Abstract
Agricultural improvement (addition of fertilizers, liming) of seminatural acidic grasslands across Ireland and the UK has resulted in significant shifts in floristic composition, soil chemistry, and microbial community structure. Although several factors have been proposed as responsible for driving shifts in microbial communities, the exact causes of such changes are not well defined. Phosphate was added to grassland microcosms to investigate the effect on fungal and bacterial communities. Plant species typical of unimproved grasslands (Agrostis capillaris, Festuca ovina) and agriculturally improved grasslands (Lolium perenne) were grown, and phosphate was added 25 days after seed germination, with harvesting after a further 50 days. Phosphate addition significantly increased root biomass (p < 0.001) and shoot biomass (p < 0.05), soil pH (by 0.1 U), and microbial activity (by 5.33 mg triphenylformazan [TPF] g−1 soil; p < 0.001). A slight decrease (by 0.257 mg biomass-C g−1 soil; p < 0.05) in microbial biomass after phosphate addition was found. The presence of plant species significantly decreased soil pH (p < 0.05; by up to 0.2 U) and increased microbial activity (by up to 6.02 mg TPF g−1 soil) but had no significant effect on microbial biomass. Microbial communities were profiled using automated ribosomal intergenic spacer analysis. Multidimensional scaling plots and canonical correspondence analysis revealed that phosphate addition and its interactions with upland grassland plant species resulted in considerable changes in the fungal and bacterial communities of upland soil. The fungal community structure was significantly affected by both phosphate (R = 0.948) and plant species (R = 0.857), and the bacterial community structure was also significantly affected by phosphate (R = 0.758) and plant species (R = 0.753). Differences in microbial community structure following P addition were also revealed by similarity percentage analysis. These data suggest that phosphate application may be an important contributor to microbial community structural change during agricultural management of upland grasslands.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Intensification of upland pastures is common across northwestern Europe, with increased fertilization, liming, and grazing resulting in agriculturally improved grasslands [8]. The most abundant upland grassland formation on acidic soils in Ireland and the UK is a species-rich but low-yielding plant community, with Agrostis capillaris as the predominant grass species [44]. Regular fertilization and liming, often combined with intensification of grazing, drives such seminatural grasslands to a species-poor, high-yielding pasture dominated by Lolium perenne. There has been some concern regarding the reduction in floristic diversity as a result of intensification, particularly since little is known about the associated impacts on belowground microbial biodiversity and the consequences for biogeochemical and decompositional processes [25, 26]. Changes in physico-chemical variables such as pH and nutrient levels [29, 30], floristic community change [22, 24], changes in soil physical structure [27, 47], and the impacts of grazing animals [4, 45] have all been proposed as principal causes of shifts in grassland microbial community structure, although at present, it is still not fully understood which environmental factors influence microbial community change [51], with much evidence suggesting complex interactions between prevailing environmental variables. In acidic upland systems [7, 9, 10, 36], phosphate deficiency often limits ecosystem functioning, as much phosphate becomes bound to immobile aluminum or iron complexes; however, no study has yet focused on the responses of soil microbial communities to phosphate addition in grasslands.
Phosphorus (P) is essential for both plant and microbial growth and can be broadly categorized into either mineral or organic P in soils, with the distribution of P forms dependent on factors including soil type and pH, vegetation type, microbial activity, and fertilizer inputs [9, 14, 15]. Although plants obtain the majority of their P from the labile inorganic phosphate pool within soils [14], bioavailability of labile P is often very low, and in acidic grasslands, cycling of P is largely dependent upon microbial transformations including the breakdown of organic matter and solubilization of insoluble phosphates [54]. Phosphorus is routinely applied to agricultural lands to maximize productivity [60], primarily through addition of phosphates, which have high water solubilities and are thus readily taken up by plants and microorganisms. Deficiencies in P often limit both plant and microbial growth [54], so its availability is a major determinant of overall grassland productivity. It is thus likely that the roles of microbes in P cycling in unfertilized soil differ to that of improved pastures [40], and could be reflected by differences in microbial community composition. Additionally, the responses of plant species to phosphate addition could also have important consequences for the distribution of microbial populations. In particular, functionally significant groups, such as mycorrhizal fungi, are known to affect P supply to higher plants [33, 48, 50] and are likely to be prominent in P nutrition in unimproved grasslands.
Although a great deal is known about phosphorus cycling in soils in general, there is little information about how phosphate application impacts soil microbial community structures. In grasslands, interactions between phosphate addition, plant species, and changes in microbial community structure may be critical in understanding grassland intensification processes. This study tests the hypothesis that phosphate addition to an unimproved grassland soil will alter fungal and bacterial community structures. We also aimed to investigate interactions between plant species and phosphate addition on the microbial community structure.
