Abstract
Sediment contaminated with polycyclic aromatic hydrocarbons (PAHs) is widely distributed in aquatic ecosystems. The microbial community structure of riverbank PAH-contaminated sediments was investigated using phospholipid-derived fatty acid (PLFA) analysis. Surface and subsurface riverbank sediment was collected from a highly contaminated site and from an uncontaminated site along the Mahoning River, OH. PAH concentrations, physical sediment characteristics, and other microbial community parameters (biomass as phospholipid phosphate (PLP) and activity) were also measured. PAHs were detected in all samples but were only quantifiable in the contaminated (250 μg/g g−1) subsurface sediment. Subsurface samples from both locations showed very similar PLP values and distribution of PLFAs, with 27–37 % of the microbial community structure being composed of sulfate reducing and other anaerobic bacteria. Principal components analysis indicated no correlation between PAH contamination and PLFA diversity. Although PLP and phospholipid fatty acid measurements of bacterial communities did not reflect the environmental differences among sites, the highly PAH-contaminated sediment showed the highest measured microbial activity (reduction of 1,200 nmol INT g−1 h−1), likely from a population adapted to environmental pollutants, rates that are much higher than measured in many uncontaminated soil and sediment systems. These data warrant further investigation into community structure at the genetic level and indicate potential for bioremediation by indigenous microbes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
During the industrial revolution, fossil fuels were extracted and burned in great amounts, and polycyclic aromatic hydrocarbons (PAHs) began accumulating in localized areas of rivers, estuaries, and harbors [8, 22]. PAHs constitute a large and diverse class of hydrophobic, organic compounds with multiple interlocking benzene rings that are formed during the incomplete combustion of organic substances such as coal, oil, gas, and wood [1]. Because PAHs are hydrophobic, they adsorb tightly to sediment particles and therefore become persistent in the environment. When introduced into aquatic environments, the sediments serve as a repository for the majority of these compounds [35]. Some PAHs are recognized as carcinogenic and mutagenic to many organisms by the US Environmental Protection Agency (www.epa.gov). The Mahoning River is considered one of the most contaminated in northeast USA. Between 1900 and the 1970s, the Mahoning River received up to and over 32,000 kg of oil and grease each day [47]. Types of industrial waste discharged to the river included petroleum, lubricating oils, and many various chemicals. Today, water quality in the Mahoning River has improved due to the decreased input of unregulated waste that also corresponds with the regional decline of the steel industry. However, the sediment remains contaminated with high concentrations of PAHs, metals, and other organic pollutants.
Pollutants as well as other chemicophysical parameters influence microbial community composition, thus selecting for microbes that live and function under those conditions. Conversely, microbes play an important role in contaminant fate through metabolic mechanisms (e.g., mineralization, biodegradation). Knowledge of microbial communities in long contaminated systems is important for determining potential for bioremediation, transformation, or persistence of pollutants. Relatively little is known about microbial community structure in PAH-contaminated soils and sediments. Microbial community structure could provide information for potential intrinsic biodegradation by elucidating relationships between PAHs and microbial communities. In this study, we investigated possible correlations between the microbial community structure and PAHs in riverbank sediment using a phospholipid-derived fatty acid (PLFA)-based fingerprinting approach.
PLFA analysis has been proven to be relatively simple, fast, sensitive, inexpensive, and reproducible for assessing the structure of the living microbial community in soil ecosystems [7, 9, 17, 40]. Contrary to molecular approaches (e.g., terminal restriction fragment length polymorphism) that do not provide community composition unless coupled to sequencing, PLFAs are useful biomarkers that give an instant profile of the bacterial community. Neutral lipids and fatty acids may persist after death and do not represent living microbes as well as PLFAs [9]. PLFAs have been used to determine differences in microbial community structure across environmental gradients such as redox (e.g., [42]), soil pore size (e.g., [39]), soil water availability (e.g., [53]), between different treatments (e.g., [29, 43]), and as temporal and spatial distribution (e.g., [18, 50]). Lipid analyses of microbial communities can provide identification at the kingdom, group, or species level depending on the community in some cases [10, 18]. For instance, fatty acids can be specific enough to be useful for source tracking fecal coliforms [13] and for identifying different strains of bacteria such as Enterobacter and Yersinia [24, 52]. The lipid extraction method used in this study [14] has the advantage of simultaneously extracting PAHs and phospholipids to determine biomass as phospholipid phosphate (PLP) and community structure (as PLFAs). The main disadvantage of this method is specificity. Individual fatty acids cannot generally be used to identify individual species since the same fatty acids can occur in more than one species, individual species may contain numerous fatty acids, and the amounts of fatty acids may vary within a species depending on growth conditions [21]. A minor drawback of this analysis is that samples must be processed shortly after sampling since profiles change when stored at −70 °C for a year [55].
Methods
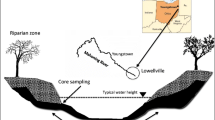
Study Site Description and Sampling Procedure
Two riverbank sites of sediment deposition located upstream of low head dams located along the Mahoning River were chosen for this study. The first site, designated by the US Army Corps of Engineers [47] as river mile 13.3, was located in Lowellville (LWV), OH, and the second site, river mile 46.3, was located in Leavittsburg (LVT), OH. The LWV site was previously shown to have PAH-contaminated sediments [34, 47] while the LVT site was located upstream of industrial sources of PAH contamination.
