Abstract
The microbial community of a Colombian high mountain hot spring, El Coquito, was analyzed using three different culture-independent assessments of 16S ribosomal RNA genes: clone libraries, pyrosequencing of the V5–V6 hypervariable region, and microarray. This acidic spring had a diverse community composed mainly of Bacteria that shared characteristics with those from other hot springs and extreme acidic environments. The microbial community was dominated by Proteobacteria, Firmicutes, and Planctomycetes and contained chemotrophic bacteria potentially involved in cycling of ferrous and sulfur-containing minerals and phototrophic organisms, most of which were eukaryotic micro-algae. Despite the presence of a large proportion of novel, unclassified sequences, the taxonomic profiles obtained with each strategy showed similarities at higher taxonomic levels. However, some groups, such as Spirochaetes and Aquificae, were identified using only one methodology, and more taxa were detected with the gene array, which also shared more groups with the pyrosequencing data. Overall, the combined use of different approaches provided a broader view of the microbial community in this acidic hot spring.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
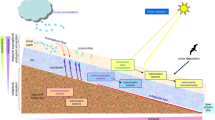
The Tropical Andes are located in the Neotropical ecoregion and are considered a hotspot for biodiversity due to the large number of endemic plant and vertebrate species [1]. The central Andean mountain range in Colombia is part of a volcanic belt characterized by geothermal activity that is made evident by the presence fumaroles and hot springs, many of which are found within the Nevados National Natural Park (Nevados NNP) [2]. These hot springs are heated mainly by the underlying magma chamber from volcanic activity and have a high sulfate content and low pH. El Coquito spring is located within protected areas of the Nevados NNP; it has little or no anthropogenic influence and constitutes a unique and extreme ecosystem due to its high elevation (3,973 m above sea level), high exposure to UV light, water temperature, and mineral composition. Furthermore, no research has been done regarding the microbial communities that populate this acidic hot spring.
The use of culture-independent methods to study microbial diversity has expanded our view of the microbial world and allowed access to extreme and difficult to study environments, such as these acidic water ecosystems [3]. High-throughput approaches, which include gene chips and novel sequencing technologies, can provide rapid detection and higher resolution of the microbial communities in a complex sample. A high-density 16S ribosomal RNA (rRNA) gene microarray for parallel, multispecies detection has been used to analyze and compare diverse communities [4], revealing greater diversity when compared with 16S rRNA gene clone libraries [5–8]. Surveys with pyrosequencing of 16S rRNA gene variable regions (pyrotags) provide greater depth of coverage, with thousands of sequence reads per sample, and have revealed the presence of rare community members that might otherwise go undetected when using more labor-intensive clone libraries [9, 10]. A recent study of the microbial communities from the human intestine using 454 pyrosequencing and a phylogenetic microarray showed similar profiles and a strong correlation at the phylum, class, and order levels [11]. While these high-throughput approaches provide a great amount of information without a priori knowledge of the community structure, they are also subject to biases that arise from methodological and technical limitations inherent to sample preparation, PCR amplifications, and sequencing [12–15]. Thus, descriptions of bacterial communities using these different approaches may result in different profiles that, even if not completely consistent with one another, can provide a more thorough overview of the community structure.
In this study, the microbial community present in El Coquito hot spring was characterized using three different approaches based on amplification and analysis of 16S rRNA genes: a high-density 16S rRNA gene microarray, 454 pyrosequencing of hypervariable regions (V5–V6), and clone libraries of near full-length genes. Our multi-approach analysis revealed great diversity and gave a more thorough assessment of the structure of this acidophilic microbial community.
Materials and Methods
Sample Collection and Analysis
Superficial running stream water (16 L) was collected in 5-L sterile plastic containers in April 10 2008 at El Coquito hot spring (04°52′27″ N; 75°15′51.4″ W) by filling the containers to the brim and capping. Samples were transported at 4°C to the laboratory and processed within 18 h for further analysis (physicochemical analysis, total microbial counts, and DNA isolation). Temperature and pH were recorded in situ using a Hach pH-meter equipped with a pH and temperature probe. Physicochemical analyses were performed according to Standard Methods [16]. Water (500 mL) was fixed with 4% (v/v) paraformaldehyde and filtered using a 0.22-μm polycarbonate filter (Millipore, Billerica, MA, USA). Filter sections were impregnated with 4′,6-diamidino-2-phenylindole (DAPI; 1 mg mL−1) for 10 min, washed with Milli-Q water for 1 min, impregnated with 70% ethanol, and air-dried. Fluorescent in situ hybridization (FISH) was done with probes (Integrated DNA Technologies, Coralville, IA, USA) EUB 338 [17] and ARCH 915 [18] labeled with Alexa Fluor® Dyes 488 and 546 (Invitrogen, San Diego, CA, USA) for Bacteria and Archaea, respectively. Dehydration with ethanol and in situ hybridization were conducted as described [19]. Cells were counted in duplicate using an epifluorescent microscope (Nikon Eclipse 50, Nikon, Melville, NY, USA) [20]. If more than 30 cells per field were observed, 20 microscope fields were counted; otherwise, 50 microscope fields were analyzed.
DNA Extraction
DNA was isolated as previously described [21] by filtering water (10 L) first through a 5.0-μm cellulose filter (Fisherbrand, Fisher, Houston, TX, USA) and then through a 0.22-μm polycarbonate filter (Millipore, Billerica, MA, USA). Cells on the filter were lysed by incubation at 37°C for 45 min in lysis buffer [lysozyme (1 mg mL−1), proteinase K (0.2 mg mL−1), and achromopeptidase (0.6 mg mL−1)]. Crude lysates were extracted twice with phenol/chloroform/isoamyl alcohol (25:24:1, pH 8.0). DNA was further cleaned using the UltraClean® GelSpin® DNA Purification Kit (MOBIO Laboratories, Inc., Carlsbad, CA, USA), quantified by NanoDrop (Thermo Scientific, Inc., Wilmington, DE, USA), and checked for quality by 1% agarose gel electrophoresis using SYBR® Safe staining (Invitrogen, San Diego, CA, USA). Images were digitized with the software Quantity One® v. 4.6.3 (BioRad Laboratories, Inc., Hercules, CA, USA). The DNA was stored at −20°C prior to amplification.
