Abstract
Biofumigation (BIOF) is carried out mainly by the incorporation of brassica plant parts into the soil, and this fumigation activity has been linked to their high glucosinolate (GSL) content. GSLs are hydrolyzed by the endogenous enzyme myrosinase to release isothiocyanates (ITCs). A microcosm study was conducted to investigate the effects induced on the soil microbial community by the incorporation of broccoli residues into soil either with (BM) or without (B) added myrosinase and of chemical fumigation, either as soil application of 2-phenylethyl ITC (PITC) or metham sodium (MS). Soil microbial activity was evaluated by measuring fluorescein diacetate hydrolysis and soil respiration. Effects on the structure of the total microbial community were assessed by phospholipid fatty acid analysis, while the impact on important fungal (ascomycetes (ASC)) and bacterial (ammonia-oxidizing bacteria (AOB)) guilds was evaluated by denaturating gradient gel electrophoresis (DGGE). Overall, B, and to a lesser extent BM, stimulated microbial activity and biomass. The diminished effect of BM compared to B was particularly evident in fungi and Gram-negative bacteria and was attributed to rapid ITC release following the myrosinase treatment. PITC did not have a significant effect, whereas an inhibitory effect was observed in the MS-treated soil. DGGE analysis showed that the ASC community was temporarily altered by BIOF treatments and more persistently by the MS treatment, while the structure of the AOB community was not affected by the treatments. Cloning of the ASC community showed that MS application had a deleterious effect on potential plant pathogens like Fusarium, Nectria, and Cladosporium compared to BIOF treatments which did not appear to inhibit them. Our findings indicate that BIOF induces changes on the structure and function of the soil microbial community that are mostly related to microbial substrate availability changes derived from the soil amendment with fresh organic materials.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Microbial community is an integral part of the soil ecosystem because it regulates significant processes such as nutrient cycling, decomposition of organic residues, formation of humic substances, and pollutant degradation. Several studies have shown that agronomic [8, 45, 56] and crop protection [33, 55] practices significantly influence both the structure and the function of the soil microbial community.
Crop protection practices aiming to control soil-borne pests and pathogens are of great importance to EU countries and worldwide, especially nowadays that several synthetic soil fumigants are to be withdrawn from the EU market according to the Directive 91/414 EEC. For that reason, the introduction and use of environment-friendly alternatives for the control of soil-borne pests and pathogens are needed. Biofumigation (BIOF), performed by the incorporation of fresh biomass from brassica plants into the soil, has been proposed as an alternative method [25, 31, 37]. It is based on the high concentrations of glucosinolates (GSLs) which are secondary metabolites found in most brassica plants [13]. Structurally, GSLs consist of a β-thioglucose moiety, a sulfonated oxime group, and a variable side chain according to which they are categorized as aliphatic, aromatic, and indolyl GSLs [60]. They are biologically inactive molecules; however after tissue disruption, they are hydrolyzed by myrosinase (EC 3.2.3.1) to several byproducts like indoles, isothiocyanates (ITCs), thiocyanates, and nitriles [40]. Among those, ITCs are the most toxic for soil-borne pathogens [35], and they are produced from aliphatic and aromatic GSLs.
The efficacy of the method in situ relies on the BIOF potential of the incorporated brassica plant species [24], which depends on the rate and the quantity of ITC formation in the soil and the toxicity of the produced ITCs on the targeted pests and pathogens [4, 53]. It has been shown in vitro that fungi are more susceptible than bacteria and that ITCs derived from aliphatic GSLs such as allyl-ITCs are more active in soil than ITCs derived from aromatic GSLs like 2-phenylethyl-ITC (PITC) [36]. The suppression of soil-borne pathogens induced by BIOF methods has also been linked to factors other than ITCs. These include the enhanced competition of soil-borne pathogens with copiotrophic soil microorganisms favored by the addition in soil of fresh organic matter [15, 30], proliferation of Streptomycetes, and enhanced nitrification leading to NO production [9, 10] or the formation of bioactive sulfur-containing compounds [3].
So far, most studies concerning BIOF have focused on its efficacy against soil-borne pathogens, while any effects on the structure and function of non-target members of the microbial community have been overlooked. Ammonia-oxidizing bacteria (AOB) are considered, along with ammonia-oxidizing archaea, the key microbial groups in soil nitrogen cycling [47]. Ammonia oxidation is the primary step in the oxidation of ammonia to nitrate via nitrite production and is considered as the rate limiting step of nitrification. Moreover, regarding soil application of brassica biomass with low GLS content, a relation between enhanced nitrification, NO production, and disease suppression has been proposed [9, 10]. Difficulties in the cultivation of AOB [27] led to the development of culture-independent methods for studying their ecology in environmental samples [26, 34, 38]. Molecular analysis of 16S rRNA sequences has placed AOB into two distinct monophyletic groups belonging to γ-proteobacteria and β-proteobacteria. The latter group encompasses all AOB inhabiting terrestrial ecosystems and comprises two genera: Nitrosospira and Nitrosomonas [48]. Several studies so far have investigated the impact of anthropogenic activities on AOB in soils [28, 59]. Previous studies by Bending and Lincoln [4] showed that pure ITCs and sulfur volatile compounds like dimethyl disulfide reduced AOB abundance and nitrification in soil. Similarly, Brown and Morra [6] suggested that GSL hydrolysis products are involved in nitrification inhibition. However, none of the above studies investigated the impact of BIOF on the AOB community structure.
Ascomycetes (ASC) is the largest group of the true fungi [29], including noxious plant pathogens belonging to the genera Fusarium, Pyrenochaeta, Sclerotinia, and Verticillium, which are expected to be controlled by BIOF or chemical fumigation methods; the group, however, also includes a wide range of saprotrophic fungi which are important for soil organic matter decomposition [44], modification, and polymerization of humic substances [16]. Only a few studies so far have investigated the effects of agricultural practices on the ASC community [21, 57, 58], while no information is available regarding the impact of BIOF on their community structure.
