Abstract
Despite the general consensus that fertilizer is the most important driver of the evolution of soil microbial communities, the specific effects of long-term fertilizer use on microbial communities remain unclear. Here, we collected soil samples from fertilized (NPK) and unfertilized (NF) plots in a subtropical farmland in southwestern China. NPK plots were consistently treated with chemical fertilizer (nitrogen, phosphorous, and potassium) for the 20 years; NF plots were left unfertilized for the same period. To explore the effects of long-term fertilizer use on soil microbial community structure, microbial community composition in the topsoil was assessed using the bacterial 16S rRNA gene and the full-length fungal ITS1 gene. In conjunction, we measured various soil chemical properties. We found that metrics associated with soil fertility (i.e., total nitrogen, total phosphorus, total potassium, available nitrogen, available phosphorus, and available potassium) were significantly greater in the NPK samples as compared to the NF samples, but that soil pH was significantly lower. We also found that long-term fertilizer use reshaped soil microbial community composition but did not alter community α-diversity. Notably, the bacterial phyla Nitrospirae and Planctomycetes and the fungal phylum Ascomycota were closely associated with the NPK plot, influenced by soil organic matter, total nitrogen, available potassium, and available phosphorus, while the bacterial phyla Bacteroidetes and Chloroflexi and the fungal phyla Glomeromycota and Basidiomycota were closely associated with the NF plot, influenced by soil pH. Our results provide long-term data clarifying the microbial regulation mechanisms underlying the response of farmland soil to long-term fertilizer use in subtropical China.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
INTRODUCTION
The soil microbial community serves as an important resource for agricultural production in the era of climate change for two main reasons [12]: First, as the most important decomposer of organic matter, the soil microbiome participates in biogeochemical nutrient cycling, which helps to regulate climate change [2]. Second, soil microbial communities are involved in various processes of nitrogen transformation, including biological nitrogen fixation, nitrification, and denitrification [26]. These transformation processes improve nutrient use efficiency and soil enzyme activity levels, thereby affecting crop growth [23] and helping to reduce the negative environmental effects of nitrogen application [10].
Generally, inorganic fertilizers, especially nitrogen, phosphorus, and potassium fertilizers, not only maintain or increase crop yields, but also directly or indirectly change the chemical, physical, and biological properties of the soil [3, 6]. Thus, these fertilizers are believed to have significant long-term effects on soil quality and productivity [1, 7]. Some studies have suggested that the continuous overuse of inorganic fertilizers may lead to a decline in soil quality and production capacity [11]. For example, fertilizer overuse leads to soil acidification and hardening [4] and may also alter soil chemical properties in a way that is harmful to plants [24]. Moreover, excessive fertilizer use changes the composition of plant communities and decreases biodiversity [5, 27]. However, other studies have indicated that inorganic fertilizers have both positive and negative effects [4] or no noticeable effects on soil quality and productive capacity. The inconsistencies among these previous reports demonstrate the importance of long-term fertilization experiments, which can be used to better evaluate the effects of agronomic management measures on soil quality and productivity.
Fertilizer use strongly affects the structure of the soil microbial community, impacting not only soil nutrients but also specific bacterial and fungal taxa [28]. In a previous study, increase of crop yield under balanced fertilizations might due to the increase of soil microbial function traits, which is associated with decreasing influence of environmental filtering [34]. A growing body of work focused on agricultural sustainability has explored changes in soil microorganismal composition to clarify the effects of fertilizer use over longer time scales. For example, the long-term use of nitrogen-containing chemical fertilizers increased soil acidification, decreased soil fertility, and significantly altered the soil bacterial community, whereas the long-term use of organic fertilizers, in conjunction with fallow management, improved soil quality and maintained bacterial diversity [33]. Furthermore, the increased application of nitrogen and phosphorus fertilizers changed the composition of the soil microbial community, reducing the abundance of phospholipid fatty acid (PLFA)-producing gram-positive bacteria such as actinomycetes and significantly increasing the fungal/bacterial PLFA ratio [8]. Similarly, phosphate fertilizer may be a key factor controlling the number and diversity of microbial colonies in dry red soils in China [6]. In addition, Wang et al. [29] showed that the application of urea significantly reduced the relative abundance of saprophytes and symbiotic bacteria. Thus, the relationship between long-term fertilization management and soil microbial community function is critical for ecological health and sustainable agricultural production. However, previous studies have mainly focused on temperate regions. Few reports of the effects of long-term fertilizer use on soil microorganisms in subtropical regions are available.