Sediment samples were collected using a soil recovery auger (AMS, Inc., American Falls, ID). Protective gloves and clothing were worn to prevent contact with contaminated sediment. The auger was cleaned between samples using Liquinox® detergent and rinsed with distilled deionized water. Sediment for analyses was removed from the coring device using a sterile steel spatula and was immediately placed into Ziploc bags. Samples were transported on ice to the laboratory for processing within a few hours of collection.
Discrete samples of approximately 500 g of sediment were collected from 5 to 25 cm below the surface and at a depth of approximately 2 m (subsurface). The visual appearance (and odor) of oily contamination was used to determine the subsurface sampling depth of contaminated riverbank sediment at LWV. From the LVT site on each sampling date (21 March, 4 April, and 25 April 2005), three surface and three subsurface sediment samples were collected and processed immediately in the laboratory with exception of 4 April when only two subsurface sediment samples were processed. Similarly from the LWV site on each sampling date (28 March, 11 April, and 2 May 2005), three surface and three subsurface sediment samples were collected and processed immediately in the laboratory with the exemption of 28 March when only two surface sediment samples were processed. Thus, a total of eight surface and eight subsurface sediment samples per site were used for PLFA, PAHs, biomass, and microbial activity measurements.
Particle Size Distribution, Moisture Content, and Total Organic Matter
Three surface and three subsurface sediment samples per site were used to determine particle size distribution, moisture content, and organic matter. Sediment was homogenized and oven-dried at 105 °C prior to analyses. The particle size distribution was measured using a hydrometer (Fisher Environmental). Proportions of major size classes, clay (<0.002 mm), silt (0.05–0.002 mm), and sand (2.0–0.05 mm) were determined from size distributions [45]. Moisture content determination followed standard methods for soil analysis. Total organic matter was determined in 5 g of dried sediment by loss of mass following ignition at 550 °C for 1 h.
Lipid and PAH Extraction of PLP, PLFA, and PAHs
Sediment was and kept on ice and processed immediately upon returning to the lab. Sediment samples were extracted as described by Fang and Findlay [14] to determine PLP biomass PLFA and PAHs. Briefly, lipids and PAHs were extracted from 0.65 g sediment in the dark at 4 °C in 50-ml screw-cap tubes with dichloromethane: methanol: 50 mM phosphate buffer (pH 7.4)–1:2:0.8 (v/v/v), followed by an organic and aqueous partitioning with dichloromethane (DCM) and water (1:1:9 v/v/v).The DCM fraction was collected, dried under nitrogen to one drop, and brought to 1 ml in chloroform. Two 100-μl samples were removed for lipid phosphate biomass analysis. The remainder of the extracted lipids and PAHs in the organic phase was fractionated using silica gel solid phase extraction (SPE) chromatography. SPE columns were prepared using 100–200-mesh Unisil (Clarkson Chromatography). The Unisil was first activated by heating at 100 °C for 2 h, dissolved in chloroform, and then transferred to a glass column. Copper filings (cleaned with HCl, methanol, DCM, and hexane) were added to the top of the column. The columns were then ready to use.
The PAH fraction was extracted with hexane from the SPE column, cleaned by passage through aminopropyl columns, eluted in hexane, and quantified by gas chromatography–mass spectrometry (GC/MS). Neutral lipids and glycolipids were eluted and discarded from the SPE column prior to recovering phospholipids in a solution of chloroform/methanol/DI water (5:5:1) which was dried under nitrogen. PLFAs were then converted to their corresponding fatty acid methyl esters (FAMEs) as described in detail by Findlay [16]. Findlay’s procedure does not produce FAMEs from free fatty acids. The PLFA-derived FAMEs were purified using reverse-phase SPE consisting of silicic acid packed in glass columns also as described by Findlay [16]. Shorthand designation of PLFAs followed the rules of widely used nomenclature.
GC–MS Analyses of PLFA FAMES and PAHs
The GC/MS used to identify and quantify PAHs and PLFA-derived FAMEs was a Hewlett Packard 5890 Gas Chromatograph/5970B Mass Spectrometer fitted with a DB-5 column 30 M, 0.32 mm ID, and 0.25 μm film thickness. Samples (1.0 μl) were injected splitless using a Finnigan-Mert A 2005 autosampler.
Determination of PLP Biomass
Microbial biomass was determined by extracting phospholipids. Phosphate was released by acid digestion and measured using colorimetric analysis with malachite green by spectrophotometer at a wavelength of 610 nm [48].