Phospholipid fatty acid (PLFA) analysis is a well-documented method used for the estimation of soil microbial biomass and for studying changes in the structure of the soil microbial community [23, 55]. However, PLFA provides information regarding community structure only at the microbial group level. Thus, in cases where a more in-depth investigation of the microbial community structure is required, complementary molecular fingerprinting techniques like denaturating gradient gel electrophoresis (DGGE) could be used. DGGE analysis has been used for the identification of changes in the structure of specific microbial groups and could identify changes at the species or genus level if supplemented with the parallel development of clone libraries [21, 22].
The scope of this study was (1) to investigate the impact of the incorporation of broccoli plant residues into the soil as a BIOF treatment on the structure and the activity of the microbial community with special focus onto the structure of two important microbial guilds, AOB and ASC and (2) to compare and contrast these effects with the effects induced by pure ITCs, which are considered responsible for BIOF activity and with a standard commercial ITC-liberating soil fumigant.
Methods
Experimental Design
The effect of broccoli residues incorporation on the soil microbial community was investigated in a pot experiment. The soil used was collected from a field site of the National Agricultural Research Foundation of Greece, Larisa, and was characterized as sandy loam (sand 64%, clay 11%, loam, 25%) with pH 7.2 and organic matter content 1.2%.
Broccoli plants (Brassica oleracea var. Italica) variety “Marathon” were transplanted at the stage of four true leaves in 12-L pots containing a soil/compost (1:1 v/v) mixture and maintained until harvest. Youngest fully expanded leaves of broccoli (B) were carefully removed from the plants and blended using a high speed blender to achieve maximum tissue disruption. The finely chopped plant residues were incorporated into soil at a rate of 15 g kg−1 dry soil. The same quantity of leaves was blended, directly treated with 10 mL myrosinase (0.32 U mL−1 Sigma-Aldrich, Germany), and mixed with the soil 3 h later (BM). Preliminary studies showed that addition of myrosinase in the blended broccoli leaves resulted in complete hydrolysis of GSLs, and this treatment was considered optimized for maximum ITCs release. Amended soil samples (1 kg) were immediately transferred to 2-L pots, and the appropriate quantity of deionized water was added to adjust soil moisture content to 45% of the water holding capacity. Moisture content was maintained constant at these levels by regular addition of deionized water when needed.
The effect of pure or commercial ITCs on the soil microbial community was comparatively assessed in the same experiment. Thus, PITC (Sigma-Aldrich, Germany), the ITC derived from the aromatic GSL gluconasturtin found in the majority of Brassica species [13], was applied to soil in the form of an aqueous solution (200 μg mL−1) to give a final soil concentration of 10 mg kg−1. The commercial ITC-liberating soil fumigant metham sodium (MS) was applied in a different way for technical and safety reasons and in order to imitate as much as possible the field application of MS. Thus, MS and extra water for moisture adjustment (45% of water holding capacity) were applied in the soil which was immediately placed in a pot (2 L) and covered with double layer plastic film to minimize fumigant emissions in the growth room and confine them over the soil headspace. Metham sodium was applied as an aqueous solution (10,000 mg L−1) prepared by VAPAM 51SL. The concentration of MS in soil after its application corresponded to its recommended dose (300 μg g−1) for control of soil pathogens and pests in protected crops. The plastic cover was maintained for 14 days, and after its removal, the soil was gently mixed to ensure the release of any residues of methyl-ITC which is formed after MS application into wet soil. Finally, soil samples (1 kg) receiving appropriate amount of water without chemicals for moisture content adjustment served as untreated controls. All five treatments were repeated three times, in a completely randomized design.
Pots were transferred in a growth room and incubated at 23 ± 2°C for the next 60 days. At each sampling date, five subsamples (5 g) were collected from the top 10 cm of each pot (14-cm diameter × 13-cm depth) using a cork borer. The subsamples collected from each pot at each sampling time were pooled to obtain one composite sample (25 g) leading to triplicate samples from each treatment at each sampling time. Soil samples used for PLFA analysis were collected at 7, 15, and 60 days after application (DAA) and were stored at 4°C until analysis, whereas soil samples used for DNA extraction were collected at 3, 15, and 60 DAA and were stored at −20°C until analyzed.
Determination of Microbial Activity and Soil Respiration
Effects on soil microbial activity were assessed by measurements of soil respiration and fluorescein diacetate (FDA) hydrolysis. Soil respiration was measured at 0, 1, 2, 3, 7, 15, and 60 DAA by CO2–C evolution using the alkali trap method as described by Ntougias et al. [42]. FDA hydrolysis was measured at 3, 15, and 60 DAA according to the method described by Adam and Duncan [1].
Phospholipid Fatty Acid Analysis
Extraction of PLFA from soil samples was performed as it was described before [55]. Briefly, triplicate samples (5 g dry weight) of each treatment were extracted for 2 h with 19 mL of chloroform/methanol/phosphate buffer mixture (pH 3.0; 1:2:0.8 v/v/v). After centrifugation, the supernatant was mixed with 5 mL of chloroform and 5 mL phosphate buffer, and after short agitation, the chloroform layer was removed, dried under N2, and resuspended at 4 × 250 μL chloroform. Phospholipids were separated from glycolipids and neutral lipids on solid phase extraction columns C18 Supelco SPE DSC-Si (Silica Tube 6 mL, 500 mg). Neutral lipids and glucolipids were eluted with 5 mL chloroform and 5 mL acetone, respectively, while phospholipids were eluted with 5 mL methanol and dried under a stream of N2. Samples were then subjected to mild alkaline methanolysis with 0.5 mL of 1:1 mixture of methanol/toluene and 0.5 mL of 0.2 M methanolic solution of KOH and heating at 37οC for 15 min. Subsequently, 0.5 mL acetic acid was added, and the resulting fatty acid methyl esters (FAME) were extracted twice with addition of 2 mL of hexane/chloroform (4:1 v/v) and 2 mL water. The hexane layer was removed, combined, and dried under N2 stream. Samples were re-dissolved in 200 μL hexane and analyzed in a Hewlett Packard HP5890 Series ΙΙ gas chromatography (GC) with FID detector [55].