Clone Libraries
Bacterial primers 8F and 915R [22] and archaeal primers ARCH 109F and ARCH 915R [23] were used to amplify 16S rRNA genes (Table 1). Each 50 μL PCR reaction contained 2 μL (40 ng) DNA, 0.1 mM of each dNTP, 1.5 mM MgCl2, 1× PCR buffer (Invitrogen), 0.2 μM of each primer, and 0.5 U DNA Polymerase (Invitrogen). Amplification using bacterial primers was accomplished by denaturation at 94°C for 3 min, 25 cycles of 92°C for 40 s, 52°C for 30 s, and 72°C for 2 min, with a final extension at 72°C for 10 min. PCR conditions using archaeal primers were identical, except that the initial denaturation was done at 96°C for 3 min. Amplicons were purified using the UltraClean® PCR Clean-Up kit (MOBIO Laboratories Inc., Carlsbad, CA, USA) and cloned using the HTP TOPO TA kit (Invitrogen). Insert DNA was amplified using primers M13F and M13R (Table 1), and randomly selected clones were sequenced on both strands (Macrogen Inc., Seoul, South Korea). Sequences of insufficient length or quality were removed using the software CLC Workbench version 5.2 and checked using the Chimera Check tool available in Greengenes [24] (http://greengenes.lbl.gov/cgi-bin/nph-bel3_interface.cgi). These sequence data have been submitted to the GenBank database under accession numbers JF280147–JF280363 (archaeal sequences) and JF280364–JF280675 (bacterial sequences). Sequence alignment was carried out using Infernal [25], and classification was done with the Ribosomal Database Project (RDP) naive Bayesian classifier [26] (http://rdp.cme.msu.edu/classifier/classifier.jsp) using an 80% confidence threshold. The DOTUR software was used to assess microbial richness and sample coverage associated with each clone library [27]. Non-parametric richness (S CHAO and S ACE) and coverage (Good’s and C ACE) estimators are widely used to estimate the total number of operational taxonomic units (OTUs) and the proportion represented in a given sample to assess if sufficient work has been done to capture most of the diversity in a sampled environment [28]. Representatives of each OTU were analyzed with the GenBank database by using the basic local alignment search tool (BLAST) at the NCBI website.
Pyrosequencing of V5–V6 Hypervariable Regions
Primers were designed based on previous reports [10] and modified to include a broader range of taxa by downloading and aligning 5,530 bacterial 16S rRNA sequences from the RDP (as of November 2008, in the range of 100–400 bp) and analyzing the V5–V6 region for sequence variation. Primers were designed according to full-alignment representation by placing 3′ degenerate nucleotides. Using the probe match tool (RDP 10.4), this primer set could theoretically anneal and amplify 99.96% of bacterial and 97.82% of archaeal sequences, allowing one mismatch, based on comparison against the entire database of sequences reported in the RDP database on October 11, 2008 (690,149 sequences). PCR amplifications were done as reported at the time [29] in a 25-μL reaction volume containing 2 μl (20 ng) DNA, 0.75 μM of each primer 807F and 1050R designed by us (Table 1), 2.5 U Pfu Turbo® DNA polymerase (Stratagene, Inc., La Jolla, CA, USA), 1× Pfu reaction buffer, 0.6 mM dNTPs, 5% v/v dimethyl sulfoxide using the following PCR conditions: 2 min at 95°C, 30 cycles consisting of denaturation for 30 s at 95°C, a temperature touch down from 60 to 51°C (2°C every six cycles), 72°C for 1 min and a final extension of 72°C for 5 min. The PCR product was cleaned using the QIAquick PCR Purification Kit (Qiagen N.V., Hilden, Germany) and used as template for a second PCR to add pyrosequencing adapter and barcode sequences (http://pyro.cme.msu.edu/) using primers 16S807F-b15 and 16S1050R-b5 (Table 1) and conditions identical to those of the first PCR, except for the number of cycles (five) and the annealing temperature (53°C). PCR products were assessed by 1% agarose gel electrophoresis stained with ethidium bromide. Pyrosequencing was carried out from the reverse primer (Engencore, University of South Carolina, Columbia, SC, USA). These sequence data are available at the NCBI Short-Read Archive (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=studies) under accession number SRA029240.1. Sequences were cleaned using tools implemented in the GeBiX diversity portal (www.gebix.org.co/gbx_diversity), taking care to minimize the effects of sequencing errors, as reported [24, 30, 31]. Sequences were evaluated based on quality scores, eliminating those that did not match perfectly the primer and barcode at the beginning of the read or that contained more than one undetermined nucleotide (N). Sequences were then trimmed using a 40-nucleotide window for analysis, shifted one nucleotide at a time, and required 90% of the bases to surpass the established threshold of 20 for the window to be considered of good quality. The size of the sequences was restricted to a minimum of 50 bases, after eliminating the first 29 bases corresponding to the tag (two bases), barcode (eight bases), and primer (19 bases). Complementary reverse sequences obtained for each clean read were aligned using Infernal v 1.0.2 [25]. Dnadist PHYLIP version 3.6 was used to generate pairwise distance matrices [32], and diversity and richness indices were obtained using DOTUR with the furthest neighbor algorithm [27]. Sequences were assigned to bacterial phyla and families with the RDP-naive Bayesian classifier, using an 80% confidence threshold.