Materials and Methods
Soil
Soil was collected from an area of unimproved Nardo-Galion grassland at Longhill, Kilmacanogue, County Wicklow, Ireland in July 2004 (National Grid Reference O 218 124). The site is 300 m above sea level and consists of peaty podzolic soil formed over granite/quartzite bedrock [19], with an annual rainfall between 2,000 and 2,800 mm [39]. Soil (pH 4.5) from the Ah horizon (5–15 cm depth) was removed, sieved to less than 4 mm, and used for microcosms.
Microcosms
Eighty grams (dry mass) of fresh soil was weighed into black polyvinylchloride pots (40 mm diameter, 110 mm height), which had been pierced to allow free drainage of water. Pots were planted with 20–25 surface-sterilized (2% sodium hypochlorite for 5 min) seeds (Emorsgate Seeds, Kings Lynn, UK) of A. capillaris, L. perenne, or Festuca ovina. A set of control pots was left unplanted. Eight pots of each of the three plant species and unplanted soil were prepared, allowing for four pots of each plant treatment to be treated with phosphate and the remaining four as untreated controls. Microcosms were incubated in a greenhouse in a randomized block design for 75 days from seed germination, from August 4 to October 17, 2004. Water content was maintained at 35% by the addition of distilled water as necessary. Twenty-five days after seed germination, a solution of K2PO4 was added to four pots from both planted and unplanted microcosms at a field application rate of 25 kg P ha−1, while the remaining control pots were treated with the same volume of deionized water. Microcosms were destructively sampled 50 days after phosphate addition. All soil was separated from plant roots and considered rhizosphere soil due to the small dimensions of the microcosm pots. Plants were divided into roots and shoots, dried at 70°C for 7 days, and weighed. Soil was removed from pots, sieved to less than 4 mm, and stored at 4°C for 48 h for pH, microbial activity, and microbial biomass analyses and at −20°C for molecular analysis. Within treatments, each pot was considered a replicate, with one analysis per pot carried out.
pH, Total Microbial Biomass, and Microbial Activity
Microcosm soil samples (one soil sample per replicate pot; four replicates per treatment) were analyzed for pH water according to Sparks et al. [49]. Soil microbial biomass carbon was measured by substrate-induced respiration, using a modification of the methods of Anderson and Domsch [2] and West and Sparling [57]. Total microbial activity was measured as triphenylformazan dehydrogenase activity [1] and was determined based on a modification of the method of Thalmann [52]. Methods for microbial biomass and activity were used as previously described by Brodie et al. [10].
Total Soil DNA Extraction and Purification
Total soil deoxyribonucleic acid (DNA; one DNA extraction per replicate pot; four replicates per treatment) was extracted as described by Brodie et al. [10]. Soil (0.5 g) was added to tubes containing glass and zirconia beads, to which a hexadecyltrimethylammonium bromide extraction buffer was added. Tubes were incubated at 70°C for 10 min, and phenol/chloroform/isoamylalcohol (25:24:1) was added. Tubes were then shaken in a Hybaid Ribolyser at 5.5 m s−1 for 30 s. Following bead beating, tubes were centrifuged, and the aqueous layer was removed and purified twice with chloroform/isoamylalcohol (24:1) and then incubated with lysozyme solution (100 mg ml−1) for 1 h at 37°C. The aqueous layer was removed and further purified using a High-pure PCR Product Clean-Up Kit (Roche Diagnostics GmbH, Penzberg, Germany), according to the manufacturer’s instructions. DNA was eluted in a final volume of 50 μl and was suitable for PCR amplification.
Fungal and bacterial community fingerprinting was carried out by automated ribosomal intergenic spacer analysis (ARISA). For fungal polymerase chain reaction (PCR) amplification, internal transcribed spacer (ITS) primer set ITS1-F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS4 (5′-TCCTCCGCTTATTGATA TGC-3′) was used [18, 59]. Amplified sequences were in the range 300–1,100 bp. Thermocycling conditions for fungal ITS amplification were as follows: a hot start at 94°C for 2 min (one cycle), after which 2.5 U Taq DNA polymerase was added; 94°C for 3 min (one cycle); 94°C for 1 min, 55°C for 30 s, 72°C for 1 min (25 cycles); and 72°C for 5 min (one cycle). The 16S–23S intergenic spacer region (ITS) from the bacterial ribosomal ribonucleic acid (rRNA) operon was amplified using forward primer S-D-Bact-1522-b-S-20 (eubacterial rRNA small subunit, 5′-TGCGGCTGGATCCCCTCCTT-3′) and the reverse primer L-D-Bact-132-a-A-18 (bacterial rRNA large subunit, 5′-CCGGGTTTCCCCAT TCGG-3′) [41], with amplicons in the range 200–1,200 bp. Thermocycling conditions for bacterial intergenic spacer region amplification were as follows: a hot start at 94°C for 2 min (one cycle), after which 2.5 U Taq DNA polymerase was added; 94°C for 3 min (one cycle); 94°C for 1 min, 55°C for 30 s, 72°C for 1 min (25 cycles); and 72°C for 5 min (one cycle). Both fungal and bacterial forward primers were labeled with Beckman Coulter fluorescent dye D4 (Proligo). PCR amplifications were performed according to Kennedy et al. [29, 30]. PCR products were first visualized on a 1.2% agarose gel, then purified using a High-pure PCR Product Clean Up Kit (Roche Diagnostics GmbH), and eluted in 50 μl of sterile water heated to 50°C. Aliquots (1 μl) were mixed with 38.4 μl deionized formamide, 0.2 μl of Beckman Coulter size standard 600 (ranging from 60 to 600 bp in intervals of 50 bp), and 0.4 μl of a custom-made marker (ranging from 600 to 1,200 bp in intervals of 20 bp and 1,000–1,200 bp in intervals of 50 bp; Bioventures, Murfreesboro, TN, USA). Size standards were labeled with Beckman Coulter dye D1. During DNA extraction, PCR amplification, and ARISA profiling, several samples were randomly selected for replicate analysis at each stage, to ensure reproducibility of the methods used.