Identification of FAME was based on retention time comparisons with commercial standard mixtures (Supelco, UK). Further confirmation of the different components of the standard FAME mixtures was achieved in a GC–mass spectrometry system [55]. Quantification of each fatty acid (nanomoles per gram) was carried out by calibration against standard solutions of nonadecanoate methyl ester (19:0). Known amounts of nonadecanoate were added before methanolysis as internal standard.
Standard nomenclature was used to describe FAME detected in the soil samples. Numbering of carbon atoms begins at the aliphatic (ω) end of the fatty acid molecule. The number of double bonds within the molecule is given after the colon. Other notations are “OH” for hydroxyl fatty acids, “cy” for cyclopropane groups, and the prefixes “i” and “α” for iso- and anteiso-branched chain fatty acid methyl esters.
DNA Extraction, Polymerase Chain Reaction Amplification, and Denaturating Gradient Gel Electrophoresis Analysis
DNA was extracted from 0.25 g of soil using the Power Soil DNA isolation kit according to the manufacturers’ instructions (MoBio Laboratories, USA). A nested polymerase chain reaction (PCR) approach was followed for DGGE analysis of ASC and AOB. In the first PCR for ASC, soil DNA was amplified with the group-specific primers ITS1F–ITS4A (600–700 bp) [29]. The PCR product obtained was semi-nested with universal fungal primers ITS1F + GC–ITS2, to give a product appropriate for DGGE analysis (ca. 300 bp). The forward primer was exactly the same as in the first PCR but with addition of a GC-clamp at its 5′ end to stabilize the melting behavior of DNA fragments. Thermocycling conditions were as described elsewhere [21].
For AOB, soil DNA was initially amplified using primer pair βΑΜΟf–βAMOr with a relaxed specificity for AOB [38]. This product (ca. 1,100 bp) was nested with primers CTO189fABC–CTO654r [26], which were designed to specifically amplify partial rRNA gene sequences (465 bp) of AOB from β-proteobacteria [27]. Finally, the product obtained was nested again with primers 357f + GC and 534r to give a product appropriate for DGGE (ca. 190 bp). Thermocycling conditions for the different PCR were as reported elsewhere [22].
The final concentrations of the components used in the PCR reactions were 10× polymerase reaction buffer, 200 μM of each dNTP, 1 U DNA polymerase (Dynazyme EXT-Finnzymes OY, Finland), and 0.2 μΜ of each of the primers (0.4 μΜ for ASC-specific primers). Bovine serum albumin at a concentration of 400 ng μL−1 was included in all first PCR amplifications to prevent polymerase inhibition by soil humic acids. In the first PCR round, 1 μL of the extracted soil DNA was added to 49 μL of PCR reaction mix, while in the subsequent PCR rounds, 1 μL of a tenfold diluted PCR product from the previous round was added to 49 μL of PCR reaction mix.
DGGE analysis was performed with an INGENYphorU-2 × 2 system (Ingeny International BV, The Netherlands). Polyacrylamide gels (8%) in 1× TAE buffer were prepared for all soil microbial communities combinations with a denaturating gradient of 30–55% and 45–62% for ASC and AOB, respectively. Denaturation conditions were obtained using a solution (100%) with 7 M urea and 40% formamide. Gels were electrophorized for 16 h at 60°C and 75 V and were silver stained as described by McCaig et al. [39]. Stained gels were photographed using a digital camera, and the images were subjected to analysis using CrossChecker 2.9v (Wageningen University, The Netherlands). Binary data for the presence/absence of bands in all samples were derived and used for statistical analysis.
Cloning and Sequencing of PCR Products
Clone libraries for AOB and ASC were constructed based on the PCR products generated with the group-specific primers CTO189f–CTO654r and ITS1F–ITS4A, respectively. Since the DGGE results showed that replicate samples of the same treatment had low variability, replicated PCR products of the same treatment were pooled and purified to a final volume of 30 μL using the Nucleospin II PCR clean-up kit (Macherey-Nagel GmbH, Germany). Cloning into the pGEM-T vector (Promega, Madison, WI, USA) was performed following the protocol proposed by the manufacturers, and the screening of the clone libraries by PCR and DGGE was carried out as described by Karpouzas et al. [21]. Briefly, 25 white colonies were selected from each treatment and were subjected to colony PCR using primers 357f + GC–534r and ITS1F + GC–ITS2 for AOB and ASC, respectively. Subsequently, PCR products were checked in agarose gels (1%), and the positive clones were screened on a DGGE gel to determine their electrophoretic mobility compared with the band pattern of the original environmental sample. Representative clones for each band type matching the migration pattern of bands in the original samples were sequenced.
Clones were subjected to plasmid DNA extraction and purification using NucleoSpin Plasmid kit (Macherey-Nagel GmbH, Germany). Sequencing reactions were performed according to the manufacturers’ instructions using a PRISM BigDye Terminator Cycle Sequence reaction kit (Applied Biosystems, UK). Sequences were deposited in the European Molecular Biology Laboratory (EMBL) database under accession numbers FN562026–FN562083. Cloning sequences were subjected to similarity searches using the Basic Local Alignment Search Tool. Multiple alignments were performed using ClustalW, evolutionary distances were calculated using the method of Jukes and Cantor [20], and the topology was inferred using the “neighbor-joining” method [50] based on bootstrap analysis of 1,000 trees.
Statistical Analysis
The data obtained from temporal measurements of soil respiration and FDA hydrolysis were subjected to a two-way analysis of variance (ANOVA). In cases were significant interactions between the main factors (time × fumigation treatment) were identified, Tukey’s multiple comparison tests (a = 0.05) were separately applied at each single factor level.
Relative abundance data from the PLFA analysis were subjected to principal component analysis (PCA). Ordination of the different samples according to their PLFA content was performed for the first two principal components (PCs) which included most of the variance of the original dataset. The scores for the first two PCs for all the samples were subjected to two-way ANOVA to identify the significance of the effects of fumigation treatments and time on the new dependent variables.
The binary data matrix obtained from DGGE profiling of the two communities were subjected to principal coordinate analysis (PCoA) to compare the effect of fumigation treatment and time on the structure of ASC and AOB communities. All statistical analyses were performed using Genstat 11.0v.