Microbial Activity
Microbial activity was determined by measuring reduction of 2-(p-iodophenyl)-3(p-nitrophenyl)-5-phenyl tetrazolium chloride (INT) to INTF resulting in a color change and detected spectrophotometrically [33]. INT reduction is microbially mediated under aerobic or anaerobic conditions and measures respiration [4, 49]. This method is quick, simple, does not use radioisotopes, and only measures viable cells [46]. Sediment from cores was homogenized, reducing variability, but may produce different values from undisturbed sediment. Triplicate samples of fresh sediment (0.2 g each) were placed into conical tubes with 0.3 ml milli-Q water. Samples were vortexed and equilibrated at room temperature for 30 min. After adding 0.5 ml of a 1.08-mM INT solution, the solution was mixed and incubated for 10 min at 25 °C under aerobic conditions, although subsurface sediment was anaerobic. The reduced product, a red colored formazan (INTF), was extracted for 10 min with 6.0 ml of acetonitrile filtered through filter paper (Whatman no. 40) using a vacuum. Samples were rinsed with 6.0 ml of acetonitrile filtered, and the total filtrate was analyzed spectrophotometrically. The amount of INT reduction in controls, essentially a time zero extraction using acetonitrile, was subtracted from INT reduction that occurred during the incubation period. This took into account a low and consistent amount of INT that was reduced during the 10-min acetonitrile extraction.
Statistical Analysis
Principal component analysis (PCA; SPSS) was used to analyze patterns of intercorrelations among variables measured. This method of data reduction was chosen to reduce the number of variables measured: PAHs, PLFAs, microbial biomass, microbial activity, percent moisture, percent total organic carbon, and particle size distribution. PCA for the Lowellville site was performed using all variables. However, because no PAHs were quantified in Leavittsburg site, PAH data were not included. The Shannon diversity index was used to estimate PLFA diversity as an indicator of microbial diversity.
Results
Sediment Characteristics
LVT surface and subsurface and LWV subsurface contaminated sediment all had similar moisture content (between 42 and 46 %). LWV surface sediment had the least amount of moisture (24 %) (Table 1). Because the subsurface sediment was collected from within the ground water saturation zone, moisture content was high as expected. LVT surface sediment moisture was high likely due to recent flooding at the site.
Total organic matter content of LWV and LVT sediment was very similar (5–6 %) among both surface and subsurface samples. LWV subsurface contaminated sediments contained the highest total organic matter (11 %) likely due to high concentrations of organic contaminants (total petroleum hydrocarbons and PAHs).
Patterns of particle size distribution were very similar among both surface and subsurface samples from LVT and from surface samples from LWV (Table 1). LWV subsurface sediment results may be a methodological artifact due to properties of industrial sludge since there were no visible sand particles.
Distribution of PAHs
The highest total PAH concentrations (250 μg/g) were detected in subsurface LWV riverbank sediment (Fig. 1), which account for as much as three times the total PAH concentrations (78 μg/g) detected in surface sediment. While LWV subsurface sediment contained 11 different PAHs (acenaphthene, acenaphthylene, anthracene, benzo(a)anthracene, benzo(b&k)fluoranthene, chrysene, fluoranthene, fluorene, naphthalene, phenanthrene, and pyrene) (Fig. 1), only seven different PAHs were detected in surface sediment (anthracene, benzo(a)anthracene, benzo(b&k)fluoranthene, chrysene, fluoranthene, phenanthrene, pyrene). Concentrations of individual PAHs at the LWV site ranged from 6.4 to 20.5 μg/g in the surface sediment samples and from 0.4 to 61.8 μg/g in subsurface sediment samples (Fig. 1). The most abundant PAHs found were fluoranthene and pyrene in both LWV sediment layers. PAHs in LVT sediment were very low. However, anthracene, fluoranthene, and pyrene were detected in a few LVT surface sediment samples and in almost all of the subsurface sediment samples, but at concentrations too low to be quantified.
Microbial Biomass and Activity
River bank surface sediment from LVT showed higher PLP biomass (350 nmol P dry weight) than was found in subsurface sediment (Fig. 2). In contrast at the LWV site, the highly contaminated riverbank subsurface sediment showed higher biomass than was found in the less contaminated surface sediment. Subsurface sediments from both locations had similar biomass values (Fig. 2). However, the contaminated subsurface riverbank sediments at LWV showed the highest microbial activity of all samples taken. Lower levels of microbial activity were found in surface sediments from both locations (Fig. 2).
Microbial Community Structure
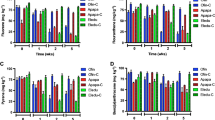
Consistent results were found during each of several separate extractions, indicating that fatty acid analyses were very reliable and reproducible. A total of 17 different PLFA biomarkers were identified in Mahoning riverbank sediments from the two sites in this study (Fig. 3a, b). Overall, markers for green algae, higher plants, heterotrophic microeukaryotes, fungi, aerobic bacteria, sulfate reducing, and other anaerobic bacteria subsurface sediment from both locations made up 84–86 % of the quantified community by PLFAs (Table 2).
