Abstract
The Grande Albergo Ausonia & Hungaria (Venice Lido, Italy) has an Art Nouveau polychrome ceramic coating on its façade, which was restored in 2007. Soon after the conservation treatment, many tiles of the façade decoration showed coloured alterations putatively attributed to the presence of microbial communities. To confirm the presence of the biological deposit and the stratigraphy of the Hungaria tiles, stereomicroscope, optical and environmental scanning electron microscope observations were made. The characterisation of the microbial community was performed using a PCR–DGGE approach. This study reported the first use of a culture-independent approach to identify the total community present in biodeteriorated artistic tiles. The case study examined here reveals that the coloured alterations on the tiles were mainly due to the presence of cryptoendolithic cyanobacteria. In addition, we proved that the microflora present on the tiles was generally greatly influenced by the environment of the Hungaria hotel. We found several microorganisms related to the alkaline environment, which is in the range of the tile pH, and related to the aquatic environment, the presence of the acrylic resin Paraloid B72® used during the 2007 treatment and the pollutants of the Venice lagoon.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Monuments can be degraded by physical, chemical, and biological factors. Microorganisms are among the principal biological agents causing biodeterioration. Microbial deterioration is related to both environmental conditions and the physico-chemical properties of construction materials [15, 27]. Microorganisms cause damage to stone surfaces through a variety of mechanisms, including chemical reactions with the materials (e.g. microbial excretion of aggressive organic or inorganic acids), physical disruption [e.g. microbial production of extracellular polymeric substances (EPS) that can cause mechanical stresses to the mineral structure], and aesthetic alterations (e.g. the production of pigments) [7, 27, 30, 42]. Dust, pollutants and, finally, synthetic polymers, widely employed both as consolidants and water repellents for the treatment of stone materials in objects and buildings, are potential additional substrates for microorganisms [7, 8].
Ceramic has been used as an ornamental material from antiquity. To create ceramics, clay is mixed with water and subsequently air-dried and subjected to fire [6]. Tiles are generally ceramic plaques glazed on one side.
Microorganisms cause physical deterioration on the external part of ceramics, leading to surface detachment and increased porosity. Biochemical deterioration is due to microbial metabolism that produces acids that can solubilise the original pottery materials [33]. The produced salts are often highly water soluble and increase the water content of the porous material. Microbiological deterioration is an underestimated problem as microbial physico-chemical attack cannot be easily distinguished from other sources of damage [33].
The Grande Albergo Ausonia & Hungaria, one of the most prestigious hotels on the Lido of Venice (Italy), has Art Nouveau polychrome ceramic tiles on its façade. After the 2007 conservation treatment, some tiles showed coloured alterations between the pottery and the glaze layers, putatively attributed by conservators to microbial growth.
The aim of this work was to confirm the microbial deterioration and to characterise the microbial community present on the ceramic tiles of the Grande Albergo Ausonia & Hungaria façade. To date, only one study concerning microflora present in artistic ceramic tiles using optical microscope observations [26] and another dealing with the effect of a lichen on sekishu glazed roof tiles [41] are available in the scientific literature.
Methods
Description of Hungaria Hotel
In 1914, the Neo-Renaissance façade of the Grande Albergo Ausonia & Hungaria was coated with Art Nouveau polychrome ceramic tiles covering approximately 800 m2. The façade is ornamented with pilasters made of white and coloured tiles, and high or low relief decorations. Hungaria tiles are painted and glazed soft stoneware; in particular, the glaze process was made through complete immersion in a bath of crushed glass dispersed in water, meaning that they were completely glazed by a vitreous layer [22]. In 2007, the façade underwent conservation treatment to consolidate severely damaged tiles, e.g. showing cracking, and to remove dark spots present on its surface. Stained tiles were cleaned using water, hydrogen peroxide and sodium hypochlorite. Finally, tiles on the first horizontal register were treated with the commercial synthetic resin Paraloid B72® (copolymer methylacrylate–ethylmethacrilate) as the consolidant and protective product. Soon after the intervention, both treated and untreated tiles showed coloured alterations between the pottery layer and the glaze.
Sampling
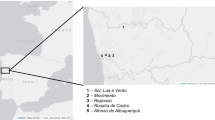
Figure 1 shows the sampling areas and the samples taken from each area. Sampling areas were selected by visual inspection of putative biological alterations in order to obtain representative samples.
All the tile samples were taken by sterile chisel and stored in sterile tubes at room temperature. Samples I to VIII and from XI to XVIII are fragments of glazed pottery showing coloured alterations (black, green-yellowish, green-greyish) below the glaze layer, putatively attributed (visual inspection) to the presence of microorganisms. Samples IX and XIX are from mortar, taken between two deteriorated tiles, and sample X is a putative biological deposit taken by scraping off the balcony surface.