Intergenic spacer lengths were determined by electrophoresis using a Beckman Coulter (CEQ8000) automated sequencer. Run conditions were 60°C separation temperature, 4 kV voltage, and 120 min separation time to allow for the separation of the larger fragments. Analysis of spacer profiles was performed using the Beckman Coulter CEQ 8000 fragment analysis software, algorithm v 2.1.3. A quadratic polynomial model rather than the recommended cubic model was used for size standard calibration as this resulted in improved correlation between expected and actual size standard fragment sizes, particularly for amplicons in the range 400–1,200 bp. Amplicons with fluorescence greater than 1% of the total peak fluorescence were selected for inclusion in the data set, based on relative abundance data. Amplicons that differed by less than 0.5 bp in different profiles were considered identical, and only those that were present in at least two out of four replicates were included in the analyses, thus reducing the risk of including false amplicons [16]. Following these standardization procedures, each amplicon was considered a unique operational taxonomic unit (OTU).
Statistical Analysis
Data sets for pH, root and shoot biomass, microbial biomass, microbial activity, and number of fungal and bacterial OTUs were analyzed by analysis of variance (ANOVA) using Genstat version 6. The significance level was set at p < 0.05. Multivariate statistical analysis was performed on ARISA profiles using Primer 6 (Primer-E) and Canoco for Windows v 4.5.
Similarities of ARISA OTU profiles (presence/absence transformed) for each treatment were examined using the Bray–Curtis index as a measure of similarity. Microbial communities were analyzed using Primer software, which created nonmetric multidimensional scaling (MDS) plots for fungal and bacterial data sets. MDS is an ordination technique that represents relationships inherent in multivariate data sets and attempts to preserve the ranked order of the similarity of two communities as an inverse function of the distance between the points representing those communities on the plot [32]. The communities with the highest similarity in the data set are represented by points that plot close together, with the least similar communities represented on the plot by points which are further apart. The degree to which the plot matches the similarity matrix can be judged by looking at stress (Kruskal’s stress) with values less than 0.15 representing good ordination plots [12]. MDS was performed using 30 random starting configurations of sample points, and in all cases, two-dimensional solutions are presented. Points on MDS plots represent an average of the four replicates used.
Analysis of similarity (ANOSIM) was used to investigate the effects of phosphate and plant species on fungal and bacterial community structure. ANOSIM was performed on Bray–Curtis similarity matrices. Comparisons of mean distances within treatments were used to calculate the ANOSIM R-statistic (R), with R = 1 indicating that the populations were dissimilar, and R = 0 indicating that populations are random. Permutational multivariate ANOVA (PERMANOVA) was also carried out to investigate the effects of phosphate and plant species on microbial community profiles, as this procedure can determine whether the interaction between phosphate and plant species was significant.
Initial analysis by detrended correspondence analysis revealed that the data exhibited a unimodal, rather than linear, response to the environmental variables (plant species and phosphate addition), so canonical correspondence analysis (CCA; Canoco for Windows v 4.5) was used. On the ordination plots, OTUs found to be significantly affected by one of the environmental factors or their interactions (according to ANOVA on individual OTUs) were numbered on the basis of their base pair length (OTUs not significantly affected are not numbered). A Monte Carlo permutation test based on 199 random permutations was used to test the null hypothesis that fungal and bacterial profiles were unrelated to environmental variables.
Similarity percentage (SIMPER) analysis was used to investigate if phosphate addition resulted in differences in fungal and bacterial community composition, based on the relative percentage abundance of OTUs present. Data sets for fungal and bacterial species within phosphate treatments (plus/no phosphate) were ranked by most abundant OTUs. Species comprising the top 30% most abundant species within fungal and bacterial profiles were selected as representatives of the respective communities and were used for SIMPER analysis.