a Distribution of PLFA biomarkers in Mahoning Riverbank surface and subsurface sediment from the LWV site. Black bars surface sediments, white bars subsurface sediments. b Distribution of PLFA biomarkers in Mahoning Riverbank surface and subsurface sediment from the LVT site. Black bars surface sediments, white bars subsurface sediments
Many of the detected PLFA biomarkers were found at both sites at both depths with some notable exceptions. General and non-specific microbial markers (16:0, 16:1ω7, 18:1ω9, and 18:1ω7; [2, 20]) were detected in high concentrations in all samples (Fig.3a, b). The PLFA 18:0, indicative of sulfate reducing and other anaerobic bacteria, was also abundant in all samples. Markers for Gram-positive bacteria, i15 and a15, were detected in all samples. A marker for eukaryotes, 18:2ω6, was also found in all samples. Mid-branched br17:0b, a marker for Gram-positive bacteria, was detected everywhere except LWV subsurface contaminated sediment. PLFA biomarkers for general bacteria (i17, 17:0, 19:0, and a19:0) were all present in surface sediment and were not detected (with one exception, 19:0) in subsurface sediment at both sites (Fig. 3). A PLFA biomarker for methanotrophic bacteria type I (16:1ω8) was detected in surface sediment but not in subsurface sediment at both sites. Subsurface sediment from LWV had the highest proportion of the PLFA biomarkers for anaerobic bacteria (37 %) relative to surface sediment and sediment from LVT (Table 2). However, 10Me16:0, a PLFA biomarker for actinomycetes and/or the sulfate reducing bacteria Desulfobacter, was also found in surface sediment and not in subsurface sediment at both sites.
Shannon–Weaver diversity indices indicated that surface samples displayed higher PLFA community diversity than found subsurface samples, regardless of site (Fig. 4). Surface riverbank sediment from the LVT site showed both the greatest number and most diverse PLFA biomarkers while conversely, LVT subsurface riverbank sediment showed the least diverse PLFA biomarkers (Fig. 4).
Principal Component Analysis
When the sediment samples (excluding PAH data) were plotted on ordination axes consisting of the first two principle components they clustered primarily by depth and then by site (Fig. 5). Since PAH concentrations in samples from LVT were present at or below the limit of quantification, they were not used for ordination analysis of LVT and LWV data (Fig. 5). PCA extracted two components that explained 64 % of the data variance for both locations (not including PAH data). High positive loadings on PC 1 among PLFAs and biomass were found within the data set (Fig. 6).
A separate analysis was conducted to evaluate the LWV data which included PAH concentrations. When LWV samples were plotted on ordination axes consisting of the first two principle components they clustered by depth (Fig. 7). In the LWV site when the PAH data were included, PCA extracted two components that explained 59 % of the data variance (Fig. 8). There were high positive loadings on PC 1 among PLFAs and percent moisture in LWV and opposite loadings of PAHs and PLFAs. Multivariate ordination (principal components analysis) indicated opposite loadings (negative vs. positive) of PAHs and PLFAs.
Discussion
This study showed that microbial communities within riverbank sediment responded to multiple environmental factors including anthropogenic PAH contamination. High microbial activity was the most distinct biological observation associated with PAH contamination within Mahoning riverbank sediment. Higher microbial activity measured as tetrazolium dye reduction in the presence of organic contaminants has also been observed for beach sediment microbial communities spiked with oil [36] or soil microbial communities spiked with oil-based nonaqueous drilling fluids [51]. In both of these systems, the authors concluded that components of the contaminating material served as a carbon source for the microbes and caused higher rates of dehydrogenase activity as is also likely in Mahoning River sediment. Rates of INT reduction in this present study ranged from 0.02 to 1.2 μmol h−1 g−1 in uncontaminated and contaminated sediment, respectively, and showed values at the low end of the range reported previously for Mahoning River benthic sediments [33]. These rates of INT reduction were all higher than found in weathering sandstone (0.002–0.008 μmol h−1 g−1) from the UK [44]. The lower rates from the Mahoning River were within the ranges in arable soil (0.2–0.6 μmol h−1 g−1) reported by Von Mersi and Schinner [49], in sandy loam soil (0.04–0.2 μmol h−1 g−1) reported by Trevors et al. [46], and in beach sediment contaminated with crude oil (0.005–0.02 μmol h−1 g−1) reported by Mathew and Obbard [32]. Previously, it was shown that when anaerobic Mahoning River sediment was incubated aerobically, the measure of INT reduction was 15 % greater [33]. Thus, rates of INT reduction from subsurface sediment in this present study likely overestimate respiration by 15 %.
Evidence that INT reduction was biologically mediated rather than an abiotic reaction includes temperature and metabolic inhibitor data. Rates of INT reduction increased from 4 to 45 °C, was a little higher at 60 °C, and was the lowest at 95 °C [33]. This temperature response is typical of biological processes and would not be expected if abiotic factors caused INT reduction. Carbonyl cyanide m-chlorophenylhydrazone (CCCP), a respiratory inhibitor that acts by shuttling electrons and collapsing the membrane potential, was shown to inhibit INT reduction in contaminated Mahoning River sediment by 75 and 94 % at concentrations of 1.6 and 3.2 g/L, respectively [26]. CCCP did not inhibit INT reduction at 160 mg/L, likely due to interference by one or more components in the complex mixture of organic contaminants and heavy metals found in this sediment. Mosher et al. [33] also reported that 20 mM sodium azide, a concentration that normally inhibits respiration, did not inhibit INT reduction in contaminated Mahoning River sediment. Overall this evidence strongly supports the hypothesis that INT reduction in Mahoning River sediment is biologically mediated and depends on respiration.
