K (2000) Desulfotomaculum alkaliphilum sp. nov., a new alkaliphilic, moderately thermophilic, sulfate-reducing bacterium. Int J Syst Evol Microbiol 50:25–33
Plugge CM, Balk M, Stams AJ (2002) Desulfotomaculum thermobenzoicum subsp. thermosyntrophicum subsp. nov., a thermophilic, syntrophic, propionate-oxidizing, spore-forming bacterium. Int J Syst Evol Microbiol 52:391–399
Postgate J (1959) A diagnostic reaction of Desulphovibrio desulphuricans. Nature 183(4659):481–482
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35(21):7188–7196
Rees GN, Grassia GS, Sheehy AJ, Dwivedi PP, Patel BKC (1995) Desulfacinum infernum gen. nov., sp. nov., a thermophilic sulfate-reducing bacterium from a petroleum reservoir. Int J Syst Bacteriol 45:85–89
Rosnes JT, Torsvik T, Lien T (1991) Spore-forming thermophilic sulfate-reducing bacteria isolated from north sea oil field waters. Appl Environ Microbiol 57(8):2302–2307
Ryu E (1938) On the gram-differentiation of bacteria by the simplest method. J Jpn Soc Vet Sci 17:58–63
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, Microbial technical note 101. MIDI Inc, Newark, DE
Stackebrandt E, Goebel BM (1997) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Tardy-Jacquenod C, Caumette P, Matheron R, Lanau C, Arnauld O, Magot M (1996) Characterization of sulfate-reducing bacteria isolated from oil-field waters. Can J Microbiol 42:259–266
Tardy-Jacquenod C, Magot M, Patel BKC, Matheron R, Caumette P (1998) Desulfotomaculum halophilum sp. now, a halophi I ic sulfate-reducing bacterium isolated from oil production facilities. Int J Syst Bacteriol 48:333–338
Tasaki M, Kamagata Y, Nakamura K, Mikami E (1991) Isolation and characterization of a thermophilic benzoate degrading, sulfate-reducing bacterium, Desulfotomaculum thermobenzoicum sp. nov. Arch Microbiol 155:348–352
Trüper HG, Schlegel HG (1964) Sulphur metabolism in Thiorhodaceae I. Quantitative measurements on growing cells of Chromatium okenii. J Microbiol Ser 30:225–238
Vandieken V, Mussmann M, Niemann H, Jorgensen BB (2006) Desulfuromonas svalbardensis sp. nov. and Desulfuromusa ferrireducens sp. nov., psychrophilic, Fe(III)-reducing bacteria isolated from Arctic sediments, Svalbard. Int J Syst Evol Microbiol 56(5):1133–1139
Weijma J, Stams AJ, Hulshoff Pol LW, Lettinga G (2000) Thermophilic sulfate reduction and methanogenesis with methanol in a high rate anaerobic reactor. Biotechnol Bioeng 67(3):354–363
Widdel F, Bak F (1992) Gram-negative mesophilic sulfate reducing bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes. Springer, New York, pp 3352–3378
Widdel F, Pfennig N (1981) Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. I. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments. Description of Desulfobacter postgatei gen. nov., sp. nov. Arch Microbiol 129(5):395–400
Wolin EA, Wolin MJ, Wolfe RS (1963) Formation of methane by bacterial extracts. J Biol Chem 238:2882–2886
Zeikus JG, Dawson MA, Thompson TE, Ingvorsen K, Hatchikian EC (1983) Microbial ecology of volcanic sulphidogenesis: isolation and characterization of Thermodesulfobacterium commune gen. nov. and sp. nov. J Gen Microbiol 129:1159–1169
Acknowledgments
This work was supported by a grant from the National Institute of Biological Resources (NIBR) funded by the Ministry of Environment (MOE) (NIBR No. 2013-02-055), the R & D project ‘Exploration of Seafloor Hydrothermal Deposits in Tongan Waters (PM57063)’ funded by the Ministry of Oceans and Fisheries of Korea, the Marine and Extreme Genome Research Center Program of the Ministry of Land, Transportation, and Maritime Affairs, Republic of Korea, and BK21 Plus program.
Author information
Authors and Affiliations
Corresponding author
Additional information
In-Tae Cha and Seong Woon Roh contributed equally to this work.
Rights and permissions
About this article
Cite this article
Cha, IT., Roh, S.W., Kim, SJ. et al. Desulfotomaculum tongense sp. nov., a moderately thermophilic sulfate-reducing bacterium isolated from a hydrothermal vent sediment collected from the Tofua Arc in the Tonga Trench. Antonie van Leeuwenhoek 104, 1185–1192 (2013). https://doi.org/10.1007/s10482-013-0040-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-013-0040-0