Results
Soil pH values from microcosms harvested after the application of phosphate are shown in Table 1. ANOVA revealed a slight but significant (p < 0.001) increase in soil pH when phosphate was added. pH was significantly (p < 0.05) higher in unplanted pots, with F. ovina-planted pots having the lowest pH at 5.3. The addition of phosphate affected plant biomass, with significantly higher root and shoot biomass (p < 0.001 and p < 0.05, respectively) recorded in plants where phosphate was added. Root and shoot biomass differed between plant species type, with L. perenne showing the greatest plant biomass, followed by F. ovina and A. capillaris. Microcosm soils were assessed for microbial activity and biomass-C using dehydrogenase- and substrate-induced respiration measurements, respectively (Table 1). The microbial activity was significantly (p < 0.001) higher in soils with phosphate added, compared to those with no phosphate. The presence of plant species also had a significant effect on microbial activity, with planted pots exhibiting greater microbial activity than the unplanted pots. On the other hand, the microbial biomass was significantly (p < 0.05) lower in pots with added phosphate but was unaffected by the presence of plant species. The extent of bacterial and fungal diversity was estimated by counting the number of OTUs present within profiles (Table 1). The addition of phosphate did not significantly alter the number of fungal or bacterial OTUs, although plant species did significantly (p < 0.05) change the bacterial OTU number, with F. ovina having the greatest number of bacterial OTUs. The fungal OTU number was not significantly affected by plant species alone. Table 2 shows soil, plant, and microbial measurements as affected by interactions between phosphate and plant species. Interactions between phosphate and plant species impacted pH, root and shoot biomass, and fungal OTU number.
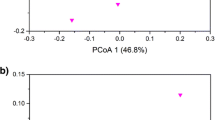
Community profiles consisted of individual ARISA amplicons present and their relative abundances, with each amplicon indicating a sequence polymorphism or OTU. A total of 536 unique bacterial OTUs and 231 fungal OTUs were detected across all samples, and multivariate statistical analysis was performed to determine which factors were contributing to variation within the microcosms. Figure 1a,b shows the MDS ordinations for fungal and bacterial communities, respectively, as affected by plant species and phosphate addition. Figure 1a indicates that the presence of plant species changes the fungal community composition, with replicates clustering together slightly apart from other groups of replicates. Phosphate addition also changed the fungal community composition, with the filled symbols (phosphate added) separating from the unfilled (no phosphate) on the MDS plot. The most notable difference in fungal species composition could be seen in F. ovina-planted pots, where the addition of phosphate resulted in marked differences in the fungal community composition. Cluster overlays were added at 60% and 80% similarity levels, which indicate some similarities between compositions across treatments. For example, all treatments except A. capillaris/no P and F. ovina/no P are clustered in the center of the plot, with an 80% similarity, perhaps indicating that phosphate addition has a greater effect on the fungal communities associated with A. capillaris- and F. ovina-planted soils, due to their separation from the centered cluster. Bacterial communities are represented in Fig. 1b, again showing changes between different treatments. For unplanted pots and pots planted with A. capillaris or L. perenne, there were distinct differences in bacterial community composition between pots to which phosphate was added and pots that had no phosphate, whereas F. ovina-planted pots did not show the same degree of separation, indicating the presence of a similar bacterial community composition, irrespective of phosphate treatment. Cluster overlays are again overlaid on Fig. 1b, indicating similarity at 60% or 80% levels.
MDS plots showing effects of phosphate and plant species on microbial community compositions across different plant treatments. a Fungi; b bacteria. Dashed circles represent 60% resemblance; Unbroken circles represent 80% resemblance. Upright triangle, unplanted soil; square, A. capillaris soil; inverted triangle, L. perenne soil; diamond, F. ovina soil. Filled symbols, phosphate added; empty symbols, no phosphate added
ANOSIM was used to determine the effects of phosphate and plant species on fungal and bacterial communities (Table 3). The fungal community structure was significantly affected by both phosphate (R = 0.948) and plant species (R = 0.857), and the bacterial community structure was also affected by phosphate (R = 0.758) and plant species (R = 0.753), with a significance level at p < 0.005. The larger R values for fungal communities indicated that phosphate and plant species had a greater effect on fungal, rather than bacterial, community structure. PERMANOVA pairwise comparisons for both fungal and bacterial community structures showed significant interactions between phosphate and plant species (p < 0.001), indicating that phosphate addition plus plant species resulted in different fungal and bacterial community profiles.