According to the species-energy theory, first developed by Wright [54] as an extension of the species-area theory from island biogeography, available energy can be used to predict species abundance. The species-energy theory can be extended to sedimentary systems where carbon is generally added during deposition. Over time, in deeper sediment, the more easily degraded organic material is consumed while more recalcitrant, harder to degrade organic carbon persists. Deeper sediment is generally associated with relatively lower concentrations of easily degraded organic matter and oxygen (i.e., lower yields of biologically available energy) and expected to have lower biomass and microbial diversity [42]. At the LVT site, both biomass and diversity were lower in subsurface than surface sediments as predicted by species-energy theory. At the LWV site, the pattern of organic carbon distribution in riverbank sediment diverges from the expected pattern due to high anthropogenic inputs of oils, greases, petroleum hydrocarbons, and PAHs that lasted for 80 years until the mid-1970s. At LWV, high concentrations of organic matter (PAHs and petroleum hydrocarbons) in subsurface sediment have persisted and may provide energy for suitable microbes, supporting a somewhat higher biomass than found in surface sediment. There was a greater difference in biomass values in PAH-contaminated vs. PAH-uncontaminated Mahoning River benthic sediment [33]. PLP biomass in PAH-contaminated benthic sediment the from the LWV site was 91 nmol P g−1 and was only 25 nmol P g−1 in relatively uncontaminated benthic sediment from the LVT site. Fang and Barcelona [15] also showed that microbial communities in subsurface hydrocarbon-contaminated anaerobic zones had higher biomass values than those in uncontaminated aerobic zones. There was no direct relation, however, between biomass and microbial activity in Mahoning River sediments. This is not unexpected since it is commonly observed that microbial biomass, diversity, and activity at different sediment or soil depths are affected by multiple factors such as amount and quality of organic carbon, redox, nutrient concentrations, and sediment grain size [6, 12, 42].
PAHs and Microbial Communities
Subsurface total PAH concentrations in riverbank sediment at LWV (250 μg/g) (Fig. 1) were within 10 % of the values reported earlier (230 μg/g) for benthic river sediment located nearby [33]. Other reported results in Mahoning River benthic sediment from near LWV (River Mile 13.3) showed total PAH concentrations of ~80 μg/g of PAHs [35]. These results indicate a high heterogeneity and distribution of PAHs within location.
In this study, any effect of PAH contamination on community structure was expected as differences in patterns of PLFAs in the subsurface sediment from LWV and LVT. Interestingly, PAH contamination did not appear to greatly influence the distribution of PLFA biomarkers apart from depth (Fig. 3). A closer ordination analysis did indicate underlying differences between PLFA diversity in the two sites (Fig. 5). PLFA subsurface data from the contaminated LWV site clustered more closely than the subsurface data from the uncontaminated LVT site (Fig. 5). In agreement with these results, Slater et al. [41], when examining microbial communities in surface harbor sediments, showed that PAH deposition did not have a major effect on PLFA distribution.
A comparable study of microbial communities in PAH-contaminated river was conducted in the Little Scioto River in Marion, OH [31]. In that study, PAH-contaminated benthic sediment with intermediate levels of PAHs (30 μg/g−1) had higher biomass and PLFA diversity than the non-to-low PAH-contaminated sediment (0.4 μg/g−1) and highly PAH-contaminated sediment (107 μg/g−1). LWV subsurface Mahoning riverbank sediment in this present study is more than twice as contaminated by PAHs as the most highly contaminated benthic sediments in the Little Scioto River, while the relatively uncontaminated Mahoning riverbank LWV surface sediment is closer to the high range for the Little Scioto River. Highly contaminated LWV subsurface sediment might be a more extreme environment selecting for a more adapted microbial community reflected by a lower PLFA diversity with higher activity.
PLFA biomarkers revealed that the overall most abundant marker was 16:0, which represents a broad phylogenetic distribution and is very often the most abundant marker found in aquatic sediments [11]. There were higher percentages of anaerobes in the subsurface sediment samples than in the surface sediment samples. However biomarkers characteristic for other sulfate reducing bacteria such as Desulfobulbus and Desulfovibrio (15:1g6c and i17:ω7c respectively; [29]) were not detected. The biomarker 10Me16:0, present in surface and absent in subsurface sediment, is often associated with Desulfobacter and other anaerobic prokaryotes [17] and/or with Actinomycetes [42]. In Mahoning sediment, 10Me16:0 was most likely associated with Actinomycete since many Actinomycetes species are aerobes. Short PLFAs (i:15, a:15, and 16:0) were observed consistently for LWV surface sediments which correlates with a typical spring community composition [5].
Ringelberg et al. [38] indicated that Gram-negative bacteria markers became less abundant over time in a PAH-contaminated sediment slurry system suggestive that there should be fewer Gram-negative bacteria within PAH-contaminated sediment. Gram-negative bacteria markers (16:0, 16:1ω7c, and 18:1ω7c) were more abundant in Mahoning River bank surface sediments than in the subsurface at both sites. However, only the LWV subsurface sediments were highly contaminated; thus, the distribution of Gram-negative likely had more to do with sediment depth rather than PAH and metal contamination. Gram-positive markers (e.g., i17) were only detected in surface sediments while i15 and a15, both considered as specific to Gram-positive bacteria, were present in contaminated and uncontaminated surface and subsurface sediments. Mid-branched fatty acids have been associated with anaerobic bacterial communities; however, in this study, mid-branched fatty acids (br17:0b and br17:0a) were more abundant in surface than in subsurface sediments (Fig. 3a, b). In LWV subsurface sediments, br17:0b and br17:0a were not detected (in multiple samples). It is possible that high concentrations of contaminants (PAHs or metals) within that layer may have inhibited bacteria that exhibit those markers.