Results
Microbial Respiration and FDA Activity
Soil chemical fumigation and the incorporation of broccoli residues had significant (p < 0.001) but contrasting effects on soil respiration and the FDA hydrolysis compared to the untreated control (Table 1). Moreover, these general microbial activity indicators were significantly affected by time (p < 0.001), and a significant interaction between fumigation treatment and time was observed (p < 0.001).
B- and BM-amended soils showed a sharp initial stimulation of soil respiration, and CO2 evolution became six and five times higher, respectively, compared to the untreated control (Fig. 1). The microbial respiration reverted to levels similar to the control 3 DAA in the BM-amended samples, whereas a more persistent effect was observed in the B-amended samples where 30 days was required for soil respiration to equal levels in the untreated control samples. In contrast to BIOF treatments, PITC- and MS-treated soils showed a significant decrease in soil respiration compared to the control and the BIOF treatments (Fig. 1). This effect was also temporal, and respiration reverted to levels similar to the untreated control 7 and 30 DAA for PITC and MS, respectively.
FDA hydrolysis was influenced in the same negative manner as soil respiration by chemical fumigation treatments, whereas the positive effects of BIOF treatments were less pronounced and lasted longer. Thus, fumigation with MS resulted in a significant decrease (p < 0.05) of the FDA hydrolysis rate at 3 and 15 DAA compared to control samples; however, it was doubled at the end of the experimental period. PITC-amended samples were not different from controls although a nonsignificant decrease trend (p > 0.05) was evident at 3 DAA only (Fig. 2). On the other hand, soils amended with B or BM showed a significant increase (p < 0.05) in FDA hydrolysis rate, which was still present at 15 DAA for BM and up to 60 DAA for B (Fig. 2).
PLFAs Analysis
A total of 16 individual fatty acids were identified in the soil samples. These were i-15:0, α-15:0, i-16:0, i-17:0, 17:0 (Gram-positive bacteria), 16:1ω7, cy17:0, cy19:0, 3-OH 14:0, 18:1ω9cis, 18:1ω9trans (Gram-negative bacteria), 16:0 (universal microbial), 18:2ω6,9, and 18:3ω3,6,9 (fungi) [14]. Two other fatty acids of microbial origin were also detected, 20:0 and the internal standard 19:0.
The total PLFA concentration depended significantly on the fumigation treatments and the interaction between time and treatment (p < 0.001, Table 1). The impact of time on the total PLFA concentration was not significant (p > 0.05), apparently depicting the average effect of the contrasting BIOF and chemical fumigation treatments on total PLFAs (Table 1, Fig. 3d). The highest concentration of total PLFAs (162 nmol g−1) was observed in the B-amended samples 3 DAA, whereas the lowest was measured at the same date in the MS-treated samples (11.5 nmol g−1). At 60 DAA, total PLFAs for the B and BM still remained higher than the controls, but total PLFAs for MS treatment had reached those of the control soils (Fig. 3d).
Effect of fumigation treatments and sampling time on the summed amounts of PLFAs characterizing different soil microbial groups: a Gram-negative bacteria, b Gram-positive bacteria, c fungi, and d total microbial PLFAs. Each value is the mean of three replicates; error bars show standard error of means. A shared letter on top of bars indicates lack of statistically significant difference between treatments at each time interval (Tukey’s HSD tests, α < 0.05)
The BIOF treatments (B, BM) showed significantly higher concentrations of total PLFAs for bacteria compared to the control soil throughout the experiment (with the exception of Gram-positive bacteria on 60 DAA in the B treatment; Fig. 3a, b). PLFA indicators for fungi were also higher throughout the experiment for the B treatment, but the addition of myrosinase-treated material (BM) appears to have suppressed this effect on fungi at least up to 15 DAA (Fig. 3c). The opposite effects were observed in the MS-treated soil where the concentrations of the PLFA indicators of the different microbial groups were significantly reduced compared to the control soils at least up to 3 DAA for the Gram-negative bacteria (Fig. 3a) and up to 15 DAA for the Gram-positive bacteria (Fig. 3b) and fungi (Fig. 3c), the latter suffering the most severe effect. For the PITC-treated soils, there was no significant difference in their PLFA indicator content compared to the control. When relative abundance of the different PLFA indicators was considered instead of measured PLFA concentrations, the picture was identical (data not shown).
The relative abundances of the 16 individual PLFAs detected in the soil samples were subjected to PCA, and the results are presented in Fig. 4. A separation of the different treatments along PC1, which described about 60% of the variance in the original dataset, was observed (Fig. 4). Thus, the B- and BM-amended samples clearly clustered on the negative side of PC1 regardless of the time of application. On the contrary, PITC-treated samples grouped together with the control samples and MS-treated samples on the positive side of PC2. A closer observation clearly shows that this separation on the PC1 is reduced with time for the contrasting ΒΙΟF and MS treatments and is minimized at 60 DAA (Fig. 4). Α time partitioning along the PC2 is also observed: At 7 DAA, all treatments apart from PITC are clustered in the upper part of the diagram compared to their lower clustering for 15 and 60 DAA (Fig. 4).
PCA ordination of the PLFA profiles of the soil samples treated either by biofumigation (B, BM) or chemical fumigation (PITC, MS). Ordination of the samples was performed for the first two principal components, which describe 57. 7% (PC1) and 11.1% (PC2) of the variance in the original dataset. Each value is the mean of three replicates with error bars showing the standard deviation of the mean for the two PCs
DGGE Analysis of the ASC Community
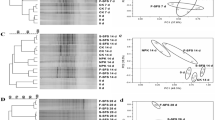
Variability in the DGGE profiles of the replicates of each treatment was negligible (data not shown); thus, one sample per treatment is presented (Fig. 5a). The DGGE fingerprints of the ASC community showed variable patterns, and the number of bands in the different profiles ranged from 10 to 24 with the lowest numbers observed in the MS-treated samples. Principal coordinate analysis on the banding patterns obtained showed a clear separation of MS-treated soils collected 15 and 60 DAA from the rest of the samples at the negative end of both PCs (Fig. 5b). In addition, B- and BM-amended samples collected 3 DAA clustered together and separated along PC2 from the control, PITC-, and MS-treated samples collected at the same time (Fig. 5b).