Here, we explored the long-term effects of fertilizer use on soil microorganisms in a subtropical monsoon climate, and assessed soil quality and production capacity. We conducted a 20-year field experiment in which soils were subjected to different fertilization regimes. We then assessed bacterial and fungal communities in each topsoil sample using 16S rRNA and ITS1. We aimed to characterize changes in the soil microbial community after long-term fertilizer use and to determine the relationship between soil microbial community composition and soil chemical properties.
OBJECTS AND METHODS
Field site and experimental design. The monitoring site, which was established in 1998, is located in Jingdong county, Yunnan Province, southwestern China (24°29′ N, 100°49′ E). This site has a subtropical monsoon climate, an altitude of 1152 m, a mean annual precipitation of 1110 mm, a mean annual average temperature of 18.3°C, a mean annual sunshine duration of 2131 h, and a frost-free period of 354 days (data from 1998–2019). The field soil is Red soil (Chinese Classification), Typic Hapludult (US Soil Taxonomy) or Ferric Acrisol (WRB Classification). In 1998, prior to experimentation, the topsoil (0–20 cm deep) had a pH of 6.0 and contained 0.68 g/kg nitrogen (N), 1.35 g/kg phosphorus (P2O5), and 9.80 g/kg potassium (K2O). This corresponded to 135.60 mg/kg, 90.36 mg/kg, and 81.00 mg/kg of available nitrogen, phosphorus, and potassium, respectively. Each was 6 meters wide and 23 meters long, with total planting area of 133 m2. Between 1998 and 2018, both plots were cultivated using a double cropping system: winter wheat (Triticum aestivum L.) and summer maize (Zea mays L.) was planted, respectively.
One of the established plots (selected randomly) was left unfertilized (NF, the control), and the other plot was fertilized using a conventional fertilization regime (NPK). Nitrogen was applied to the NPK plot as urea (N ≥ 46%), phosphorus was applied as super phosphate (P2O5 ≥ 16%), and potassium was applied as K2SO4 (K2O ≥ 50%). Nitrogen, phosphorus, and potassium usage was 300, 225, and 108 kg/ha, respectively, for winter wheat, and 375, 225, and 108 kg/ha, respectively, for summer maize.
Sample collection and chemical analysis. Soil samples were collected from both plots in October 2019, after the maize harvest but before soil plowing in preparation for the subsequent wheat crop. Ten soil cores (0–20 cm deep) were collected from each plot using an auger (5 cm in diameter by 20 cm long). And the pooled samples were then sieved through a 2-mm mesh. One part was stored at –80°C for subsequent soil microbe DNA extraction.
In the sieved, air-dried samples we measured several soil properties [3, 13]. In brief, SOM was measured using the K2Cr2O7–H2O oxidation method, and TN was measured using an automatic Kjeldahl apparatus (K-355, Buchi, Switzerland). AN was extracted with 2 M KCl and determined using a Skalar SANplus Segmented Flow Analyzer (Skalar Analytic B.V., De Breda, The Netherlands). AP was extracted by sodium bicarbonate and determined using the molybdenum-antimony anti-colorimetric method. AK was extracted by CH3COONH4 and determined on a flame photometer with a detection limit of 0.004 mM (Model 410, Sherwood, UK). Soil pH was measured using a pH meter (Mettler FE28, Mettler Toledo, Switzerland) in a slurry composed of 1 part soil to 5 parts carbon dioxide-free water.