16S rRNA Gene Microarray
We used the PhyloChip (Affymetrix Inc., Santa Clara, CA, USA), as described previously using universal primers 27F and 1492R (Table 1) [5, 33], by pooling eight PCR reactions, each carried out with 10 ng of DNA per PCR. Briefly, full-length 16S rRNA gene PCR was fragmented, biotin-labeled, and hybridized to the array. The microarray was scanned and recorded as a pixel image, and initial data acquisition and intensity determination were performed using custom Affymetrix software. Data analysis was performed as described [33]. In order to assign the same taxonomy to 16S rRNA gene sequences, the reference Greengenes sequence (rep-prokMSA-id) for each OTU detected on the microarray was classified using the online RDP II classifier with an 80% confidence threshold. Array taxa were analyzed for the presence of 454 primer sequences using 78 sequences belonging to orders detected only with the PhyloChip (two or more sequences for each order). Sequences were aligned using Muscle 3.6 with the parameter, −maxiters 2 [34], and aligned sequences were manually inspected for the presence of the 454 primers used to amplify the V5–V6 regions.
Phylogenetic Reconstruction
Phylogenetic reconstruction was made using the ARB 5.1 software [35] equipped with SILVA 100 database [36], with the representative sequences of each OTU obtained at 97% identity for both clone libraries and 454 pyrotags and the reference sequences from Greengenes for each OTU in the microarray. Sequences were aligned using the SILVA aligner tool (http://beta.arb-silva.de), imported into ARB, and inserted into the existing reference neighbor-joining tree using the parsimony insertion tool [35].
Results
Site Description and Physicochemical Features
Water at the hot spring El Coquito emerges at 3,973 m above sea level and becomes a flowing 2–3 m wide, shallow stream (30 cm) surrounded by the plant Calamagrostis effusa and different species of moss; the area in general is dominated by tussock grasses and Espeletia spp. Chemical analysis of the sample indicated that SO 2−4 (1,003 mg SO 2−4 L−1) and Ca2+ (320 mg L−1) were the most abundant ions, followed by Mg2+ (55.3 mg L−1), Na+ (45.2 mg L−1), K+ (9.25 mg L−1), and total Fe (Fe2+ and Fe3+) (8.27 mg L−1). The water had a pH of 2.7 and temperature of 29°C at the source, while the ambient air temperature was 9°C. DAPI staining to enumerate total counts revealed 2.35 (±0.07) × 105 cells/mL, and phase contrast microscopy showed the presence of bacilli with predominance of coccoid bacteria and some bacilli. FISH analysis revealed a community dominated by Bacteria (4.32 × 103 cells/mL) with a smaller number of Archaea (1.3 × 103 cells/mL).
Microbial Composition Based on Clone Libraries
From a total of 351 non-archaeal and 297 archaeal 16S rRNA gene sequences obtained from PCR clone libraries, those of insufficient quality or that did not pass the chimera check analysis were removed, leaving 315 and 247 non-archaeal and archaeal sequences, respectively. More OTUs, defined using a distance of 3%, were found for non-Archaea than for Archaea and the estimated coverage for each of the libraries indicated good representation, with slightly better coverage for the Archaea (Table 2). The non-parametric richness estimators S CHAO1 and S ACE gave similar values and indicated greater richness than observed (number of OTUs) (Table 2). A rarefaction analysis for archaeal (Fig. 1a) and bacterial (Fig. 1b) sequences showed that at 3% distance more sampling might still be required to cover the prokaryotic diversity in this sample.
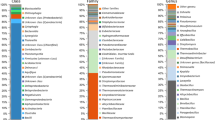
Out of the total number of 315 sequences obtained, 143 (45%) were affiliated to chloroplasts (Eukaryotes) and were not analyzed in this study, leaving 172 and 247 sequences belonging to the Bacteria and Archaea, respectively. The predominant phylum present in the bacterial library was Proteobacteria (69%), followed by Actinobacteria and Nitrospira (Supplementary Table 1, Fig. 2). Betaproteobacteria was the dominant class among the Proteobacteria, although almost half of these sequences could not be further classified. The most abundant order was Burkholderiales, represented by sequences similar to Thiomonascuprica (97% identity), based on BLAST analysis (data not shown). Gammaproteobacteria was the next most abundant group, with sequences related to Legionella (98% identity), the iron oxidizer Acidithiobacillus ferroxidans (99% identity), and uncultured gammaproteobacterial clones (96–98% identity) from acid impact lakes [37]. The next largest group was Alphaproteobacteria that included the orders Rickettsiales and Rhodospirillales, with sequences affiliated to the acidophilic heterotrophic bacteria of the genera Acidisphaera, Acidiphilium, and Acidocella (96–99% identity). Within the phyla Actinobacteria and Nitrospira, clones were affiliated to the iron-oxidizing mixotrophic acidophilic genus Ferrithrix [38] and to Leptospirillum ferriphilum (99% identity) that can use ferrous iron or pyrite as energy sources [39]. Phyla recovered in smaller numbers included Planctomycetes, Firmicutes, Spirochaetes, Acidobacteria, and Bacteroidetes (Supplementary Table 1). Six of the Planctomycetes clones had 90–100% identity to Zavarzinella, an aerobic bacterium isolated recently from an acidic Sphagnum peat bog [40], and two of the Acidobacteria clones had 100% identity with sequences obtained from metal-enriched environments [41, 42].