CCA was also used to further explain data for fungal and bacterial communities. For both fungal and bacterial analyses, the species–environment correlations were all greater than 0.9, indicating a strong correlation between species data and environmental variables. The Monte Carlo significance tests indicated that for both data sets, both the first axis and all axes combined explained a significant amount of variation (p value <0.002). CCA ordinations are presented in Fig. 2a,b (fungal and bacterial species data, respectively). On the CCA plots, environmental variables are represented as arrows, with the relative length of arrows indicating the relative importance of that variable with the angle in between arrows indicating the degree to which these two factors were correlated. The most abundant OTUs (representing around 80% of total abundance for both fungal and bacterial species sets) are represented by triangles, with the significantly affected OTUs labeled according to their length (bp). The relative position of OTUs in relation to the environmental arrows indicates how a given OTU is affected by an environmental factor or variable. Both ordinations include data from the unplanted control. The CCA ordination plot for fungal species data (Fig. 2a) showed that phosphate, A. capillaris, and L. perenne were the most influential in determining the orientation of fungal OTUs on the ordination. Figure 2a also highlights the importance of interactions between variables, with the L. perenne × phosphate arrow almost as influential as the L. perenne arrow. The arrow for F. ovina was considerably shorter, with few fungal OTUs positively correlated with this plant species. Most fungal OTUs were positively correlated with phosphate, A. capillaris, L. perenne, and their interactions. The CCA ordination for bacterial OTUs (Fig. 2b) differed considerably from the fungal CCA plot. In this case, the environmental factors that were most influential were phosphate and F. ovina (represented by the longest arrows). The interactions between A. capillaris × phosphate and L. perenne × phosphate were more important than A. capillaris or L. perenne alone. Some bacterial OTUs were negatively orientated with respect to the phosphate arrow, indicating that phosphate addition had a negative effect on these particular OTUs. Most bacterial OTUs were positively associated with phosphate, plant species, and their interactions.
SIMPER analysis revealed different fungal and bacterial communities associated with phosphate addition (Fig. 3). For fungal communities, phosphate treatment resulted in markedly different community composition (of the top ranked 30% most abundant species), with no common species present across phosphate treatments (Fig. 3a). For bacterial communities, the only species (of the top ranked 30% most abundant species) common to both phosphate and no phosphate treatments was OTU 506 (Fig. 3b).
Discussion
This study indicated that the addition of phosphate to grassland microcosms significantly influenced both fungal and bacterial communities and that the type of plant species present is significant in determining fungal and bacterial responses to phosphate addition. These observations lend support to other studies where it has been suggested that microbial community structure in grassland systems is controlled by several linked factors, rather than any one environmental parameter [38].
Previous studies regarding the effects of agricultural management on grasslands have shown that the improvement of acidic grasslands (addition of fertilizers, liming) leads to a noticeable increase in soil pH [4, 10], often by up to 2 pH units. The pH rise recorded as a result of phosphate addition in this experiment was substantially smaller, however, and although the difference was found to be significant, it is unlikely that this finding can be regarded as ecologically significant in terms of overall grassland microbial ecology, as most microorganisms will have a wider pH range. The increase in microbial activity as a result of phosphate addition may reflect P limitations in natural acidic grasslands. The soil used for this study was removed from an unfertilized acidic pasture, which was low in available nutrients, including P [10, 11]. The addition of an available nutrient source such as phosphate is perhaps likely to stimulate soil microbial activity in soil that has previously been P deprived. In a study carried out by Treseder and Allen [53], the addition of N and P to nutrient-limited soils resulted in a significant increase in microbial activity but not if applied to previously fertilized soils. Additionally, Lovell et al. [36] noted that after an input of N fertilizer, microbial N and C measurements were substantially higher in previously unfertilized soil when compared to soil that had been fertilized.
Plant species alone also had a significant effect on microbial activity, which is perhaps not surprising, as exudates and deposits from plant roots are known to stimulate microbial growth [3]. The increase in microbial activity observed when plant species were present was therefore likely the result of greater rhizosphere microbial activity [22], possibly including mycorrhizal fungi [25, 42], which form intimate associations with the majority of plant roots and are integral components of the plant–soil interface. As the method used in this study for determining microbial activity did not discriminate between bacterial and fungal populations, it was difficult to assess whether both bacterial and fungal populations responded positively to the addition of phosphate or whether the increase in microbial activity reflected the response of just one. The addition of phosphate in this study resulted in a slight decrease in microbial biomass. In acidic upland soils, where nutrients are limiting, fungal biomass is known to be higher than that for bacteria [5, 6, 11] and is much less responsive to environmental change in comparison to bacterial communities [29, 30]. Grayston et al. [23] also found that fungi dominate in unmanaged grasslands—such as upland acidic pastures—while the main contributor to microbial biomass in managed grasslands was bacteria. While no conclusive evidence can be presented from this study regarding the distribution of biomass in these soils, phosphate addition may result in a decreased fungal biomass fraction. Fungal communities can be affected by P manipulation [35] and may perhaps be due to a negative growth response by mycorrhizal fungal communities in particular, which are known to proliferate in P-limited soils. The addition of phosphate may therefore result in a markedly different fungal community profile in the presence of a labile P source such as that used in this study. Fungal and bacterial communities were assessed using ARISA, a sensitive community-fingerprinting technique, which has recently become a widely used method for microbial community profiling in a range of environments, including soil [29], aquatic environments [61], and weathered surfaces [20]. Care must be taken in interpreting results derived from ARISA (a PCR-based approach) due to species differences in rRNA gene copy number and biases generated during DNA extraction and PCR amplification [13, 17]. On the other hand, recent studies on community profiling found that terminal restriction fragment length polymorphism, a similar method to ARISA, gave a quantitative view of microbial communities not affected by the PCR bias [37]. Although some potential bias may have been introduced during PCR, by preferentially amplifying shorter amplicons, it is thought that a higher diversity of organisms have ITS regions in the range 150–600 bp [43]; thereby, a higher number of amplicons in this range may have been revealed. Using ARISA profiles, it was possible to estimate fungal and bacterial diversity, using OTU numbers (Tables 1 and 2), which, when used in relative terms as in this study, can be a useful tool to compare any possible effects of treatment on microbial diversity within the confines of the experimental microcosms. Of particular interest from this study were the differences in fungal OTU numbers when interactions between phosphate and plant species occurred, with plant species responded remarkably differently to phosphate addition in terms of their associated fungal communities.