DNA extraction and sequencing. Genomic DNA were extracted from 0.25 g of each homogenized soil sample using a PowerSoil DNA Isolation Kit (MO BIO Laboratories: Carlsbad, CA, USA), following the manufacturer’s instructions. We amplified the V1–V9 region of the bacterial 16S rRNA gene using the primers 27F (5′-AGRGTTTGATYNTGGCTCAG-3′) and 1492R (5′-TASGGHTACCTTGTTASGACTT-3′) [14], and the full-length fungal ITS1 gene using the primers Euk-A (5′AACCTGGTTGATCCTGCCAGT-3′) and Euk-B (5′-GATCCTTCTGCAGGTTCACCTAC-3′) [24]. Equal molar amounts of the PCR products from each sample were mixed and pair-end sequenced using the PacBio Sequel platform (Sequel II, Pacific Biosciences, USA).
Statistical analyses. Soil chemical properties data were analyzed with SPSS software (version 25, IBM, Chicago, IL, USA). The remaining high-quality clean sequences were processed and analyzed using the QIIME v.1.8.0 pipeline. We taxonomically identified 16S OTUs (Operational taxonomic units) clusters using the Silva database release 132 (http://www.arb-silva.de), and we identified the 18S OTU clusters using the ITS1 : Unite database release 8.1 (http://unite.ut.ee/index.php). The PCoAs, Unweighted pair-group method with arithmetic mean (UPGMA) trees, linear discriminant analysis (LDA) effect size (LEfSe) analysis, and redundancy analyses (RDAs) were selected using the bioEnv function in the vegan R package. All sequencing data were deposited in the National Center for Biotechnology Information (NCBI) GenBank database under accession number PRJNA657484.
RESULTS
Effects of long-term fertilizer use on soil chemical properties. Our results indicated that soil chemical properties were significantly affected by long-term fertilizer use. In the NPK plot, SOM, TN, AN, AP, and AK levels were significantly higher than those in the NF plot (P < 0.001), while pH level was significantly lower (P < 0.001; Table 1). In addition, the annual yield of crops in NPK was significantly higher than that of NF treatment (P < 0.01). Among them, the crop yields of NPK treatment than that of NF treatment in 2007, 2016, 2017, 2018 and 2019 were higher 164.2, 60.3, 72.6, 250.0 and 214.9%, respectively.
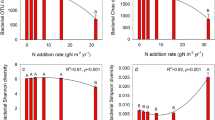
Bacterial and fungal alpha diversity. The NF sequences clustered into 138–161 OTUs (149 on average), while the NPK sequences clustered into 138–146 OTUs (144 on average). Coverage for all samples was >95%, and OTU rarefaction curves for the sequencing libraries showed that saturation had plateaued (Figs. 1a, 1b). This reflects the reliability of our results. The Shannon, Simpson, Ace, and Chao 1 indices indicated that bacterial and fungal α-diversity was similar between the NF and NPK samples (Table 2), suggesting that long-term fertilizer use had little effect on soil microbial diversity.
Soil microbial community composition. In both the NF and the NPK samples, the bacterial community was dominated by four phyla: Proteobacteria, Planctomycetes, Bacteroidetes, and Verrucomicrobia (Fig. 2a). However, the relative abundances of Planctomycetes, Acidobacteria, Nitrospirae, and Firmicutes were higher in the NPK samples as compared to the NF samples, while the relative abundances of Proteobacteria, Bacteroidetes, Gemmatimonadetes, Chloroflexi, and Actinobacteria were lower in the NPK samples as compared to the NF samples; the relative abundance of the Verrucomicrobia was similar between the NPK and NF samples (Fig. 2a). The bacterial species Tepidisphaera mucosa, Pseudolabrys taiwanensis, Acidobacteria sp., Xanthomonadaceae sp., Nostocoida limicola and Gemmata_like.str. were more abundant in the NPK samples than in the NF samples (Fig. 2b).