The majority of the archaeal sequences (65%) could not be classified (Supplementary Table 1). The rest of the sequences belonged to the phyla Crenarchaeota (5%), specifically to the class Thermoprotei that contains several thermoacidophilic microorganisms, and Euryarchaeota (30%) that includes some of the most acidophilic microorganisms known [43, 44]. In general, the classified archaeal sequences were most similar to sequences obtained from acidic hot springs, acid mine drainage, or environments with high concentrations of heavy metals [45–47].
Microbial Composition Based on Pyrosequencing of the V5–V6 Region
From an initial 5,439 sequence reads, 5,095 sequences with an average length of 186 bases were retained for analysis after cleaning. At 3% distance, the diversity indices indicated a high microbial diversity (Table 2), and rarefaction analysis showed that more sequences would still be required to cover the diversity in this sample (Fig. 1c). Consistent with previous reports [10, 48], there was a large number of rare OTUs represented by single sequences (33%) and the predominance of few sequences. There were also a few predominant OTUs, the most abundant one affiliated to an unclassified bacterium represented by 12% of the sequences. Only 23% of the OTUs were represented by more than ten sequences, and the remaining 44% of the OTUs contained between two and ten sequences. Among these sequences, 293 (6%) were assigned to chloroplasts (Eukaryotes) and not further analyzed here. The discrepancy between the number of chloroplast sequences identified by pyrosequencing and clone libraries could be due to the difference in strategies and the ability to capture different populations with the primers used.
Of the remaining 4,800 sequences, 4,776 were classified as Bacteria (99.5%), and only 24 sequences were classified as Archaea (0.5%). Approximately half of both archaeal and bacterial sequences could not be classified further (Supplementary Table 1). This low number of Archaea contrasts with results for clone libraries and could be due to differences in PCR conditions and the fact that amplification for pyrosequencing was done in a single reaction and with primers designed for coverage of >99% bacterial and archaeal sequences (see methods), whereas separate reactions were done for clone libraries. Thus, clone libraries give information on the phylotypes present, but not necessarily about the relative proportion of Archaea and Bacteria in the community, especially when more sampling is required as indicated by rarefaction.
The predominant bacterial phylum was Proteobacteria (42.5% of all sequences) with affiliations, in decreasing order, to the Beta-, Gamma-, and Alphaproteobacteria (Fig. 2). The predominant genera were Thiomonas (Betaproteobacteria), Aquicella, and Legionella (Gammaproteobacteria). Other abundant phyla in the sample were candidate division TM7 (5.5%) and Planctomycetes (2.4%). All remaining bacterial phyla identified represented <2% of the sequences (Supplementary Table 1). A small number of sequences (0.08%) were not classified as either Bacteria or Archaea (unclassified).
Phylogenetic Profile Obtained with the Microarray
Due to the small amount of DNA available, amplification was done only with Bacteria-specific primers. Of the 8,434 bacterial taxa represented on the array, 366 were identified in our sample, with OTU hybridization intensities ranging between 0.02% and 0.86% of the total signal intensity detected. To compare these results with those obtained with the other two strategies, the reference sequences corresponding to positive signals in the microarray were classified using the RDP classifier. The relative abundance at each level (phylum, class, or order) was calculated by adding the hybridization intensities of the corresponding OTUs identified by the gene array. The array detected 22 different phyla that covered a wide range of abundances from the dominant Proteobacteria (55%) to groups present in low numbers such as the Chlamydiae (0.1%) and Tenericutes (0.08%). The most abundant orders were the Burkholderiales (Betaproteobacteria) (13.8%) and Campylobacterales (Epsilonproteobacteria) (9.1%), followed by the Actinomycetales (7.6%), Clostridiales (7.2%), and Rhizobiales (5.4%) (Fig. 2). Some of the genera detected are reported to be iron-oxidizing bacteria, such as Thiobacillus and Acidimicrobium, while others are sulfur-oxidizers (Thiomonas) or sulfate reducers (Desulfovibrio and Desulfomicrobium) [49–53]. Some taxa, such as candidate divisions OP10 and BRC1, were identified only with the 16S rRNA gene array and at low abundances (Supplementary Table 1).
Comparative Analysis of the Prokaryotic Community
Despite the methodological differences inherent to each strategy used, a comparison of the results can nevertheless provide valuable information in complex environmental samples [4–8, 11]. This comparison showed that 12 out of a total of 25 bacterial phyla were shared by at least two of the strategies used (Supplementary Table 1). Despite the fact that the microarray detects only known taxa, it detected more unique groups, such as the phyla Aquificae and Deinococcus-Thermus. Some phyla were detected only by the other strategies, such as Spirochaetes and candidate divisions Ktedonobacteria (found only in gene libraries) and OD1 (only by 454 pyrosequencing). To more easily see the overlap and differences among strategies, a Venn diagram was constructed based on the orders detected (Fig. 3). Proteobacterial sequences were the most abundant by all strategies, with the Betaproteobacteria, and specifically the order Burkholderiales being predominant (Fig. 3). Although differences in abundance were also seen for each technique, as was the case for the Deltaproteobacteria and the Gammaproteobacteria, it is difficult to assess the real significance of this variation in the absence of technical replicates.
Venn diagram at the order level. Classification at the rank of order shows shared and unique taxa (numbers in parenthesis inside circles) identified with each strategy, as well as the total number identified for each method (in parenthesis outside circles). Letters c, p, and m indicate the five most abundant taxa identified by each strategy: c for clone libraries, p for pyrotags, and m for microarray
Greater differences were evident at other taxonomic levels. A total of 56 bacterial and archaeal orders were found overall, most of which belonged to the Proteobacteria (Fig. 3). The microarray alone detected 27 orders not detected by the other strategies, while only three and four orders were identified only by 454 pyrotags and libraries, respectively. Many of the orders shared by at least two strategies included those that were most abundant either by 454 pyrotags or by gene libraries (Fig. 3). Some groups, such as the Rhodocyclales, were identified by all strategies even though they were not necessarily the most abundant. Other shared taxa were abundant only in two of the three methods, such as Legionellales and Rhodospirillales (gene libraries and 454 pyrotags) and Clostridiales (PhyloChip and 454 pyrotags) (Fig. 3). Some highly abundant organisms found by one strategy, such as Acidimicrobiales in gene libraries, were less prominent in the microarray data and not observed at all with 454 pyrotags (Fig. 3). Thus, it is evident that each of the three strategies showed differences in terms of detecting certain groups and providing information regarding their abundance.