PLFA analyses in soils contaminated with petroleum hydrocarbons [27] and in PAH-contaminated river sediment [30] indicate that eukaryotes such as fungi make up a greater portion of the microbial community in contaminated vs. uncontaminated soils or sediments. Specifically, eukaryotic biomarkers (18:2ω6 or 18:2 ω9,12) were found in greater abundance in contaminated vs. uncontaminated soil or sediment. In contrast, in this study, there was on average lower levels of 18:2ω6 (fungi biomarker) in Mahoning contaminated vs. uncontaminated sediment; however, standard deviations were too large for statistical significance. Fatty acids indicators of fungi, mycorrhizal fungi, and actinomycetes are usually found in lower numbers in environments closed to smelters and with metal contamination. Kelly et al. [28] reported low presence of 18:2ω6 in soils with metal contamination. Others (e.g., [23, 37]) have suggested that this trend might be related to root tree damage by metal contamination.
The association of unique PLFAs as biomarkers for taxa came from work with pure cultures and is not always valid in situ [19]. Changes in PLFA biomarkers in environmental samples may be from either shifts in species composition or from changes in membrane composition within the same species. There are discrepancies and overlaps between some reported associations of PLFAs with particular taxa which can make interpretation difficult. A large number of researchers (e.g., [3, 25]) have suggested indexing a PLFA community structure as PLFA phenotypic microbial community structure. Because phospholipids are essential membranes of cells, bacteria can change their composition as response to environmental stresses, i.e., high pollutant contamination. Therefore, it is challenging to assess if PLFA community structure is the result of changed species or is the result of phenotypic responses in an unchanged community.
Conclusions
This study shows that the PAH fingerprint the Mahoning River was consistent with previous reports, but highly heterogeneous. PAH concentration was not an important factor in estimating PLFA diversity. PLFA analysis of subsurface sediments revealed that a majority of the community corresponded to anaerobic bacteria. Fungal biomarkers appeared to be influenced by PAH contamination, while Gram-negative bacteria did not correlate with sediment contamination but with depth. Riverine and marine PAH-contaminated systems show a wide diversity of PAH degrading bacteria and/or bacteria with PAH degrading genes, so it is likely that they are also present in PAH-contaminated Mahoning River bank sediments. PAH contamination is associated with other parameters such as humic compounds, petroleum hydrocarbons, and high heavy metal concentrations, which may also be confounding factors for microbial diversity. The upstream LVT uncontaminated site is the best available control for the contaminated LWV site, but it (like all other sites) is not an exact match for all environmental parameters on the micro scale. Further work at the molecular level will be needed to conclusively determine the effect of PAH contamination on the microbial community structure and diversity and to determine the presence of PAH degrading genes.
References
Agency for Toxic Substances and Disease Registry (ATSDR) Division of Toxicology (2010) ToxFAQs for polycyclic aromatic hydrocarbons. ATSDR, Atlanta. http://www.atsdr.cdc.gov/tfacts69.html. Accessed Nov 2010
Baniulyte D, Favila E, Kelly J (2009) Shifts in microbial community composition following surface application of dredged river sediments. Microb Ecol 57:160–169
Bausenwein U, Gattinger A, Langer U, Embacher A, Hartmann H-P, Sommer M, Munch JC, Schloter M (2008) Exploring soil microbial communities and soil organic matter: variability and interactions in arable soils under minimum tillage practice. Appl Soil Ecol 40:67–77
Bhupathirajua VK, Hernandezb M, Landfeara D, Alvarez-Cohen L (1999) Application of a tetrazolium dye as an indicator of viability in anaerobic bacteria. J Microbiol Methods 37:231–243
Björk R, Björkmand M, Andersson M, Klemedtsson L (2008) Temporal variation in soil microbial communities in Alpine tundra. Soil Biology & Biochemistry 40:266–268
Blume E, Bischoff M, Reichert JM, Moormanc T, Konopka A, Turco RF (2002) Surface and subsurface microbial biomass, community structure and metabolic activity as a function of soil depth and season. Appl Soil Ecol 20:171–181
Boschker HTS, Middelburg JJ (2002) Stable isotopes and biomarkers in microbial ecology. FEMS Microbiol Ecol 40:85–95
DeBruyn J, Mead T, Wilhelm S, Sayler G (2009) PAH biodegradative genotypes in Lake Erie sediments: evidence for broad geographical distribution of pyrene-degrading Mycobacteria. Environ Sci Technol 43:3467–3473