In both the NPK and NF samples, the fungal community was dominated by the phylum Ascomycota (>65% relative abundance; Fig. 2c). However, the relative abundances of Basidiomycota and Glomeromycota were greater in the NPK samples than in the NF samples, while the relative abundances of Ascomycota and Mucoromycota were lower in the NPK samples than in the NF samples; the relative abundance of the Chytridiomycota was similar between the NPK and NF samples (Fig. 2c). The fungal species Chalara heteroderae, Cercophora samala, Podospora bulbillosa, Penicillium raperi, and Thanatephorus cucumeris were significantly more abundant in the NPK samples than in the NF samples (Fig. 2d). These results suggested that long-term fertilizer use altered the structures of both the bacterial and the fungal communities in the topsoil.
Bacterial and fungal beta diversity. Principal coordinate analysis (PCoAs) showed that fungal and bacterial communities were more similar among soil samples subjected to the same fertilizer treatment regime; that is, there were noticeable distinctions between the NF and NPK microbial communities (Figs. 3a, 3b). In the bacterial analysis, principal component 1 (PC1) and principal component 2 (PC2) explained 68.68 and 16.65%, respectively, of the variation in the data (Fig. 3a). In the fungal analysis, PC1 and PC2 explained 41.33 and 23.59%, respectively, of the variation in the data (Fig. 3b). Consistent with the PCoAs results, UPGMA hierarchical clustering analyses of bacterial community composition (Fig. 3c) and fungal community composition (Fig. 3d) showed that communities were more similar within treatment type than between treatment type. Thus, our results indicated that long-term fertilizer use affected soil microbial beta diversity.
Linear discriminant analysis effect size (LEfSe) results for the microbial community LDA cladograms indicated that there were significant shifts in bacterial and fungal community composition between the NPK and NF samples (P < 0.01, LDA score >3.0; Fig. 4). In total, 105 bacterial taxa were differentially enriched between the two treatments: the relative abundances of 57 taxa were greater in the NF samples, while the relative abundances of 48 taxa were greater in the NPK samples. For example, the relative abundances of Planctomycetes, Nitrospirae, Proteobacteria, and Acidobacteria were significantly greater in the NPK samples, and the relative abundances of Bacteroidetes and Chloroflexi were significantly greater in the NF samples (Fig. 4a). Similarly, 84 fungal taxa were differentially enriched between the NPK and NF samples: the relative abundances of 25 fungal taxa, in particular Ascomycetes, were significantly greater in the NPK samples, while the relative abundances of 59 fungal taxa were significantly greater in the NF samples, including Basidiomycotes, Glomeromycotes, and Mortierellomycotes (Fig. 4b).
LEfSe cladograms showing the (a) bacterial and (b) fungal taxa that were significantly differentially abundant (LDA score ≥ 3.0, P ≤ 0.05) between the soil samples from the NPK and NF plots. The cladograms are organized in concentric circles as follows: domain (innermost), phylum, class, order, family, genus, and species (outermost).
Associations between soil properties and soil microbial community structure. The RDA_CCA indicated that five soil properties (SOM, TN, pH, AK, and AP) significantly affected the phyla level structures of the bacterial and fungal communities at the study site (Figs. 5a, 5b). Of the bacterial phyla, Nitrospirae and Planctomycetes were closely associated with the NPK samples and were most influenced by SOM, TN, AK, and AP, while Bacteroidetes, Chloroflexi, and Gemmatimonadetes were closely associated with the NF samples and were most influenced by soil pH. Of the fungal phyla, Ascomycota was closely associated with the NPK samples and was most influenced by SOM, TN, AK, and AP, while Glomeromycota and Basidiomycota were closely associated with the NF samples and were most influenced by soil pH.
DISCUSSION
Soils harbor a huge diversity of microorganisms that participate in various bio-geochemistry cycles and influence soil fertility [31]. However, the complexity and heterogeneity of soil habitats and soil microbial communities limit our understanding of the relationships between soil microbes and soil health. Here, we found that long-term fertilizer use reshaped soil bacterial and fungal community structures, but had little effect on the alpha and beta diversity of the overall soil microbial community.