Tile pH Measurement
Sample pH was measured according to the concrete surface field test [16] except for the quantity of sample, which was of few milligrams, and the quantity of deionised water (two drops).
Stereomicroscope, Optical and Epifluorescence Microscope Observation
Samples were observed using a Wild Makroskop M5A stereomicroscope (Heerbrugg, Switzerland) equipped with a Nikon E5600 camera (Chicago, IL, USA).
Polished cross-sections were obtained after including samples in a polyester resin (New Basic) and observed under a Wild M3 stereomicroscope and a Leitz Orthoplan microscope equipped with a Leica DC300 camera.
Optical and epifluorescence observations of tile samples were carried out with a Leica DM4000B digital epifluorescence microscope equipped with Cool-Snap CF camera (Photometrics, Roper Scientific); pictures were acquired using RS Image ver. 1.7.3 software (Roper Scientific) after sample grinding. DAPI staining was performed without sample fixation according to Villa [40] in order to confirm the biological origin of the patina.
ESEM–EDX Observation
Observations of the glass surface and polished cross-sections were performed by a Fei Quanta 200 FEG–ESEM instrument to evaluate the polymer morphology and distribution, and the cell size and cell location in tile samples. The semi-quantitative elemental compositions were obtained by an EDAX Genesys energy-dispersive X-ray spectrometer, using an accelerating voltage of 25 keV. The samples were observed directly, without any preliminary conductive coating.
MicroFTIR Analysis
The samples collected were placed on a gold plate and treated with a few drops of CHCl3 to extract any potentially present soluble organic fraction. After gentle solvent evaporation, the soluble residue, distributed as a halo around the sample, was analysed using microFTIR. A Nicolet microscope connected to a Nicolet 560 FTIR system, equipped with a mercury cadmium telluride (MCT) detector and OMNIC32 software, was used for spectra collection. The size of the sample area investigated was about 50 × 50 μm. The IR spectra were recorded in reflectance mode in the range of 4,000–650 cm−1, with a resolution of 4 cm−1.
DNA Extraction
Before DNA extraction, all the samples were ground using a sterile mortar and pestle. Total DNA was extracted directly from the samples, as described by Polo et al. [27].
Analysis of the Bacterial Community
The 16S rRNA gene fragment extracted from the samples (tiles, mortar and putative biological deposit) were amplified with primers GC-357 F and 907 R with chemical conditions and a thermal cycling program as reported by Polo et al. [27], except for the dNTP mix concentration of 0.2 μM.
Analysis of Fungal Community
The internal transcribed spacer (ITS) region fragments were amplified by a semi-nested PCR performed as follows: a first amplification step using the combination of primers NS5 and ITS4 [43] with 1× of PCR buffer, 1.8 mM of MgCl2, 0.2 mM of dNTP mix, 0.5 μM of each primer and 0.625 U of Taq DNA polymerase (GoTaq, Promega) in 25 μl PCR reaction; the cycling program consisted in an initial denaturation at 95°C for 3 min followed by 30 cycles of denaturation at 95°C for 45 s, annealing at 52°C for 45 s and extension at 72°C for 2 min, and a final extension at 72°C for 10 min. The first PCR product was used as template for a second amplification step performed with the primers ITS4 and GC clamped ITS1 [14] (GC clamp = 5′-CCGGCGCCGCGGCGGGCGGGGCGGGGGCACGGG-3′). The reaction mixture was identical to first-step PCR except for 0.12 mM of dNTP mix and 0.3 μM of each primer. The cycling program consisted in an initial denaturation at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 58°C for 45 s and extension at 72°C for 2 min, and a final extension at 72°C for 10 min.
Analysis of the Phototrophic Community
The fifth dominium of 23S gene fragments were amplified by DGGE–PCR performed with primers p23SrV-F GC clamped (GC clamp = 5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG-3′) and p23SrV-R [34] with the following chemical conditions: 1× of PCR buffer, 1.8 mM of MgCl2, 0.2 mM of dNTP mix, 0.5 μM of each primer and 2 U of Taq DNA polymerase (Invitrogen) in 50 μl PCR reaction. The thermal cycling program included an initial denaturation at 95°C for 2 min, followed by 35 cycles consisting of denaturation at 94°C for 30 s, annealing at 57°C for 30 s and extension at 72°C for 30 s, and a final extension step at 72°C for 10 min.