Changes in plant species have often been singled out as important drivers of microbial community composition during the intensification of acidic upland grasslands [7, 22–24]. Unplanted soil should differ from planted soil in terms of microbial communities, mostly due to the effects of plant growth patterns [34] and rhizodeposition [31, 58], through altering rhizosphere conditions, thereby influencing localized microbial populations. Several plant species, including the grasses used in this study, have been shown to demonstrate clear arbuscular mycorrhizal fungal host–plant preferences [21, 46, 55, 56], which may explain some of the differences observed in the fungal community composition. Although the fungal community analysis used in this study did not focus specifically on mycorrhizal populations, the differences in fungal community compositions in our study may represent a selection for distinct mycorrhizal associations by the plant species present [28] and perhaps also reflect changes brought about by plant-specific responses to the presence or absence of available phosphate within their environment.
It is interesting that there appears to be some division in the microbial community structure between grassland species characteristic of seminatural acidic grasslands (both A. capillaris and F. ovina) and L. perenne, which dominates improved pastures. MDS similarity clusters revealed a high degree of similarity (80%) between all treatments except A. capillaris/no P and F. ovina/no P. While there was some degree of similarity (60%) between all treatments, there were nevertheless some treatment effects driving compositional differences within these microcosms. In general, CCA demonstrated that the phosphate was an important factor governing overall microbial community structure in these microcosms, while plant species exerted differential influences over fungal and bacterial communities. A. capillaris is the predominant grass species in unimproved acidic grasslands, which support significant fungal biomass, and may be a reason why A. capillaris exerts such an influence over fungal community structure. Conversely, F. ovina, which is also a grass species typical of unimproved pastures, appears to have little influence over fungal community structure, according to the CCA ordination. F. ovina, on the other hand, appears to be very influential in governing the bacterial community structure. The orientation of bacterial and fungal OTUs in relation to the environmental factors (Fig. 2) highlights the complexity of the microbial community dynamics in changing environmental conditions, with distinct responses of subgroups of the community to different external factors. Individual responses to one or more environmental factors may reflect different physiological or functional characteristics of fungal and bacterial populations. SIMPER analysis confirmed that phosphate addition to grassland soil influenced both fungal and bacterial communities, notably through the markedly different community compositions when comparing phosphate addition to the control treatment (no phosphate), implying that phosphate can select for specific microbial populations in soil.
In conclusion, while it appears that phosphate alone was influential in governing the structures of both fungal and bacterial communities in upland grassland soils, it was not possible to generalize any phosphate-induced effects on microbial community structure. It is clear, however, that no single factor can be isolated as a determinant of the microbial community structure; rather, a number of interacting factors are responsible. Fungal and bacterial communities differed in their responses to environmental parameters and their interactions, emphasizing the difficulties faced when attempting to generalize the impacts of agricultural improvement processes on the soil microbial community structure.