One possible explanation for the overall greater detection obtained with the microarray could be that the primers used for amplification for 454 pyrosequencing might not pick up these sequences. To test this, the PhyloChip sequences (two or more sequences for each order) were analyzed in silico for the presence of the V5–V6 amplification primers. In all cases, there was a perfect match, except for four sequences with <79% match. This indicated that the primers used here should be able to amplify those sequences observed with the microarray. As expected, only few of the total microarray sequences were unclassified (4%) given that it detects only known taxa, while larger proportions of both 454 pyrotags (46%) and clone libraries sequences (65% and 6.4% for Archaea and Bacteria, respectively) could not be classified (Supplementary Table 1).
In order to see how these sequences were distributed, sequences representative of each OTU were inserted into the neighbor joining phylogenetic tree in ARB (Supplementary Fig. 1). Again, the microarray sequences represented more taxa, and similarities were evident at higher phylogenetic levels (phylum and class). Clusters of sequences obtained by a single strategy were also observed within the Actinobacteria, Verrucomicrobia, and Synergistetes. Interestingly, some of the previously unclassified 454 pyrotags sequences were placed within clusters of known taxa, such as the phyla Crenarchaeota, Planctomycetes, and Nitrospira (Supplementary Fig. 1). Some groups like Nitrospira and Planctomycetes contained many of these unclassified pyrosequences, which could indicate great sequence diversity and few reported sequences similar to the ones found in this ecosystem. A closer examination of these unclassified pyrosequences showed that using the RDP classifier at a confidence threshold lower than the one used previously (50–79% instead of 80%) resulted in a taxonomic placement consistent with the phylogenetic reconstruction. Using a confidence threshold of 50% to classify 454 reads, the overall results were similar, although six new orders were detected for 454 pyrosequences, including two detected previously only by microarray and clones, and taxa belonging to the Planctomycetes, Nitrospira, and Crenarchaeota (data not shown). Additionally, a distinct, deep-branching clade was also observed (Supplementary Fig. 1), which, however, should be further analyzed and verified using near-full length sequences.
Discussion
Microbial Community Structure
El Coquito is an acidic thermal spring with a high sulfate content, which makes it similar to other thermal springs located in this Andean volcanic belt [2]. Although the water temperature is not very high (29°C), it contrasts greatly with the ambient temperature, which can oscillate between−4°C to 50°C in 1 day. The microbial community is diverse and dominated by Bacteria rather than Archaea, even though Archaea tend to predominate in more extreme environmental conditions [54]. Although the time taken prior to processing samples and isolating DNA could generate biases, this result was also consistent with FISH analysis. The discrepancy between DAPI counts and the much lower counts obtained using FISH probes could be due to low coverage of FISH probes, since only one EUB338 probe was used and, therefore, much of the diversity present could have been missed [55]. It could also be due to the presence of viruses, which could have been included in DAPI counts or of eukaryotic micro-algae in the sample. It is also possible that a portion of this microbial community was not targeted with the primers used in the three molecular approaches. However, the large proportion of unclassified sequences from Archaea in clone libraries (65%) and Bacteria using 454 pyrotags (46%) indicates that many novel microorganisms were in fact detected that are different from those previously identified in other acidic environments and thermal springs [45–47, 50]. This could be due to differences in physicochemical parameters and geographical isolation of this spring. Future work will involve comparison of this community with those present in other hot springs in this area.
The prokaryotic community is composed predominantly of Proteobacteria, consistent with the assessment of mesophilic acidophilic microbial communities (temperature for growth <40°C) [56]. The dominance of the Betaproteobacteria, a group containing microbes with a broad distribution of functions that can be considered as ecological generalists [37, 57], is similar to results from other environmental surveys [58, 59]. In general, the community is reminiscent of those found in hot and acidic environments with mesophilic organisms (Acidithiobacillus, Leptospirillum, Thiomonas, Acidocella, Acidisphaera, and Epsilonproteobacteria) as well as thermophilic microorganisms (Acidiphilium, Acidithiobacillus, Leptospirillum, Acidocella, and Acidisphaera) that are indicative of microhabitats with different temperature gradients within the hot spring. The presence of generalists that can grow under different environmental conditions and use diverse carbon and energy sources, together with specialists (e.g., sulfur and iron oxidizers and sulfate reducers), is also indicative of metabolic diversity. The high abundance of the sulfur-oxidizing bacteria Thiomonas and Acidithiobacillus that can oxidize reduced sulfur to sulfuric acid could contribute to the extreme conditions in this hot spring by dramatically lowering the pH [51, 56]. Thus, the presence of these microorganisms, together with the high concentrations of sulfate and iron present in this hot spring, are suggestive of microbial activity associated with the cycling of ferrous and sulfur-containing minerals.