Denaturing Gradient Gel Electrophoresis (DGGE) and Sequencing
The DGGE analysis was performed with 6% polyacrylamide [6% of a 37:1 acrylamide–bisacrylamide mixture (Sigma) in a Tris acetate EDTA (TAE) 1× buffer (Sigma), 0.75 mm thick, 16 × 10 cm] gels prepared according to Polo et al. [27]. Denaturant gradients were 40–70% for bacteria and the phototrophic community, and 30–60% for the fungal community. The DNA fragments were separated by electrophoresis run for 17 h at 90 V, performed by the D-Code Universal Mutation Detection system (Bio-Rad). The gels were stained by SYBR Green (Amersham Pharmacia Biotech) and the results observed by a GelDoc (Bio-Rad) apparatus. Individual lanes of the gel images were straightened and aligned using Adobe Photoshop (Adobe Systems Incorporated). The excised bands were eluted in 50 μl milli-Q water by incubation at 37°C for 5 h, re-amplified and identified by sequencing (Primm, Milan). The sequences were analysed using the BLASTN software (www.ncbi.nlm.nih.gov/BLAST) and the Classifier tool by Ribosomal Data Project (http://rdp.cme.msu.edu/classifier/classifier.jsp).
Results
Stereomicroscope, Optical, Epifluorescence and Electronic Microscope Observation of the Coloured Alterations
Using a stereomicroscope, we observed in samples I to VII and XIII to XVIII the coloured alteration at the interface glaze–pottery (see Fig. 2a–b and Fig. 4a) with an average thickness of about 250 μm with a maximum of 800 μm in sample XVI (Fig. 2b). In samples XI and XII, the coloured alteration was mainly in the pottery layer (see Fig. 2f–g and Fig. 4b).
Top view of the samples XVI (a) and XII (f) under the stereomicroscope; thin sections of samples XVI (b) and XII (g); yellow arrows indicate coloured alterations. Optical microscope images of biological deposits obtained after grinding samples XVI (c) and XII (h); autofluorescence of samples XVI (d) and XII (i) under Texas Red filter cube; ESEM observation of sample XVI (e); DAPI staining of sample XII (l) under DAPI filter cube
The optical, DAPI and electronic microscope (ESEM) observations confirmed the presence of microbiological cells forming a biofilm, which often included tile material (Fig. 2c, h, e, l). We mainly observed smaller (diameter 1–2 μm) and bigger (diameter 3–4 μm) coccoid cells (Fig. 2c, h). Due to the small size, and the autofluorescence, the presence of cyanobacteria was hypothesised (Fig. 2d, i).
pH Measurement, ESEM–EDX Observation and MicroFTIR Analysis of the Tile Materials
Tile pH was 9–9.5 for all samples.
The polymer was found only in samples XI and XII (Figs. 3b–c and 4b), and was identified as Paraloid B72® by microFTIR analysis (Fig. 3d–f). In samples I, XI, XII and XIII, we observed a superficial chemical deposit by ESEM–EDX that, in samples I, XI and XII, was identified as air particulate matter (data not shown), and in sample XIII (Fig. 3h), as guano, due to the presence of high value of calcium and phosphor, absent in sample XVI (Fig. 3g).
Backscattered electron images collected from the surfaces of samples I (a), XI (b) and XII (c) at magnification ×50, ×80 and ×100, respectively. Pottery is the grey layer; the glaze layer is in white, while the white arrows clearly indicate the dark amorphous coating corresponding to the applied polymer. IR spectra of the amorphous material collected from sample XI (d), sample XII (e) and reference spectra of commercial product Paraloid B72 (f). The good fitting of absorbance patterns collected from the samples XI and XII with the reference spectra confirms the acrylic polymer nature of the coating on the tiles surfaces. EDX spectra collected from the outmost surface of samples XVI (g) and XIII (h). g EDX spectrum without any presence of deposits on the surface. The presence of Ca3(PO4)2 is suggested by the elements Ca and P, highlighted by the black arrows on spectra (h)
Schematic representation of samples I–VIII, which are characterised by the absence of the acrylic resin (a) and samples XI–XVIII, which were treated with the synthetic polymer (b). PB72 stands for the acrylic polymer-based Paraloid B72. The star indicates a richer microbial community, with the presence of black fungi, such as Phoma, and bacterial species as Methylibium sp. absent in samples I–VIII
Analysis of Total Community
The composition of the total community was determined by DGGE analysis coupled with a partial sequencing of the 16S rRNA genes, ITS region and the fifth dominium of the 23S gene fragment for bacteria, fungi and both prokaryotic and eukaryotic algae, respectively.