References
Alef K, Nannipieri P (1995) Methods in applied soil microbiology and biochemistry. Academic, London, UK
Anderson JPE, Domsch KH (1978) A physiological method for the quantitative measurement of microbial biomass in soils. Soil Biol Biochem 10:215–221
Atlas RM, Bartha R (1997) Microbial ecology: fundamentals and applications. Addison-Wesley, Reading, MA
Bardgett RD, Frankland JC, Whittaker JB (1993) The effects of agricultural management on the soil biota of some upland grasslands. Agric Ecosyst Environ 45:25–45
Bardgett RD, Hobbs PJ, Frostegard A (1996) Changes in soil fungal:bacterial biomass ratios following reductions in the intensity of management of an upland grassland. Biol Fertil Soils 22:261–264
Bardgett RD, McAlister E (1999) The measurement of soil fungal:bacterial biomass ratios as an indicator of ecosystem self-regulation in temperate meadow grasslands. Biol Fertil Soils 29:282–290
Bardgett RD, Mawdsley JL, Edwards S, Hobbs PJ, Rodwell JS, Davies WJ (1999) Plant species and nitrogen effects on soil biological properties of temperate upland grasslands. Funct Ecol 13:650–660
Blackstock TH, Rimes CA, Stevens DP, Jefferson RG, Robertson HG, Mackintosh J, Hopkins JJ (1999) The extent of semi-natural grassland communities in lowland England and Wales: a review of conservation surveys 1978–96. Grass Forage Sci 54:1–18
Bolan NS (1991) A critical review on the role of mycorrhizal fungi in the uptake of phosphorus by plants. Plant Soil 134:189–207
Brodie E, Edwards S, Clipson N (2002) Bacterial community dynamics across a floristic gradient in a temperate upland grassland ecosystem. Microb Ecol 44:260–270
Brodie E, Edwards S, Clipson N (2003) Soil fungal community structure in a temperate upland grassland soil. FEMS Microbiol Ecol 45:105–114
Clarke J (1993) Non-parametric multivariate analysis of changes in community structure. Aust J Ecol 18:117–143
Crosby LD, Criddle CS (2003) Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. Bio Techniques 34:790–802
Curtin D, Smillie GW (1984) Influence of liming on soluble and labile P in fertilised soil. Commun Soil Sci Plant Ann 15:177–188
Daly K, Jeffrey D, Tunney H (2001) The effect of soil type on phosphorus sorption and capacity and desorption dynamics in Irish grassland soils. Soil Use Manage 17:12–20
Dunbar J, Ticknor LO, Kuske CR (2001) Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl Environ Microbiol 67:190–197
Egert M, Friedrich MW (2003) Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction length polymorphism analysis of microbial community structure. Appl Environ Microbiol 69:2555–2562
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Geological Survey of Ireland (2000) http://www.gsi.ie
Gleeson DB, Mc Dermott F, Clipson N (2006) Structural diversity of bacterial communities in a heavy metal mineralized granite outcrop. Environ Microbiol 8:383–393
Gollotte A, van Tuinen D, Atkinson D (2004) Diversity of arbuscular mycorrhizal fungi colonising roots of the grass species Agrostis capillaris and Lolium perenne in a field experiment. Mycorrhiza 14:111–117
Grayston SJ, Wang S, Campbell C, Edwards A (1998) Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem 30:369–378
Grayston SJ, Griffith GS, Mawdsley JL, Campbell CD, Bardgett RD (2001) Accounting for variability in soil microbial communities of temperate upland grassland ecosystems. Soil Biol Biochem 33:533–551
Grayston SJ, Campbell CD, Bardgett RD, Mawdsley JL, Clegg CD, Ritz K, Griffiths BS, Rodwell JS, Edwards SJ, Davies WJ (2004) Assessing shifts in microbial community structure across a range of grasslands of differing management intensity using CLPP, PLFA and community DNA techniques. Appl Soil Ecol 25:63–84
Green BH (1990) Agricultural intensification and the loss of habitat, species and amenity in British grasslands: a review of historical change and assessment of future prospects. Grass Forage Sci 45:365–372
Grime JP (1998) Benefits of plant diversity to ecosystems: immediate, filter and founder effects. J Ecol 86:902–910
Ibekwe AM, Kennedy AM, Frohne PS, Papiernik SK, Yang CH, Crowley DE (2002) Microbial diversity along a transect of agronomic zones. FEMS Microbiol Ecol 26:151–163
Johnson D, Vandenkoornhuyse PJ, Leake JR, Gilbert L, Booth RE, Grime JP, Young JPW, Read DJ (2003) Plant communities affect arbuscular mycorrhizal fungal diversity and community composition in grassland microcosms. New Phytol 161:503–515
Kennedy N, Connolly J, Clipson N (2004) Impact of lime, nitrogen and plant species on fungal community structure in grassland microcosms. Environ Microbiol 7:780–788
Kennedy NM, Gleeson DE, Connolly J, Clipson NJW (2005) Seasonal and management influences on bacterial community structure in an upland grassland soil. FEMS Microbiol Ecol 53:329–337
Klein DA, Paschke MW (2000) A soil microbial community structural-functional index: the microscopy-based total/active fungal/bacterial (TA/AFB) biovolumes ratio. Appl Soil Ecol 14:257–268
Kruskal JB (1964) Multidimensional scaling by optimizing goodness of fit to a non-metric hypothesis. Psychometrika 29:1–27