There were few phototrophic bacteria (Chlorobi, Cyanobacteria, and Chloroflexi) in this spring, in contrast to other hot spring communities [49, 52, 60]. Despite receiving high levels of solar energy at this high elevation, this is consistent with the fact that water emerges from underground and with the notion that Cyanobacteria do not grow well at acidic pHs and are more sensitive to metals and solutes found in acidic waters [50, 61]. In this location, the eukaryotic microalgae could be driving primary production using solar energy at the surface, similar to what occurs in surface acid streamers and other acidic extreme environments [38, 56]. Subsequent studies will include analysis of chloroplast 16S rRNA gene sequences recovered here and 18S rRNA gene sequences to further analyze the eukaryotic community. The high abundance of chemolithoautotrophic acidophiles (Leptospirillum, Acidithiobacillus, Thiobacillus, Thiomonas, and Aquicella) also indicates that there is primary production driven by chemical energy. There are also heterotrophic acidophilic bacteria such as Acidiphilium, Acidisphaera, Acidocella, and Alicyclobacillus. Thus, it appears that in this community, primary production can be driven by both solar energy at the surface and by inorganic chemicals that affect the biogeochemistry of iron and sulfur in the water.
Complementing Culture-Independent Approaches
The three strategies used to analyze the diversity of this ecosystem show dominance of Proteobacteria, and specifically of Burkholderiales (Betaproteobacteria), with organisms closely related to Thiomonas and Thiobacillus. The large number of sequences obtained from both gene libraries and 454 pyrotags that could not be classified also suggests novel sequences and not merely overestimation of phylotypes or sequence errors, as has been reported for both strategies, and in particular for pyrosequencing [10, 62]. In fact, the pipeline for cleaning and analyzing 454 sequences takes into account previous reports to eliminate possible sequencing errors and overestimation of OTUs [63, 64]. This also highlights the importance of amplifying and sequencing 16S rRNA genes directly from an environmental sample.
Differences among the three approaches used are also evident. The microarray alone detected approximately one third of the total phyla, consistent with previous studies comparing it with clone libraries [4, 6, 8, 65]. It was surprising, however, that it detected more groups than 454 pyrosequencing, which can provide great depth of coverage. Our results could have been affected by the relatively modest number of pyrosequences and the high number of unclassified sequences, which could hamper detection of low abundance community members. It has recently been shown that low abundance taxa are harder to classify because they are infrequent and tend to be less represented in databases [66, 67]. In addition, the use of different sets of primers and amplification conditions can result in different PCR pools. This might explain why some taxa were not detected with pyrosequencing despite a bioinformatics analysis, indicating that amplification should be possible with the primer set used. It could also account for the similar estimated coverage levels obtained for both clone libraries and 454 pyrotags, despite differences in OTUs detected. Coverage of diversity and accurate taxonomic assignment can also be more sensitive to the region of the 16S rRNA gene being sequenced than to the fragment size [68–70]. Despite differences, however, several primer sets have been shown to give stable estimates of abundances and consistent taxonomic assignation [69]. In our case, the short size of the pyrosequence reads obtained (average of 186 nt) could have affected the accuracy of taxonomic classification and resulted in higher richness estimates at the OTU level [71]. However, the RDP II classifier, one of the choice algorithms for classification of short reads that produces highly stable and accurate results even for fragments of disparate sizes [67, 69], gave results consistent with the phylogenetic reconstruction of all sequences and even improved when a lower confidence threshold was used. This suggests that the original analysis of these hypervariable regions was quite strict and that a lower threshold for classification might be a useful alternative for assignment of reads in datasets with many novel and short sequence reads that are difficult to classify, as has been suggested [11].
Despite these differences, array and pyrotag data are similar in terms of the dominant groups obtained and the community profiles at the phylum and class levels, consistent with what has been reported using the HITChip array and pyrosequencing for V4 and V6 regions of microbial communities in human distal intestine samples [11]. In our case, this correlation is not observed at the OTU level where the most abundant OTU by 454 pyrosequencing represented 12% of the total sequences, while the maximum relative intensity obtained with the microarray was only 0.86% of the total. This could be explained by differences in the relative proportions of probes on the microarray for some groups [65] or by cross-hybridization of array probes with many of the novel, unclassified sequences in the dataset [72, 73]. The outcome of hybridization against these unclassified sequences in our dataset could not be further analyzed, however, in the absence of probe sequence information. Amplification bias could also be influencing the results since there was little DNA available for the PhyloChip, which prevented us from doing duplicate analyses to test reproducibility of our results. Running replicate analyses, which have been shown previously to be reproducible, represent one of the strengths of using high-density arrays [5, 7]. In addition, both the primer sets and the number of PCR cycles varied for each approach. These methodological differences can therefore affect the populations analyzed in each case, as was evident for the Archaea and chloroplasts sequences.
Although a comprehensive comparison of the three methodologies is beyond the scope of this study, it is evident that the combination of techniques improved detection of community members. These strategies share common steps in sample preparation and also involve methodological differences that can lead to differences in estimations of microbial community diversity and structure [15]. While 16S rRNA gene clone libraries analyze almost full-length genes, 454 pyrosequencing and microarrays provide greater depth of coverage but are limited in turn by the short length of sequence reads and identification of known taxa, respectively. Importantly, these results strongly suggest that no single methodology of community diversity assessment is completely reliable and that combination approaches should be followed.