Amplification with 16S rRNA bacterial primers showed result for all samples except for XV and XVI. In the fungal community study, all the samples were amplified. Finally, amplification products were found for samples I–VI, IX–X, XIII–XV, XVII and XIX using the plastid primers.
Figure 5 shows the DGGE profiles of amplified samples for all the microbial communities investigated.
DGGE analysis of the microbial community of Hungaria hotel. a, b Bacteria, denaturing gradient 40–70%. Samples X (biological patina) and XVII (glazed pottery) were used as makers and loaded in the first and latter wells in both gels. c Prokaryotic and eukaryotic algae, denaturing gradient 40–70% and (d) fungi, denaturing gradient 30–60%
Based on identical mobility within the gel, bands 25A, 16 and 11 (Fig. 5a, b) were the most dominant DGGE bands in all samples except in sample X (biological patina) and XIX (mortar). These DGGE bands were identified as to two cyanobacteria (100% from RDP). In Fig. 5c, bands 17, 19 and 20 were related to cyanobacteria of the order Chroococcales that were present and dominant in all samples except in samples X and XIX, where Rosenvingiella was found (bands 12 and 13).
Bands 6A (84% Flexibacteriaceae from RDP), 10A (84% Methylibium from RDP) and 18A (100% Pedobacter from RDP) in Fig. 5a were present in many samples, and they were dominant in samples XI and XII (samples that showed the presence of Paraloid B72®).
Bands 17B and 25 in Fig. 5d, corresponding to Sporobolomyces coprosmae, were present in samples III, IX, XI–XVI and XVIII. Band 28 (Fig. 5d), related to Phoma, was present in all samples that showed Paraloid B72® and in sample XIII, collected very closely to them.
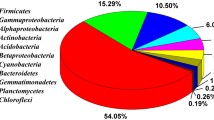
DGGE bands showed in Fig. 5 were excised, re-amplified and sequenced. We could not obtain sequences of good quality for few bands of low intensity, but likely these originated from non-dominant species in the population, and were due to PCR biases derived from the complex matrix of the tiles, rich in salts. Good sequences were reported in Tables 1, 2 and 3. Twenty-four sequences were obtained from the study of the bacterial community by BLAST in the RDP and NCBI databases. Sequence homologies to sequences of known bacteria in the NCBI database ranged from 91% to 100%. Phylogenetic affiliations of excised DGGE bands were represented by phylum Proteobacteria (27.3%), Bacteroidetes (27.3%), Actinobacteria (27.3%) and Cyanobacteria (4.5%). Five sequences were obtained from the study of the fungal community by BLAST in the NCBI database. Sequence homologies to sequences of known fungi in the NCBI database ranged from 88% to 100%. Two sequences were obtained from the study of the phototrophic community by BLAST in the NCBI database. Sequence homologies to sequences of known phototrophic microorganisms in the NCBI database ranged from 90% to 95%.
Discussion
To date, no molecular studies concerning microflora on artistic ceramics are available. A pioneer study on this topic was carried out by Oliveira et al. [26], who made optical microscopic observations of glazed ceramic tiles on the façade of Salvador and Belèm buildings (Brazil). However, the above-mentioned author has reported no further investigations apart from the documenting of the presence of Cyanophyta and Bacilliarophyta. Therefore, we present here the first molecular characterisation of a microbial community present on artistic ceramic.
The molecular analysis of the total community showed that bacteria, eukaryotic algae and fungi were present. Since the principal deterioration appearing on the Hungaria hotel is a coloured deposit, most likely due to the growth of both prokaryotic and eukaryotic algae, we employed, for the characterisation of the phototrophic community, a recent set of primers (p23SrV) designed by Sherwood and Presting [34]. Here we present the first use of p23SrV primers for DGGE analysis.
Combining the results obtained from sequencing of bacterial 16S and phototrophic 23S fragments, we found cyanobacteria in all tile samples—independently on the position of biofilms either below the glaze or in the pottery—but not in the biological deposit taken from the balcony.
Cyanobacteria are very common on monuments; for example, they have been detected on Ca’ d’Oro (Venice) [29], on glazed ceramic tiles from Portugal and Brazil [26], and, in general, on sculptures and buildings [20, 27, 30] exposed to high humidity, running-off water, high and low temperatures, as well as wetting/drying cycles and high UV exposure [28]. Using primers specific for the phototrophic community, bands 17, 19 and 20 were identified as a cyanobacterium of the order Chroococcales, which is the most widespread order present on stone monuments [20, 31]. In addition to the production of extracellular polymeric substances and pigments, some species of this order are able to precipitate magnesium and calcium [31].