Kucey RMN, Janzen HH, Leggett ME (1989) Microbiologically mediated increases in plant-available phosphorus. In: Brady NC (ed) Advances in agronomy. Academic, New York, pp 199–228
Kuske CR, Ticknor LO, Miller ME, Dunbar JM, Davis JA, Barns JA, Belnap J (2002) Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl Environ Microbiol 68:1854–1863
Lima JA, Nahas E, Gomes AC (1996) Microbial populations and activities in sewage sludge and phosphate fertilizer-amended soil. Appl Soil Ecol 4:75–82
Lovell RD, Jarvis SC, Bardgett RD (1995) Soil microbial biomass and activity in long-term grassland: effects of management changes. Soil Biol Biochem 27:969–975
Lueders T, Friedrich MW (2003) Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl Environ Microbiol 69:320–326
Marschner P, Yang CH, Lieberei R, Crowley DE (2001) Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem 33:1437–1445
Met Eireann (2003) http://www.met.ie
Molla MAZ, Chowdhury AA (1984) Microbial mineralization of organic phosphate in soil. Plant and Soil 78:393–399
Normand P, Ponsonnet C, Nesme X, Neyra M, Simonet P (1996) ITS analysis of prokaryotes. In: Akkermans AD, van Elsas JD, de Bruijn EI (eds) Molecular microbial ecology manual. Kluwer, Amsterdam, The Netherlands
Paterson E (2003) Importance of rhizodeposition in the coupling of plant and microbial productivity. Eur J Soil Sci 54:741–750
Ranjard L, Poly F, Lata JC, Mougel C, Thioulouse J, Nazaret S (2001) Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints, biological and methodological variability. Appl Environ Microbiol 67:4479–4487
Rodwell JS (1992) British plant communities: grasslands and montane communities. Cambridge University Press, Cambridge
Rooney DC, Kennedy N, Deering L, Gleeson D, Clipson N (2006) Effect of sheep urine deposition on bacterial community structure in an upland acidic grassland soil. Appl Environ Microbiol 72(11):7231–7237
Scheublin TR, Ridgway KP, Young JP, van der Heijden MGA (2004) Nonlegumes, legumes, and root nodules harbor different arbuscular mycorrhizal fungal communities. Appl Environ Microbiol 70:6240–6246
Sessitch A, Weilharter A, Gerzabek MH, Kirchmann H, Kandeler E (2006) Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl Environ Microbiol 67:4215–4224
Smith SE, Read DJ (eds) (1997) Mycorrhizal symbiosis. Academic, San Diego, CA
Sparks DL, Hlemke PA, Loeppert RH, Soltanpour PN, Tabatabai MA (eds) (1996) Methods of soil analysis: chemical methods, part 3. Soil Science Society of America, Madison, WI
Stevenson FJ, Cole MA (1999) Cycles of soil: carbon, nitrogen, phosphorus, sulfur, micronutrients. Wiley, New York
Stoate C, Boatman ND, Borralho RJ, Carvalho CR, Snoo GR, Eden P (2001) Ecological impacts of arable intensification in Europe. J Environ Manag 63:337–365
Thalmann A (1968) Zur methodik ber Bestimmung der dehydrogenaseactivitat im Boden mittels triphenyltetrazoliumchlorid (TTC). Landwirtsch Forsch 21:249–258
Treseder KK, Allen MF (2002) Direct nitrogen and phosphorus limitation of arbuscular mycorrhizal fungi: a model and field test. New Phytol 155:507–515
Troeh FR, Thompson LM (eds) (1993) Soils and soil fertility. Oxford University Press, New York
Vandenkoornhuyse P, Husband R, Daniell TJ, Watson IJ, Duck JM, Fitter AH, Young JPW (2002) Arbuscular mycorrhizal community composition associated with two plant species in a grassland ecosystem. Mol Ecol 11:1555–1564
Vandenkoornhuyse P, Ridgway KP, Watson IJ, Fitter AH, Young JPW (2003) Co-existing grass species have distinctive arbuscular mycorrhizal communities. Mol Ecol 12:3085–3095
West AW, Starling GP (1986) Modifications to the substrate-induced respiration method to permit measurement of microbial biomass in soils of differing water contents. J Microbiol Methods 5:177–189
Westover KM, Kennedy AC, Kelley SE (1997) Patterns of rhizosphere microbial community structure associated with co-occurring plant species. J Ecol 85:863–873
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, New York, pp 315–322
Withers PJA, Edwards AC, Foy RH (2001) Phosphorus cycling in UK agriculture and implications for phosphorus loss from soil. Soil Use Manage 17:139–149
Yannerell AC, Kent AD, Lauster GH, Kratz TK, Triplett EW (2003) Temporal patterns in bacterial communities in three temperate lakes of different trophic status. Microb Ecol 46:391–405
Acknowledgments
This project was funded by the Environmental Protection Agency grant 2002-PHD2-16 under the Environmental Research Technological Development and Innovation (ERTDI) program. The authors would like to thank Deirdre Gleeson for statistical guidance and Mary Murphy for technical assistance and acknowledge valuable discussions with Nabla Kennedy and Lorraine Muckian. The authors also thank three anonymous reviewers for helpful comments and advice on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rooney, D.C., Clipson, N.J.W. Phosphate Addition and Plant Species Alters Microbial Community Structure in Acidic Upland Grassland Soil. Microb Ecol 57, 4–13 (2009). https://doi.org/10.1007/s00248-008-9399-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-008-9399-2