References
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Alfaro C, Aguirre A, Jaramillo LF (2002) In: Inventario de fuentes termales en el Parque Nacional Natural de los Nevados INGEOMINAS, Bogotá, pp 101
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL (2007) High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol 53:371–383
Brodie EL, Desantis TZ, Joyner DC, Baek SM, Larsen JT, Andersen GL, Hazen TC, Richardson PM, Herman DJ, Tokunaga TK, Wan JM, Firestone MK (2006) Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol 72:6288–6298
Brodie EL, DeSantis TZ, Parker JP, Zubietta IX, Piceno YM, Andersen GL (2007) Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci USA 104:299–304
DeSantis TZ, Stone CE, Murray SR, Moberg JP, Andersen GL (2005) Rapid quantification and taxonomic classification of environmental DNA from both prokaryotic and eukaryotic origins using a microarray. FEMS Microbiol Lett 245:271–278
Yergeau E, Schoondermark-Stolk SA, Brodie EL, Dejean S, DeSantis TZ, Goncalves O, Piceno YM, Andersen GL, Kowalchuk GA (2009) Environmental microarray analyses of Antarctic soil microbial communities. ISME J 3:340–351
Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, Sogin ML (2007) Microbial population structures in the deep marine biosphere. Science 318:97–100
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA 103:12115–12120
Claesson MJ, O’Sullivan O, Wang Q, Nikkila J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O’Toole PW (2009) Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669
Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF (2005) PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl Environ Microbiol 71:8966–8969
Polz MF, Cavanaugh CM (1998) Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol 64:3724–3730
Qiu X, Wu L, Huang H, McDonel PE, Palumbo AV, Tiedje JM, Zhou J (2001) Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl Environ Microbiol 67:880–887
Roh SW, Abell GC, Kim KH, Nam YD, Bae JW (2010) Comparing microarrays and next-generation sequencing technologies for microbial ecology research. Trends Biotechnol 28:291–299
Eaton AD, Franson MAH, American Public Health Association, American Water Works Association, Water Environment Federation (2005) Standard methods for the examination of water & wastewater. American Public Health Association, Washington
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925
Stahl DA, Amann R (1991) Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 205–248
Pernthaler J, Glöckner F-O, Schönhuber W, Amann R (2001) Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. In: John HP (ed) Methods in microbiology, vol 30. Academic, New York, pp 207–226
Kirchman D, Sigda J, Kapuscinski R, Mitchell R (1982) Statistical analysis of the direct count method for enumerating bacteria. Appl Environ Microbiol 44:376–382
Schauer M, Massana R, Pedros-Alio C (2000) Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol Ecol 33:51–59
Wang Y, Zhang ZS, Ruan JS, Wang YM, Ali SM (1999) Investigation of actinomycete diversity in the tropical rainforests of Singapore. J Indust Microbiol Biotechnol 23:178–187
Chan OC, Claus P, Casper P, Ulrich A, Lueders T, Conrad R (2005) Vertical distribution of structure and function of the methanogenic archaeal community in Lake Dagow sediment. Environ Microbiol 7:1139–1149
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072
Nawrocki EP, Kolbe DL, Eddy SR (2009) Infernal 1.0: inference of RNA alignments. Bioinformatics 25:1335–1337
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Kemp PF, Aller JY (2004) Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol Ecol 47:161–177
Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, Daroub SH, Camargo FA, Farmerie WG, Triplett EW (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290
Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM (2007) Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8:R143
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123
Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5:164–166
Klepac-Ceraj V, Lemon KP, Martin TR, Allgaier M, Kembel SW, Knapp AA, Lory S, Brodie EL, Lynch SV, Bohannan BJ, Green JL, Maurer BA, Kolter R (2010) Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ Microbiol 12:1293–1303
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar BA, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Percent SF, Frischer ME, Vescio PA, Duffy EB, Milano V, McLellan M, Stevens BM, Boylen CW, Nierzwicki-Bauer SA (2008) Bacterial community structure of acid-impacted lakes: what controls diversity? Appl Environ Microbiol 74:1856–1868
Rowe OF, Sanchez-Espana J, Hallberg KB, Johnson DB (2007) Microbial communities and geochemical dynamics in an extremely acidic, metal-rich stream at an abandoned sulfide mine (Huelva, Spain) underpinned by two functional primary production systems. Environ Microbiol 9:1761–1771
Mendez MO, Neilson JW, Maier RM (2008) Characterization of a bacterial community in an abandoned semiarid lead-zinc mine tailing site. Appl Environ Microbiol 74:3899–3907
Kulichevskaya IS, Baulina OI, Bodelier PL, Rijpstra WI, Damste JS, Dedysh SN (2009) Zavarzinella formosa gen. nov., sp. nov., a novel stalked, Gemmata-like planctomycete from a Siberian peat bog. Int J Syst Evol Microbiol 59:357–364
Barns SM, Cain EC, Sommerville L, Kuske CR (2007) Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl Environ Microbiol 73:3113–3116
Mirete S, de Figueras CG, Gonzalez-Pastor JE (2007) Novel nickel resistance genes from the rhizosphere metagenome of plants adapted to acid mine drainage. Appl Environ Microbiol 73:6001–6011
Golyshina OV, Pivovarova TA, Karavaiko GI, Kondrateva TF, Moore ER, Abraham WR, Lunsdorf H, Timmis KN, Yakimov MM, Golyshin PN (2000) Ferroplasma acidiphilum gen. nov., sp. nov., an acidophilic, autotrophic, ferrous-iron-oxidizing, cell-wall-lacking, mesophilic member of the Ferroplasmaceae fam. nov., comprising a distinct lineage of the Archaea. Int J Syst Evol Microbiol 50(Pt 3):997–1006
Schleper C, Pühler G, Klenk H-P, Zillig W (1996) Picrophilus oshimae and Picrophilus torridus fam. nov., gen. nov., sp. nov., two species of hyperacidophilic, thermophilic, heterotrophic, aerobic Archaea. Int J Syst Bacteriol 46:814–816
Baker BJ, Tyson GW, Webb RI, Flanagan J, Hugenholtz P, Allen EE, Banfield JF (2006) Lineages of acidophilic archaea revealed by community genomic analysis. Science 314:1933–1935