Dijkman NA, Boschker HT, Stal LJ, Kromkamp JC (2010) Composition and heterogeneity of the microbial community in a coastal microbial mat a as revealed by the analysis of pigments and phospholipid-derived fatty acids. J Sea Research 63:62–70
Drenovsky RE, Elliot GN, Graham KJ, Scow KM (2004) Comparison of phospholipid fatty acid (PLFA) and total soil fatty acid methyl esters (TSFAME) for characterizing soil microbial communities. Soil Biology & Biochemistry 36:1793–1800
Drijber RA, Doran JW, Parkhurst AM, Lyon DJ (2000) Changes in soil microbial community structure with tillage under long-term wheat-fallow management. Soil Biology & Biochemistry 32:1419–1430
Dunn RM, Mikola J, Bol R, Bardgett RD (2006) Influence of microbial activity on plant-microbial competition for organic and inorganic nitrogen. Plant Soil 289:321–334
Duran M, Haznedarog BZ, Zitomer DH (2006) Microbial source tracking using host specific FAME profiles of fecal coliforms. Water Res 40:67–74
Fang J, Findlay RH (1996) The use of a classic lipid extraction method for simultaneous recovery of organic pollutants and microbial lipids from sediments. J Microbiol Meth 27:63–71
Fang J, Barcelona MJ (1998) Biogeochemical evidence for microbial community change in a jet fuel hydrocarbons-contaminated aquifer. Org Geochem 29:4899–4907
Findlay RH (2004) Determination of microbial community structure using phospholipid fatty acid profiles. In: Kowalchuk GA, Bruijn FJ, Head IM, Akkermans AD, van Elsas JD (eds) Molecular microbial ecology manual, 2nd edn. Kluwer Academic, Norwell, pp 983–1005
Findlay RH, Trexler MB, Guckert JB, White DC (1989) Laboratory study of disturbance in marine sediments: response of a microbial community. Mar Ecol Prog Ser 62:121–133
Foster AL, Munk L, Koski RA, Shanks WC III, Stillings LL (2008) Relationships between microbial communities and environmental parameters at sites impacted by mining of volcanogenic massive sulfide deposits, Prince William Sound, Alaska. Appl Geochem 23:279–307
Frostegård Å, Tunlid A, Bååth E (2011) Use and misuse of PLFA measurements in soils. Soil Biology and Biochemistry 43(8):1621–1625
Glucksman AM, Skipper HD, Brigmon RL, Doming JW (2000) Use of MIDI-FAME technique to characterize groundwater communities. J Appl Microbiol 88:711–719
Haack SK, Garchow H, Odelson DA, Forney LJ, Klug MJ (1994) Accuracy, reproducibility, and interpretation of fatty acid methyl ester profiles of model bacteria communities. Appl Environ Microbiol 60:2483–2493
Hayes L, Nevin K, Lovley D (1999) Role of prior exposure on anaerobic degradation of naphthalene and phenanthrene in marine harbor sediments. Org Geochem 30:937–945
Helmisaari HS, Derome J, Fritze H, Nieminen T, Palmgren K, Salemaa M, Vanha-Majamaa I (1995) Copper in Scots pine forests around a heavy metal smelter in south-western Finland. Water Air Soil Pollut 85:1727–1732
Hoffman M, Keys DE, Song K-Y, Brown EW, Fry FS, Whittaker P (2008) Evaluation of multiple strains of Enterobacter sakazakii using fatty acid profiles. Food Chem 107:1623–1628
Jeffery S, Harris JA, Rickson RJ, Ritz K (2007) Microbial community phenotypic profiles change markedly with depth within the first centimeter of the arable soil surface. Soil Biol Biochem 39:1226–1229
Johnston GP, Kalik Z, Johnston CG (2009) Effect on carbonyl cyanide m-chlorophenylhydrazone (CCCP) on microbial activity in historically contaminated river sediment. American Society for Microbiology General Meeting, Boston
Joynt J, Bischoff M, Turco R, Konopka A (2006) Microbial community analysis of soils contaminated with lead, chromium and petroleum hydrocarbons. Microb Ecol 51:209–219
Kelly J, Häggblom M, Tate R (2003) Effects of heavy metal contamination and remediation on soil microbial communities in the vicinity of a zinc smelter as indicated by analysis of microbial community phospholipid fatty acid profiles. Biol Fertil Soils 38:65–71
Kleikemper J, Pelz O, Schroth MH, Zeyer J (2002) Sulfate-reducing bacterial community response to carbon source amendments in contaminated aquifer microcosms. Microb Ecol 42:109–118
Langworthy DE, Stapleton RD, Sayler GS, Findlay RH (1998) Genotypic and phenotypic responses of a riverine microbial community to polycyclic aromatic hydrocarbon contamination. Appl Environ Microbiol 64:3422–3428
Langworthy DE, Stapleton RD, Sayler GS, Findlay RH (2002) Lipid analysis of the response of a sedimentary microbial community to polycyclic aromatic hydrocarbons. Microb Ecol 43:189–198
Mathew M, Obbard JP (2001) Optimisation of the dehydrogenase assay for measurement of indigenous microbial activity in beach sediments contaminated with petroleum. Biotechnol Lett 23:227–230
Mosher JJ, Levison BS, Johnston CG (2003) A simplified dehydrogenase enzyme assay in contaminated sediment using 2-(p-iodophenyl)-3(p-nitrophenyl)-5-phenyl tetrazolium chloride. J Microb Meth 53:411–415