a DGGE analysis of partial ITS region sequences of the community of ASC. Lanes designated with L correspond to fungal marker which contained 20 ng μL−1 of the ITS-PCR products of the following fungi with the sequence they appear on the gel from top to bottom: Pleurotus djamor, Fusarium oxysporum f. sp. radici-lycopersici, Fusarium solani, Pleurotus eryngii, Pleurotus ostreatus, Pleurotus cystidiosus. Arrows indicate bands corresponding to clones that were sequenced. b Principle coordinate analysis (group average, Jaccard similarity index) of the DGGE banding patterns for the ASC community in the different fumigation treatments. Each value is the mean of three replicates with error bars indicating the standard deviation of the mean for the two coordinates
A visual inspection of the DGGE profiles in the different treatments revealed that certain bands were associated with specific treatments. Thus, clone libraries were developed to identify members of the ASC community which were responsive to the different fumigation treatments. Overall, 47 clones corresponding to bands in the different treatments were sequenced (Fig. 5a). Several bands (5, 10, 16, 24, 26, 3, 6, and 45) that were present in the control, B, BM, and PITC samples were absent in the MS-amended samples particularly on 15 and 60 DAA (Fig. 5a). All the clones associated with those bands showed highest sequence homology (91–100%) to potential plant pathogenic fungi of the genera Fusarium, Nectria, Phaeoacremonium, and Cladosporium (see the Electronic Supplementary Material). On the other hand, bands 32, 33, and 37 became dominant at the profile of the MS-amended samples at 15 and 60 DAA (Fig. 5a), and the associated clones showed highest sequence homology to a Eurotium strain (100%), a Hypocreales isolate (92%), and a Fusarium proliferatum isolate (100%), respectively. Finally, bands 17 and 18 appeared only in the B-amended samples collected on 15 and 60 DAA (Fig. 5a). The associated clones showed generally low similarity with sequences in the database and had higher sequence homology (85–92%) to Pyrenochaeta strains (see the Electronic Supplementary Material).
DGGE Analysis of the AOB Community
The DGGE profiles of the AOB showed little variation between the different treatments and sampling times, and the number of bands in the different profiles ranged from 14 to 17 (Fig. 6a). Indeed, PCoA of the banding patterns obtained showed that the structure of the AOB community was not significantly altered by the different treatments (Fig. 6b). Clone libraries for the AOB resulted in the sequencing of 11 bands/clones, which all showed highest sequence homology to AOB of the genus of Nitrosospira, and the majority of them were placed within cluster 3 (Fig. 7). Certain bands were associated with particular treatments. Thus, band 6 found only in PITC-treated soils 60 DAA showed highest sequence homology to a β-proteobacterium AOB environmental clone. On the contrary, band 8 which disappeared from the DGGE profile of the MS-treated samples collected at 15 and 60 DAA (Fig. 6a), was associated with clones showing highest sequence homology to Nitrosospira B6 isolate.
a DGGE analysis of partial 16S rRNA gene sequences of the community of AOB in the treated soil. Lanes designated with L correspond to bacterial marker which contained 20 ng μL−1 of the 16S rRNA-PCR products of each of the following bacteria presented on the gel from top to bottom: Deinococcus radiodurans, Flavobacterium sp., Pseudomonas entomophila, Sphingomonas paucimobilis, Rhizobium sp., Mesorhizobium loti, Agrobacterium sp. Arrows indicate bands corresponding to clones that were sequenced. b Principle coordinate analysis (group average, Jaccard similarity index) of the DGGE banding patterns for the AOB community in the different fumigation treatments. Each value is the mean of three replicates with error bars indicating the standard deviation of the mean for the two coordinates
Phylogenetic tree for β-proteobacterial 16S RNA gene sequence (465 bp) related to ammonia-oxidizing bacteria in the different fumigated and nonfumigated soils. Evolution distances were calculated using the method of Jukes and Cantor [20], and the topology was inferred using the “neighbor-joining” method [55]. Values on branches denote percent bootstrap support (analysis of 1,000 trees—bootstrap support below 50% is not presented). Bar represents 0.01 inferred substitutions per nucleotide position
Discussion
This study investigated the effects of BIOF (B and BM) and chemical fumigation in the form of commercial ITC-liberating products (MS) or pure ITCs (PITC) on the structure and function of the soil microbial community.
Effects of Broccoli Residues
Incorporation of broccoli residues with or without myrosinase pretreatment resulted in an increase of FDA hydrolysis rate and soil respiration. This indicates a direct response to substrate-C addition in line with previous studies showing similar effects after the incorporation of organic materials into soil, including residues from the Brassica species [43, 51]. The drastic and lasting increase in the concentration of total microbial PLFAs observed in both BIOF treatments indicates a concomitant increase in the size of the active microbial biomass [2, 23], which is also in line with previous reports where a rapid increase of bacterial biomass was observed within 5 to 30 days after incorporation of fresh plant residues [15, 32, 61].
Interestingly, FDA hydrolysis for the BIOF treatments did not show the initial peak observed for microbial respiration but followed more closely the PLFA patterns. FDA hydrolysis measurements are estimates of potential rather than of actual microbial activity [41], and microbial esterases may persist in the soil environment. This explains the closer correlation of FDA hydrolysis data to total soil microbial biomass rather than to soil respiration also observed in the past [51, 54].
The application of broccoli residues (B) stimulated the growth of both bacterial and fungal biomass, as this was illustrated by the increased concentration of fungal and bacterial indicator PLFAs, but this effect was clearly reduced at early incubation stages when the myrosinase-treated residues (BM) were applied. This indicates that BIOF activity was initially enhanced by this treatment, probably due to rapid liberation of ITCs, counterbalancing the effects of C-substrate addition. The narrower microbial activity peak induced by BM compared to the B treatment is in line with this assumption indicating that microbial responses to C-substrates may override fumigation effects when low-GSL biomass from brassica plants is applied into soils. Suppressive effects to pathogens may also be related to such responses of broad microbial consortia rather than to ITC toxicity [10, 15]. Indeed, previous studies showed that Verticillium dahliae in potatoes was suppressed after the incorporation of broccoli plant residues, and the effect was attributed to the addition of fresh organic matter into soil rather than to the release of ITCs [43]. Additionally, Berbegal et al. [5] suggested that other mechanisms besides ITCs are involved in V. dahliae suppression following soil incorporation of cauliflower residues.