Jackson CR, Langner HW, Donahoe-Christiansen J, Inskeep WP, McDermott TR (2001) Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ Microbiol 3:532–542
Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK (2001) Archaeal diversity in waters from deep South African gold mines. Appl Environ Microbiol 67:5750–5760
Preston FW (1948) The commonness, and rarity, of species. Ecology 29:254–283
Elshahed MS, Senko JM, Najar FZ, Kenton SM, Roe BA, Dewers TA, Spear JR, Krumholz LR (2003) Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl Environ Microbiol 69:5609–5621
Gonzalez-Toril E, Llobet-Brossa E, Casamayor EO, Amann R, Amils R (2003) Microbial ecology of an extreme acidic environment, the Tinto River. Appl Environ Microbiol 69:4853–4865
Johnson DB (1998) Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiol Ecol 27:307–317
Lau MC, Aitchison JC, Pointing SB (2009) Bacterial community composition in thermophilic microbial mats from five hot springs in central Tibet. Extremophiles 13:139–149
Zhou H, Li J, Peng X, Meng J, Wang F, Ai Y (2009) Microbial diversity of a sulfide black smoker in main endeavour hydrothermal vent field, Juan de Fuca Ridge. J Microbiol 47:235–247
Chapelle FH, O’Neill K, Bradley PM, Methe BA, Ciufo SA, Knobel LL, Lovley DR (2002) A hydrogen-based subsurface microbial community dominated by methanogens. Nature 415:312–315
Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M (1999) The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444
Johnson DB, Hallberg KB (2009) Carbon, iron and sulfur metabolism in acidophilic micro-organisms. Academic, New York
Cruz-Martinez K, Suttle KB, Brodie EL, Power ME, Andersen GL, Banfield JF (2009) Despite strong seasonal responses, soil microbial consortia are more resilient to long-term changes in rainfall than overlying grassland. ISME J 3:738–744
Marcial Gomes NC, Borges LR, Paranhos R, Pinto FN, Mendonca-Hagler LC, Smalla K (2008) Exploring the diversity of bacterial communities in sediments of urban mangrove forests. FEMS Microbiol Ecol 66:96–109
Bowers RM, Lauber CL, Wiedinmyer C, Hamady M, Hallar AG, Fall R, Knight R, Fierer N (2009) Characterization of airborne microbial communities at a high-elevation site and their potential to act as atmospheric ice nuclei. Appl Environ Microbiol 75:5121–5130
van der Meer MT, Schouten S, de Leeuw JW, Ward DM (2000) Autotrophy of green non-sulphur bacteria in hot spring microbial mats: biological explanations for isotopically heavy organic carbon in the geological record. Environ Microbiol 2:428–435
Nancucheo I, Johnson DB (2010) Production of glycolic acid by chemolithotrophic iron- and sulfur-oxidizing bacteria and its role in delineating and sustaining acidophilic sulfide mineral-oxidizing consortia. Appl Environ Microbiol 76:461–467
Reeder J, Knight R (2009) The ‘rare biosphere’: a reality check. Nat Methods 6:636–637
Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898
Reeder J, Knight R (2010) Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7:668–669
Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R (2010) Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio 1:e00129-10
Dethlefsen L, Huse S, Sogin ML, Relman DA (2008) The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280
Hamp TJ, Jones WJ, Fodor AA (2009) Effects of experimental choices and analysis noise on surveys of the “rare biosphere”. Appl Environ Microbiol 75:3263–3270
Huse SM, Dethlefsen L, Huber JA, Mark Welch D, Relman DA, Sogin ML (2008) Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4:e1000255
Liu Z, DeSantis TZ, Andersen GL, Knight R (2008) Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 36:e120
Youssef N, Sheik CS, Krumholz LR, Najar FZ, Roe BA, Elshahed MS (2009) Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl Environ Microbiol 75:5227–5236
Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P (2010) Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J 4:642–647
Liu WT, Mirzabekov AD, Stahl DA (2001) Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ Microbiol 3:619–629
Chandler DP, Newton GJ, Small JA, Daly DS (2003) Sequence versus structure for the direct detection of 16S rRNA on planar oligonucleotide microarrays. Appl Environ Microbiol 69:2950–2958
Acknowledgments
We would like to thank to José Ricardo Bustos, Luis Miguel Rodríguez and Silvia Restrepo for technical assistance with setting up local server installations of bioinformatics programs, Luis Miguel Alvarez for sampling, Jose Salvador Montaña for sampling and processing DNA, and Yvette Piceno, Eoin Brodie and Gary Andersen for providing us access to the PhyloChip. This work was financed by Colciencias—SENA (project no. 6570-392-19990) and was done under MAVDT contract no. 15, 2008 for access to genetic resources and UAESPNN Research permit no. DTNO-N-20/2007.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at http://dx.doi.org/10.1007/s00248-011-9977-6
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Abundance of taxa from phylum to order for the three methods used. The number of OTUs is shown for Phylochip, the number of total sequences for 454 pyrotags, and the number of sequences for each separate clone library (Bacteria and Archaea). Abundance was determined as the percentage of the total for each strategy. In bold are the subtotals for each group (DOC 170 kb)
Supplementary Figure 1
Phylogenetic reconstruction of sequences from El Coquito hot spring. Green branches indicate unique sequences obtained by Phylochip, dark blue branches indicate unique sequences obtained by 454 pyrotags, light blue indicates unclassified sequences of 454 pyrotags, red branches indicate unique sequences obtained by clone libraries, and black branches indicate reference sequences. The asterisk indicates a novel deepbranching cluster. The scale bar shows the distance of 0.08 nucleotides substitutions per hundred nucleotides (PDF 252 kb)
Rights and permissions
About this article
Cite this article
Bohorquez, L.C., Delgado-Serrano, L., López, G. et al. In-depth Characterization via Complementing Culture-Independent Approaches of the Microbial Community in an Acidic Hot Spring of the Colombian Andes. Microb Ecol 63, 103–115 (2012). https://doi.org/10.1007/s00248-011-9943-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9943-3