The cyanobacteria (100% from RDP) detected on the Hungaria tiles using 16S rRNA primers showed high similarity to uncultured bacteria detected in a sample from the Tibetan tundra in which the community was mainly composed by coccoid cyanobacterial cells [44].
The presence of coloured deposit in depth into the pottery layer detected by microscope observations of Hungaria tiles with cracks and fractures is indicative of a cryptoendolithic niche. The fact that the microbiological deposit was located between the glaze and the pottery layer made the collect the entire biomass and estimate the quantity of cyanobacteria against the whole community not possible. Endolithic microorganisms, the most widespread of them are cyanobacteria, occur in various habitats such as hot and cold deserts, and were reported to exist also in monuments [10, 17, 23, 25, 36]. In extreme environments, endolithic growth provides protection from low temperature, UV radiation and desiccation, and provides mineral nutrients [17, 20, 23, 25, 36]. In Hungaria samples, we found small coccoid cells (from 1–2 to 3–4 μm). De los Ríos [10] found spherical to oval shaped cells of 1.1–1.5 μm in size in granite rocks collected in Antarctica, while in dolomite rock in central Switzerland, Horat [17] detected coccoid cyanobacteria not only as single 3–6 μm cells, but also as multicellular aggregates. On external stone and building surfaces, high light levels may favour endolithic growth by cyanobacteria. Del Monte and Sabbioni [11] reported a perforating activity of endolithic cyanobacteria inhabiting cracks and fissures in the marble of monuments in Torcello Island, near Venice. Especially when endolithic microorganisms are present, water absorption by the biofilm matrix causes mechanical stress that opens cracks and fissures in the material [10, 23].
The eukaryotic algae of the Hungaria hotel were found only in the mortar samples and on the first floor stone balcony, never on the ceramic tiles, and they were epilithic. In this study, chloroplasts of the phylum Chlorophyta were detected, which was represented by the genus Chlorella (99% of similarity). Green algae have already been found in Venetian buildings made of stone and marble [29] and, in particular, Chlorella sp. was found on Ca’ d’Oro [32]. The other eukaryotic alga found in this work bands 12 and 13, is a green alga of the order Prasiolales that is widespread in temperate regions, in a wide range of terrestrial and littoral habitats [31]. Microorganisms of the order of Prasiolales have been detected in very high amounts on cement and bricks in Galway (Ireland), and at the base of walls, in corners and on protrusions of buildings in Oviedo and León (Spain) [31]. Moreover, the abundance of these algae in places affected by bird guano is well documented [31]. We found guano in sample XIII that was collected very close to the balcony where Rosenvingiella was detected (band 13).
Another microorganism present mainly in the sampling area d was the red-coloured yeast Sporobolomyces coprosmae [42].
In conclusion, the colour of the deposit present in the tile samples and the greenish alteration on the balcony are most likely mainly due to the presence of cyanobacteria and eukaryotic algae, respectively.
Paraloid B72® was detected in samples XI and XII. This synthetic resin, frequently used in the conservation of ceramic tiles [39], was applied during the 2007 restoration of the hotel façade. The fact that no polymers were found on the surface of other samples (XIII–XIX), which had most likely been treated with the resin, could be due to photooxidative depolymerisation and the washout of resin from the tile surfaces [12, 13]. Phoma was found in all the samples in which we detected Paraloid B72®, and also in one without any evidence of polymer but collected in sampling area d, which was treated in the 2007 restoration. Melanised fungi, such as Phoma, are among the most damaging fungi, attacking and penetrating the surfaces of stone monuments [8]. In the study by Cappitelli et al. [8] on melanin-producing fungi that attack synthetic polymers used in cultural heritage, the Phoma genus was found. The presence of an uncultured Bacteroidetes (band 18A) and Methylibium sp. (bands 15 and 10A) can be due to the fact that Bacteroidetes show hydrolytic activity toward polymeric substances and Methylibium can grow on organic pollutants [5]. In addition, when Paraloid B72 was present, the biofilm was located in a deeper position than in samples without any evidence of the resin, as also suggested by Ariño and Saiz-Jimenez [3].