Multivariate analysis of the PLFA data showed that BIOF treatments, apart from increasing microbial activity, significantly altered the structure of the soil microbial community. However, specific DGGE analysis of the ASC and AOB soil communities showed a transient response to BIOF treatments for the ASC community, which was evident only at 3 DAA. It should be stated that our findings are based on DNA fingerprinting analysis which might have failed to identify subtle or transient effects that could have been detected by the use of RNA fingerprinting analysis. Still, certain bands which were found to be potentially associated with Pyrenochaeta ribotypes were dominant in the profile of the ASC community following broccoli residue application alone (B). Species of the genus Pyrenochaeta are common soil fungi which have been associated with plant diseases [19]. However, the low homology shown by the respective clones to sequences in the databases (maximum 92% to Pyrenochaeta sp.) precluded any further conclusions to be reached.
BIOF (B and BM) did not have any effect on the structure of the AOB community. In a recent study, Brown and Morra [6] showed that incorporation of brassica tissue into soil resulted in inhibition of nitrification and suggested that GSL hydrolysis products were responsible for this. However, no information on concurrent effects on the community structure of AOB was provided. Several previous studies have shown that the inhibition of nitrification in soils after exposure to exogenous stressors does not always lead to changes at the community structure of AOB [7, 18, 46]. Further work is under way to investigate if the absence of any effects of BIOF on the AOB community concurs with nitrification discrepancy. Our study provides first evidence that BIOF does not appear to exert a direct toxic effect on important microbial guilds; instead, the changes induced reflect a general microbial response to the addition in the soil of fresh organic matter.
Effects of Chemical Fumigants
Soil application of pure PITC, an ITC commonly formed upon tissue disruption of most brassicas [13], resulted in a transient decrease in soil respiration, while no significant effects on FDA activity and total microbial biomass were observed. Multivariate analysis of PLFA and DGGE data also suggested that PITC had no significant impact on the structure of the soil microbial community as well as on the community of ASC and AOB. Although PITC is highly toxic to soil microorganisms in vitro [53], it seems to be deactivated after its application to soil [36]. This was attributed to its increased adsorption which results in low availability in the solution or air phase in the soil [36]. Indeed, Rumberger and Marschner [49] showed that PITC released from brassica plant roots does not affect the total rhizosphere bacterial and eukaryotic community and that it does not appear to affect microbial community function and may explain only a small part only of the variance regarding the active total bacterial and eukaryotic community. Although this is in line with our results, it was also shown by these authors that the continuous application of PITC to 10-g soil samples led to noticeable structure changes in the soil microbial community, especially regarding eukaryotes. Previous laboratory studies have also reported that applications of PITC into 10-g soil samples also caused a small decrease on nitrification and on the number of culturable AOB [4]. These apparent discrepancies are probably related to the single PITC application and the large microcosm environments used in our study. Our study provides initial evidence that the concentrations of PITC liberated by the broccoli residues incorporated in soil under field conditions are unlikely to induce significant direct toxicity effects.
In contrast to broccoli residue applications, soil chemical fumigation with MS resulted in a dramatic decrease in the size (concentration of total microbial PLFAs) and activity (soil respiration and FDA activity) of the soil microbial community which persisted for at least 30 days. Our results are in line with previous studies which have also reported that MS application resulted in persistent reductions in the size and activity of soil microorganisms [11, 17, 33]. Multivariate analysis of PLFA data showed that MS soil application significantly altered the structure of the soil microbial community, an effect also observed in the past [33, 55]. Multivariate analysis of DGGE profiles showed that MS soil application induced significant changes in the structure of the ASC community and resulted in a significant reduction in the richness of the ASC community, in line with previous studies [17]. Cloning analysis of the ASC community in the MS-treated samples revealed that several bands which were present in the other soil treatments disappeared in the MS-treated samples particularly at 15 and 60 DAA. The majority of these bands were associated with phylotypes belonging to the Fusarium–Nectria complex, while other potential plant pathogenic fungi including Phaeoacremonium and Cladosporium were also suppressed. Several previous studies have documented the high eradication efficacy of MS on soil-borne fungal plant pathogens like Fusarium and others [12, 52].
Conclusions
This study was the first to comparatively assess the toxicity of biofumigation and chemical fumigation on the structure and function of the soil microbial community. We suggest that the changes induced on the soil microbial community upon biofumigation with broccoli plant residues reflected the response of the microbial community to soil enrichment with fresh and decomposable organic matter rather than a direct toxicity effect by the release of ITCs from hydrolyzed plant-derived GSLs. This is further supported by the lack of any adverse effects induced by PITC, an ITC commonly released by brassicas in soil, on the structure and activity of the soil microbial community. A transient change in the structure of ascomycetes, which are involved in soil organic matter decomposition, was observed during biofumigation, whereas the structure of the ammonia-oxidizing bacterial community, which are slow-growing microbes with limited ability to respond to altered C-substrate availability, did not change. In contrast, soil chemical fumigation with a commercial ITC-based fumigant (metham sodium) inhibited microbial activity, reduced the biomass of all microbial groups, and induced significant and persistent changes on the structure of the ascomycete community, while the ammonia-oxidizing bacterial community structure remained unaltered. Although a temporary effect on fungal biomass may have been induced by the myrosinase-treated residue applications, clone libraries further revealed that biofumigation generally failed to suppress common plant pathogenic ascomycetes which were drastically eradicated by metham sodium.