Mosher JJ, Findlay RH, Johnston CG (2006) Physical and chemical factors affecting microbial biomass and activity in contaminated subsurface sediment. Can J Microbiol 52:397–403
Ohio Environmental Protection Agency (1996) Biological and water quality study of the Mahoning River Basin. OEPA technical report MAS/1995-12-14, pp 1–249
Obbard JP, Ng KL, Xu R (2004) Bioremediation of petroleum contaminated beach sediments: use of crude palm oil and fatty acids to enhance indigenous biodegradation. Water Air Soil Poll 157:149–161
Pennanen T, Frosetgård A, Fritze H, Bååth E (1996) Phospholipid fatty acid composition and heavy metal tolerance of soil microbial communities along two heavy metal polluted gradients in coniferous forests. Appl Environ Microbiol 62:420–428
Ringelberg DB, Talley JW, Perkins EJ, Tucker SG, Luthy RG, Bouwer EJ, Fredickson HL (2001) Succession of phenotypic, genotypic, and metabolic community characteristics during in vitro bioslurry treatment of polycyclic aromatic hydrocarbon-contaminated sediments. Appl Env Microbiol 67:1542–1550
Ruamps LS, Nunan N, Chenu C (2011) Microbial biogeography at the soil pore scale. Soil Biol Biochem 43:280–286
Ruess L, Chamberlain PM (2010) The fat that matters: soil food web analysis using fatty acids and their carbon stable isotope signature. Soil Biology & Biochemistry 42:1898–1910
Slater G, Cowie B, Harper N, Droppo I (2008) Variation in PAH input and microbial community in surface sediments of Hamilton Harbour: implications to remediation and monitoring. Environ Pollut 153:60–70
Song Y, Deng SP, Acosta-Martínez V, Katsaliro E (2008) Characterization of redox-related soil microbial communities along a river floodplain continuum by fatty acid methyl ester (FAME) and 16S rRNA genes. Appl Soil Ecol 40:499–509
Syakti AD, Mazzella N, Nerini D, Guiliano M, Bertrand JC, Doumenq P (2006) Phospholipid fatty acids of a marine sedimentary microbial community in a laboratory microcosm: responses to petroleum hydrocarbon contamination. Org Geochem 37:1617–1628
Tayler S, May E (2000) Investigations of the localization of bacterial activity on sandstone from ancient monuments. Int Biodeter Biodegr 46:327–333
Tiessen H, Moir JO (1993) Total organic carbon. In: Carter ME (ed) Soil sampling and methods of analysis. Lewis, Ann Arbor, pp 187–211
Trevors JT, Mayfield CI, Innis WE (1982) Measurement of electron transport system (ETS) activity in soil. Microb Ecol 8:163–168
United States Army Corps of Engineers (USACE) (1999) Mahoning River Dredging Study (Ohio). Energy and Water Development Appropriations Act (Public Law 104–303)
Vestal JR, White DC (1989) Lipid analysis in microbial ecology: quantitative approaches to the study of microbial communities. Biosci 39:535–541
Von Mersi V, Schinner F (1991) An improved and accurate method for determining the dehydrogenase activity of soils with iodonitrotetrazolium chloride. Biol Fertil Soils 11:216–220
Villanueva L, Navarrete A, Urmeneta J, White DC, Guerrero R (2007) Analysis of diurnal and vertical microbial diversity of a hypersaline microbial mat. Arch Microbiol 188:137–146
Wakadikar K, Sil A, Kolekar N, Tandon S, Kumar R (2011) Effect of non-aqueous drilling fluid and its synthetic base oil on soil health as indicated by its dehydrogenase activity. Environ Earth Sci 64:25–28
Whittaker P (2009) Comparison of Yersinia pestis to other closely related Yersinia species using fatty acid profiles. Food Chem 116:629–632
Williams MA, Rice CW (2007) Seven years of enhanced water availability influences the physiological, structural, and functional attributes of a soil microbial community. Appl Soil Ecol 35:535–545
Wright D (1983) Species-energy theory, an extension of species-area theory. Oikos 41:496–506
Wu Y, Ding N, Wang G, Xu J, Wu J, Brookes PC (2009) Effects of different soil weights, storage times and extraction methods on soil phospholipid fatty acid analyses. Geoderma 150:171–178
Acknowledgments
This work was supported by a Presidential Academic Center for Excellence in Research grant from Youngstown State University. The authors are grateful for useful discussions about statistics from T Diggins from the Department of Biological Sciences, Youngstown State University and J Kearns from the Department of Mathematics and Statistics, Youngstown State University and for editorial suggestions by GP Johnston from the Biology Department, Kent State University.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(PPT 775 kb)
Rights and permissions
About this article
Cite this article
Pratt, B., Riesen, R. & Johnston, C.G. PLFA Analyses of Microbial Communities Associated with PAH-Contaminated Riverbank Sediment. Microb Ecol 64, 680–691 (2012). https://doi.org/10.1007/s00248-012-0060-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-012-0060-8