During this study, several microorganisms related to an alkaline environment, which is the range of the tile pH, were found. Among the Bacteroidetes, we found two uncultured bacteria (bands 8 and 32A) that were previously found in the Roman Necropolis of the Carmona tomb, carved in a calcarenite bed, an alkaline material. Both microorganisms are taxonomically related to Hymenobacter (100% from RDP) and were detected in the sample taken from the first-floor balcony. Many Hymenobacter species have been isolated from air and alkaline soil, indicating a possible relationship based on desiccation and alkaline tolerance [1]. In another study, Pedobacter, the most probable taxon (100% from RDP) for the band 18A, was detected in alkaline painted and unpainted concrete structures by Giannantonio et al. [15]. Finally, Bacteroidetes were also detected in the highly alkaline saline soil of the former lake Texcoco in Mexico [38]. Among the Betaproteobacteria found, one is Yonghaparkia alkaliphila (bands 13 and 13A), bacteria that live in natural alkaline environments [45]. From the study by Hyvärinen et al. [18], Actinobacteria were found especially on ceramic products, which may be due to their capability to tolerate alkaline conditions. Related to this, Actinobacteria were also detected in frescoes from two different churches in Siena (Italy), both of them showing an alkaline environment [24]. Among this phylum, we found one uncultured Actinobacteria (band 17A) that was previously found in the Roman Necropolis of Carmona tomb, carved in a calcarenite bed and the most related bacteria to the band 10, Microcella putealis, bacteria that live in natural alkaline environments [37].
Taking into account the natural environment in which the hotel is built, the Venice Lagoon, it is not surprising to find microorganisms correlated with aquatic ecosystems and halophilic microorganisms. As an example, Aureobasidium, which was detected in two samples near the ground, is from a study on the diversity of marine yeasts, and was previously found in building and ceramic materials and in natural salterns [18, 19]. Also, black fungi were detected on hypersaline salt pans [9]. Bacteroidetes have frequently been found to be dominant in marine ecosystems [38]; moreover, Borin et al. [5] reported that this phylum was widespread in the Venice Lagoon. Another Bacteroidetes that was detected in the biological deposit present on the balcony is Spirosoma aquatica (bands 2, 3, 5, 28A and 29A), a bacterium isolated from freshwater. Alpha- and Betaproteobacteria were detected during a study about biofilm formation and succession, occurring on the surface of unglazed ceramic tiles in an artificial reef deployed in the northern Gulf of Eilat [35]. Finally, also the order of Chroococcales (one of the cyanobacteria detected in tile samples) is abundant in marine and freshwater areas [31]. Therefore, the high salt concentration of the environment near the Hungaria Hotel is an important factor in the selection of the microflora present on its façade.
The Venice Lagoon is contaminated by heavy metals, hydrocarbons, polycyclic aromatic hydrocarbons and polychlorinated biphenyls from industrial, urban and agricultural sources [5]. Some of the bacteria detected in this study (without substratum preferences) are correlated with pollutants. The uncultured Microcella sp. (band 14) was found in shoreline environments (Spain) affected by the Prestige oil spill [2], two uncultured Betaproteobacteria and an uncultured Bacteroidetes (bands 1, 27A, 7, 31A, 1A) were found in studies about the microbial community in sites contaminated by trichloroethene and uranium, and Methylibium sp. (band 10A) was found in filters used in water treatment for the removal of natural organic matter and organic micropollutants [21].
The massive growth of microorganisms soon after the restoration might be explained by the water content of the tiles. Presence and increase of water inside the ceramic tiles completely glazed by a vitreous layer partially cracked, as for instance in the case of Hungaria hotel, can be physically associated to the phenomena of capillarity interesting masonry buildings. If the walls are covered by a partially permeable or impermeable layer as for instance the vitreous mosaics covering the walls of San Marco Basilica in Venice, the wet zone reaches up to several metres above the ground [4].
In conclusion, it was proved that the deterioration of the tiles was mainly due to the presence of microorganisms. The rapid reappearing of coloured alterations soon after the 2007 restoration, which included the use of biocides, was likely favoured by the water content and organic substances, some of which added during the conservation treatment. For a long-term maintenance plan, an immediate series of corrective measures, finalised to a more efficient conservation of the Hungaria façade, is strongly suggested.