Abbreviations
- BIOF:
-
Biofumigation
- GSLs:
-
Glucosinolates
- B:
-
Broccoli
- BM:
-
Broccoli + myrosinase
- ITC:
-
Isothiocyanate
- PITC:
-
2-phenylethyl isothiocyanate
- MS:
-
Metham sodium
- PLFA:
-
Phospholipid fatty acids
- FAME:
-
Fatty acid methyl esters
- DGGE:
-
Denaturating gradient gel electrophoresis
- ASC:
-
Ascomycetes
- AOB:
-
Ammonia-oxidizing bacteria
- DAA:
-
Days after application
- FDA:
-
Fluorescein diacetate
References
Adam G, Duncan H (2001) Development of a sensitive and rapid method for the measurement of total microbial activity using fluorescein diacetate (FDA) in a range of soils. Soil Biol Biochem 33:943–951
Bailey VL, Peacock AD, Smith JL, Bolton H Jr (2002) Relationships between soil microbial biomass determined by chloroform fumigation–extraction, substrate-induced respiration, and phospholipid fatty acid analysis. Soil Biol Biochem 34:1385–1389
Bending GD, Lincoln SD (1999) Characterization of volatile sulphur-containing compounds produced during decomposition of Brassica juncea tissues in soil. Soil Biol Biochem 31:695–703
Bending GD, Lincoln SD (2000) Inhibition of soil nitrifying bacteria communities and their activities by glucosinolate hydrolysis products. Soil Biol Biochem 32:1261–1269
Berbegal M, Garcia-Jimenez J, Armengol J (2008) Effect of cauliflower residue amendments and soil solarization on Verticillium wilt control in artichoke. Plant Dis 92:595–600
Brown PD, Morra MJ (2009) Brassicaceae tissues as inhibitors of nitrification in soil. J Agric Food Chem 57:7706–7711
Chang Y-J, Anwar Hussain AKM, Stephen JR, Mullens MD, White DC, Peackock A (2001) Impact of herbicides on the abundance and structure of indigenous β-subgroup ammonia-oxidizer communities in soil microcosms. Environ Toxicol Chem 20:2462–2468
Clegg CD, Lovell RDL, Hobbs PJ (2003) The impact of grassland management regime on the community structure of selected bacterial groups in soils. FEMS Microbiol Ecol 43:263–270
Cohen MF, Yamasaki H, Mazzola M (2005) Brassica napus seed meal soil amendment modifies microbial community structure, nitric oxide production and incidence of Rhizoctonia root rot. Soil Biol Biochem 37:1215–1227
Cohen MF, Mazzola M (2006) Resident bacteria, nitric oxide emission and particle size modulate the effect of Brassica napus seed meal on disease incited by Rhizoctonia solani and Pythium spp. Plant Soil 286:75–86
Collins HP, Alva A, Boydston RA, Cochran RI, Hamm PB, McGuire A, Riga E (2006) Soil microbial, fungal, and nematode responses to soil fumigation and cover crops under potato production. Biol Fertil Soils 42:247–257
De Cal A, Martinez-Treceno A, Salto T, Lopez-Aranda JM, Melgarejo P (2005) Effect of chemical fumigation on soil fungal communities in Spanish strawberry nurseries. Appl Soil Ecol 28:47–56
Fahey WJ, Zalcmann TA, Talalay P (2001) The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56:5–51
Federle TW, (1986) Microbial distribution in soil-new techniques. In: Megusar F, Gantar M (Eds) Perspectives in microbial ecology. Slovene Society for Microbiology, International Symposium on Microbial Ecology IV, pp 493–498
Friberg H, Edel-Herman V, Faivre C, Gautheron N, Fayolle L, Faloya V, Montfort F, Christian S (2009) Cause and duration of mustard incorporation effects on soil-borne plant pathogenic fungi. Soil Biol Biochem 41:205–2084
Grinhut T, Hadar Y, Chen Y (2007) Degradation and transformation of humic substances by saprotrophic fungi: processes and mechanisms. Fung Biol Rev 21:179–189
Ibekwe AM, Papiernik SK, Gan J, Yates SR, Yang C-H, Crowley DE (2001) Impact of fumigants on soil microbial communities. Appl Environ Microbiol 67:3245–3257
Ibekwe AM, Kennedy AC, Frohne PS, Papiernik SK, Yang CH, Crowley DE (2002) Microbial diversity along transect of agronomic zones. FEMS Microbiol Ecol 39:183–191
Infantino A, Aragona M, Brunneti A, Laroz E, Oliva A, Porta-Puglia A (2003) Molecular and physiological characterization of Italian isolates of Pyrenochaeta lycopersici. Mycol Res 107:707–716
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic, NY, pp 21–132
Karpouzas DG, Rousidou C, Papadopoulou KK, Bekris F, Zervakis G, Singh BK, Ehaliotis C (2009) Effect of continuous olive mill wastewater applications, in the presence and absence of N fertilization, on the structure of rhizopshere soil fungal communities. FEMS Microbiol Ecol 70:56–69
Karpouzas DG, Ntougias S, Iskidou E, Rousidou C, Papadopoulou KK, Zervakis G, Ehaliotis C (2010) Olive mill wastewaters affects the structure of soil bacterial communities. Appl Soil Ecol 45:101–111
Kaur A, Chaudhary A, Kaur A, Choudhary R, Kaushik R (2005) Phospholipid fatty acid—a bioindicator of environmental monitoring and assessment in soil ecosystem. Curr Sci 89:1103–1112
Kirkegaard JA, Sarwar M (1998) Biofumigation potential of brassicas—I. Variation in glucosinolate profiles of diverse field-grown brassicas. Plant Soil 201:71–89
Kirkegaard JA, Sarwar M, Wong PTW, Mead A, Howe G, Newell M (2000) Field studies on the biofumigation of take-all by Brassica break crops. Aust J Agric Res 51:445–456
Kowalchuk GA, Stephen JR, DeBoer W, Prosser JI, Embley TM, Woldendorp JW (1997) Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol 63:1489–1497
Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Ann Rev Microbiol 55:485–529
Kurola J, Salkinoja-Salonen M, Aarnio T, Hultman J, Romantschuk M (2005) Activity, diversity and population size of ammonia-oxidizing bacteria in oil-contaminated landfarming soil. FEMS Microbiol Lett 250:33–38
Larena I, Salazar O, González V, Julián MC, Rubio V (1999) Design of a primer for ribosomal DNA internal transcribed spacer with enhanced specificity for ascomycetes. J Biotechnol 75:187–194