References
Aislabie JM, Jordan S, Barker JM (2008) Relation between soil classification and bacterial diversity in soils of the Ross Sea region, Antarctica. Geoderma 144:9–20
Alonso-Gutiérrez J, Figueras A, Albaigés J, Jiménez N, Viñas M, Solanas AM, Novoa B (2009) Bacterial communities from shoreline environments (Costa da Morte, Northwestern Spain) affected by the Prestige oil spill. Appl Environ Microbiol 75(11):3407–3418
Ariño X, Saiz-Jimenez C (1996) Lichen deterioration of consolidants used in the conservation of stone monuments. Lichenologist 28(4):391–394
Bakolas A, Biscontin G, Contardi V, Franceschi E, Moropoulou A, Palazzi D, Zendri E (1995) Thermoanalytical research on traditional mortars in Venice. Thermochim Acta 269(270):817–828
Borin S, Brusetti L, Daffonchio D, Delaney E, Baldi F (2009) Biodiversity of prokaryotic communities in sediments of different sub-basins of the Venice lagoon. Res Microbiol 160:307–314
Buys S, Oakley V (ed) (1996) Conservation and restoration of ceramics (Conservation and Museology), Butterworth-Heinemann, Woburn, pp. 3–28
Cappitelli F, Nosanchuk JD, Casadevall A, Toniolo L, Brusetti L, Florio S, Principi P, Borin S, Sorlini C (2007) Synthetic consolidants attacked by melanin-producing fungi: case study of the biodeterioration of Milan (Italy) cathedral marble treated with acrylics. Appl Environ Microbiol 73(1):271–277
Cappitelli F, Principi P, Pedrazzani R, Toniolo L, Sorlini C (2007) Bacterial and fungal deterioration of the Milan Cathedral marble treated with protective synthetic resins. Sci Total Environ 385:172–181
Chertov O, Gorbushina A, Deventer B (2004) A model for microcolonial fungi growth on rock surfaces. Ecol Model 177:415–426
De los Ríos A, Grube M, Sancho LG, Ascaso C (2007) Ultrastructural and genetic characteristics of endolithic cyanobacterial biofilms colonizing Antarctic granite rocks. FEMS Microbiol Ecol 59:386–395
Del Monte M, Sabbioni C (1983) Weddellite on limestone in the Venice environment. Environ Sci Technol 17:518–522
Favaro M, Mendichi R, Ossola F, Russo U, Simon S, Tomasin P, Vigato PA (2006) Evaluation of polymers for conservation treatments of outdoor exposed stone monuments. Part I: photo-oxidative weathering. Polym Degrad Stabil 91:3083–3096
Favaro M, Simon S, Menichelli C, Fassina V, Vigato PA (2005) The four virtues of the Porta della Carta, Ducal Palace, Venice—assessment of state of preservation and re-evaluation of the 1979 restoration. Stud Conserv 50(2):109–127
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Giannantonio DJ, Kurth JC, Kurtis KE, Sobecky PA (2009) Molecular characterizations of microbial communities fouling painted and unpainted concrete structures. Int Biodeterior Biodegrad 63:30–40
Grubb JA, Hemant SL, Kakade AM (2007) Testing pH of concrete. Concrete International 29(4):78–83
Horath T, Neu TR, Bachofen R (2006) An endolithic microbial community in dolomite rock in central Switzerland: characterization by reflection spectroscopy, pigment analyses, scanning electron microscopy, and laser scanning microscopy. Microb Ecol 51:353–364
Hyvärinen A, Meklin T, Vepsäläinen A, Nevalainen A (2002) Fungi and actinobacteria in moisture-damaged building materials—concentrations and diversity. Int Biodeterior Biodegrad 49:27–37
Jurado V, Sanchez-Moral S, Saiz-Jimenez C (2008) Entomogenous fungi and the conservation of the cultural heritage: a review. Int Biodeterior Biodegrad 62:325–330
Macedo MF, Miller AZ, Dionísio A, Saiz-Jimenez C (2009) Biodiversity of cyanobacteria and green algae on monuments in the Mediterranean Basin: an overview. Microbiology 155:3476–3490
Magic-Knezev A, Wullings B, Van der Kooij D (2009) Polaromonas and Hydrogenophaga species are the predominant bacteria cultured from granular activated carbon filters in water treatment. J Appl Microbiol 107:1457–1467
Marata A (ed) (2008) Se i Muri potessero parlare. I Libri di Damoli, Arbizzano di Negrar (VR) Italy
McNamara CJ, Perry TD, Bearce KA, Hernandez-Duque G, Mitchell R (2006) Epilithic and endolithic bacterial communities in limestone from a Maya archaeological site. Microb Ecol 51:51–64
Milanesi C, Baldi F, Borin S, Brusetti L, Ciampolini F, Iacopini F, Cresti M (2009) Deterioration of medieval painting in the chapel of the Holy Nail, Siena (Italy) partially treated with Paraloid B72®. Int Biodeterior Biodegrad 63:844–850