Larkin PR, Honeycut WC (2006) Effects of different 3-year cropping systems on soil microbial communities and Rhizoctonia diseases of potato. Phytopathology 96:68–79
Larkin RP, Griffin ST (2007) Control of soilborne potato diseases using Brassica green manures. Crop Prot 26:1067–1077
Lundquist EJ, Jackson LE, Scow KM, Hsu C (1999) Changes in microbial biomass and community composition, and soil carbon and nitrogen pools after incorporation of rye into three California agricultural soils. Soil Biol Biochem 31:221–236
Macalady JL, Fuller ME, Scow KM (1998) Effects of metam sodium fumigation on soil microbial activity and community structure. J Environ Qual 27:54–63
Mahmood S, Freitag ET, Prosser IJ (2006) Comparison of PCR primer-based strategies for characterization of ammonia oxidizer communities in environmental samples. FEMS Microbiol Ecol 56:482–493
Manicci ML, Lazzeri L, Palmieri S (1997) In vitro fungitoxic activity of some glucosinolates and their enzyme-derived products toward plant pathogenic fungi. J Agric Food Chem 45:2768–2773
Matthiessen J, Shackleton MA (2005) Biofumigation: environmental impacts on the biological activity of diverse pure and plant-derived isothiocyanates. Pest Manag Sci 61:1043–1051
Matthiessen J, Kirkegaard J (2006) Biofumigation and enhanced biodegradation: opportunity and challenge in soilborne pest and disease management. Crit Rev Plant Sci 25:235–265
McCaig AE, Embley TM, Prosser JI (1994) Molecular analysis of enrichment cultures of marine ammonia oxidisers. FEMS Microbiol Lett 120:363–367
McCaig AE, Glover LA, Prosser JI (2001) Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturating gradient gel electrophoresis banding patterns. Appl Environ Microbiol 67:4554–4559
Mithen RF (2001) Glucosinolates and their degradation products. Adv Bot Res 35:213–262
Nannipieri P, Ascher J, Ceccherini MT, Landi L, Pietramellara G, Renella G (2003) Microbial diversity and soil functions. Eur J Soil Sci 54:655–670
Ntougias S, Ehaliotis C, Papadopoulou KK, Zervakis G (2006) Application of respiration and FDA hydrolysis measurements for estimating microbial activity during composting processes. Biol Fertil Soil 42:330–337
Ochiai N, Powelson ML, Crowe FJ, Dick RP (2008) Green manure effects on soil quality in relation to suppression of Verticillium wilt of potatoes. Biol Fertil Soil 44:1013–1023
Osono T, Fukasawa Y, Takeda H (2003) Roles of diverse fungi in larch needle-litter decomposition. Mycologia 95:820–826
Peacock AD, Mullen MD, Ringelberg DB, Tyler DD, Hedrick DB, Gale PM, White DC (2001) Soil microbial community responses to dairy manure or ammonium nitrate applications. Soil Biol Biochem 33:1011–1019
Philips CJ, Harris D, Dollhopf SL, Gross KL, Prosser JI, Eldor AP (2000) Effects of agronomic treatments on structure and function of ammonia-oxidizing communities. Appl Environ Microbiol 66:5410–5418
Prosser JI, Nicol GW (2008) Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10:2931–2941
Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops HP, Wagner M (2003) Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66:5368–5382
Rumberger A, Marschner P (2003) 2-Phenylethylisothiocyanate concentration and microbial community composition in the rhizosphere of canola. Soil Biol Biochem 35:445–452
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molec Biol Evol 4:406–425
Sánchez-Monedero MA, Mondini C, Cayuela L, Roig A, Contin M, De Nobili M (2008) Fluorescein diacetate hydrolysis, respiration and microbial biomass in freshly amended soils. Biol Fertil Soils 44:885–890
Sinha AP, Agnihotri VP, Singh V (1979) Effect of soil fumigation with vapam on the dynamics of soil microflora and their related biochemical activity. Plant Soil 53:89–98
Smith BJ, Kirkegaard JA (2002) In-vitro inhibition of soil microorganisms by 2-phenyl ethyl isothiocyanate. Plant Pathol 51:585–593
Son Y, Seo KY, Kim RH, Kim J (2006) Soil respiration and FDA hydrolysis following conversion of abandoned agricultural lands to natural vegetation in central Korean. J Plant Biol 49:231–236
Spyrou IM, Karpouzas DG, Menkissoglu-Spiroudi U (2009) Do botanical pesticides alter the structure of the soil microbial community. Microb Ecol 58:715–727
Tu C, Ristaino BJ, Hu S (2006) Soil microbial biomass and activity in organic tomato farming systems: effect of organic inputs and straw mulching. Soil Biol Biochem 38:247–255
Viebahn M, Veenman C, Wernars K, van Loon LC, Smit E, Bakker PAHM (2005) Assessment of differences in ascomycete communities in the rhizosphere of field-grown wheat and potato. FEMS Microbiol Ecol 53:245–253
Wang G, Xu Y, Jin J, Liu J, Zhang Q, Liu X (2009) Effect of soil type and soybean genotype on fungal community in soybean rhizosphere during reproductive growth stages. Plant Soil 144:135–144
Webster G, Embley TM, Prosser JI (2002) Grassland management regimens reduce small-scale heterogeneity and species diversity of [beta]-proteobacterial ammonia oxidizer populations. Appl Environ Microbiol 68:20–30
Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7:263–270
Zelenev VV, van Bruggen AHC, Semenov AM (2005) Short-term wavelike dynamics of bacterial populations in response to nutrient input from fresh plant residues. Microb Ecol 49:83–93
Acknowledgments
This work was financially supported by the Research Promotion Foundation in the Republic of Cyprus within the frame of the project BIOFUME 0506/07.
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 60 kb)
Rights and permissions
About this article
Cite this article
Omirou, M., Rousidou, C., Bekris, F. et al. The Impact of Biofumigation and Chemical Fumigation Methods on the Structure and Function of the Soil Microbial Community. Microb Ecol 61, 201–213 (2011). https://doi.org/10.1007/s00248-010-9740-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-010-9740-4