Norris TB, Castenholz RW (2006) Endolithic photosynthetic communities with in ancient and recent travertine deposits in Yellowstone National Park. FEMS Microbiol Ecol 57:470–483
Oliveira MM, Sanjad TBC, Bastos CJP (2001) Biological degradation of glazed ceramic tiles. In: Lourenço PB, Roca P (eds) Historical constructions. University of Minho, Guimarães, pp 337–342
Polo A, Cappitelli F, Brusetti L, Principi P, Villa F, Giacomucci L, Ranalli G, Sorlini C (2010) Feasibility of removing surface deposits on stone using biological and chemical remediation methods. Microb Ecol 60(1):1–14
Portillo MC, Alloza R, Gonzalez JM (2009) Three different phototrophic microbial communities colonizing a single natural shelter containing prehistoric paintings. Sci Total Environ 407:4876–4881
Praderio G, Schiraldi A, Sorlini C, Stassi A, Zanardini E (1993) Microbiological and calorimetric investigations on degraded marbles from the Ca’ d’Oro facade (Venice). Thermochim Acta 227:205–213
Ramírez M, Hernández-Mariné M, Novelo E, Roldán M (2010) Cyanobacteria-containing biofilms from a Mayan monument in Palenque, Mexico. Biofouling 26(4):399–409
Rindi F (2007) Diversity, distribution and ecology of green algae and cyanobacteria in urban habitats. In: Seckbach J (ed) Algae and Cyanobacteria in extreme environments. Springer, Dordrecht, pp 621–640
Salvadori O, Sorlini C, Zanardini E (1994) Microbiological and biochemical investigations on stone of the Ca’ Oro façade (Venice). In: III International Symposium on the Conservation of Monuments in the Mediterranean Basin, 22–25 June 1994, Venice, pp 343–347
Sand W (1997) Microbial mechanisms of deterioration of inorganic substrates—a general mechanistic overview. Int Biodeterior Biodegrad 40(24):183–190
Sherwood AR, Presting GG (2007) Universal primers amplify a 23S rDNA plastid marker in eukaryotic algae and cyanobacteria. J Phycol 43:605–608
Siboni N, Lidor M, Kramarsky-Winter E, Kushmaro A (2007) Conditioning film and initial biofilm formation on ceramics tiles in the marine environment. FEMS Microbiol Lett 274:24–29
Sigler WV, Bachofen R, Zeyer J (2003) Molecular characterization of endolithic cyanobacteria inhabiting exposed dolomite in central Switzerland. Environ Microbiol 5(7):618–627
Tiago I, Chung AP, Veríssimo A (2004) Bacterial diversity in a nonsaline alkaline environment: heterotrophic aerobic populations. Appl Environ Microbiol 70(12):7378–7387
Valenzuela-Encinas C, Neria-González I, Alcántara-Hernández RJ, Estrada-Alvarado I, Javier Zavala-Díaz de la Serna F, Dendooven L, Marsch R (2009) Changes in the bacterial populations of the highly alkaline saline soil of the former lake Texcoco (Mexico) following flooding. Extremophiles 13:609–621
Vaz MF, Pires J, Carvalho AP (2008) Effect of the impregnation treatment with Paraloid B72® on the properties of old Portuguese ceramic tiles. J Cult Herit 9:269–276
Villa F, Giacomucci L, Polo A, Principi P, Toniolo L, Levi M, Turri S, Cappitelli F (2009) N-vanillylnonanamide tested as a non-toxic antifoulant, applied to surfaces in a polyurethane coating. Biotechnol Lett 31:1407–1413
Watanabe K, Ohfuji H, Ando J, Kitagawa R (2006) Elemental behaviour during the process of corrosion of sekishu glazed roof-tiles affected by Lecidea s. lat. sp. (crustose lichen). Clay Miner 41(4):819–826
Weber RWS, Madhour A, Anke H, Mucci A, Davoli P (2005) 2-Hydroxytorularhodin, a new xanthophyll from the red yeast Sporobolomyces coprosmae. Helv Chim Acta 88:2960–2966
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic, San Diego, pp 315–322
Wong FKY, Lecap DC, Lau MCY, Aitchison JC, Cowan DA, Pointing SB (2010) Hypolithic microbial community of quartz pavement in the high-altitude tundra of central Tibet. Microb Ecol 60:730–739
Yoon JH, Kang SJ, Schumann P, Oh TK (2006) Yonghaparkia alkaliphila gen. nov., sp. nov., a novel member of the family Microbacteriaceae isolated from an alkaline soil. Int J Syst Evol Microbiol 56:2415–2420
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Giacomucci, L., Bertoncello, R., Salvadori, O. et al. Microbial Deterioration of Artistic Tiles from the Façade of the Grande Albergo Ausonia & Hungaria (Venice, Italy). Microb Ecol 62, 287–298 (2011). https://doi.org/10.1007/s00248-011-9812-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9812-0