The factors driving soil microbiota composition may be very complex, possible including both environmental factors (e.g., temporal variation, spatial heterogeneity, soil nutrient composition, and soil disturbances due to agricultural activity) [13] and biotic factors (e.g., reproductive behavior, dispersal ability, competition, niche differentiation, extinction, genetic diversity, and plant growth periods [22, 30]. Previous studies have shown that soil pH is the principal predictor of bacterial community structure at the ecosystem level [35]. In addition, long-term fertilization was shown to decrease bacterial diversity, primarily because long-term nitrogen input decreases soil pH, and soil microbial diversity decreases with soil pH [20, 32]. Consistent with this, the relative abundances of the dominant bacterial and fungal phyla (Proteobacteria, Bacteroidetes, and Ascomycota) were lower in the NPK samples as compared to the NF samples, as was soil pH (Figs. 3a, 3b).
Furthermore, soils with high pH values and abundant plant mulch tend to have greater microbial diversity, while acidic soils have lower microbial diversity [18]. These pH-linked differences may further impact plant-microbial interactions and plant productivity. However, although pH values were lower in the NPK samples as compared to the NF samples (Table 1), this difference appeared to have little effect on the diversity of the soil microbial community. It may be that our results differ from previous studies of the effects of pH on soil microbial communities because we focused specifically on subtropical soils in southwest China; that is, this discrepancy may be due to spatio-temporal and soil heterogeneities. Alternatively, after 20 years of exposure, the microbial community may have evolved a new homeostasis in the topsoil at the study site. Thus, it remains unclear whether long-term fertilization has a significant effect on soil microbial diversity.
Alternatively, eutrophic bacteria are abundant than oligotrophic bacteria in environments with high nutrient input [20]. Consistent with this, our results showed that long-term high nutrient input (i.e., in the NPK-fertilized plot) increased the abundance of eutrophic bacterial phyla including Planctomycetes, Acidobacteria, and Nitrospirae, while lack of fertilization (i.e., in the NF plot) promoted the growth of oligotrophic bacteria such as Proteobacteria, Bacteroidetes, and Gemmatimonadetes (Fig. S1) [17]. Thus, these taxa may be important competitors after long-term environmental filtration.
The principle of oligotrophic-eutrophication states that nutrient enrichment usually increases the growth of eutrophic bacteria but decreases the growth of oligotrophic bacteria; conversely, oligotrophic bacterial growth continues under conditions of malnutrition [21]. Thus, eutrophic bacteria are abundant than oligotrophic bacteria in environments with high nutrient input [13]. Consistent with this, our results showed that long-term high nutrient input (i.e., in the NPK-fertilized plot) increased the abundance of eutrophic bacterial phyla including Planctomycetes, Acidobacteria, and Nitrospirae, while lack of fertilization (i.e., in the NF plot) promoted the growth of oligotrophic bacteria such as Proteobacteria, Bacteroidetes, and Gemmatimonadetes (Fig. 3a). Thus, these taxa may be important competitors after long-term environmental filtration [34].
Ultimately, long-term fertilization may reduce the heterogeneity of nutrient utilization, allowing species that are adapted to higher nutrient levels to colonize and dominate the fertilized area [19]. Alternatively, fertilization may amplify initial differences unevenly, resulting in a more divergent spatial pattern of species distributions [16, 29]. Finally, the response of soil microorganisms to fertilizers may vary depending on environmental conditions [9]. Therefore, further multiscale and integrative analysis are required to assess the complexities of soil microbial communities and surrounding resources at different points in the crop growth cycle.
CONCLUSIONS
Our 20-year fertilizer experiment showed that NPK significantly increased indices associated with soil fertility (i.e., TN, TP, TK, AN, AP and AK), but decreased soil pH. In addition, long-term fertilizer use reshaped the soil microbial community without changing α-diversity. The bacterial phyla Nitrospirae and Planctomycetes and the fungal phylum Ascomycota were closely associated with the NPK plot, influenced by SOM, TN, AK, and AP, while the bacterial phyla Bacteroidetes and Chloroflexi and the fungal phyla Glomeromycota and Basidiomycota were closely associated with the NF plot, influenced by soil pH. Therefore, our results provide insights into the response of soil microorganisms to long-term fertilizer use in subtropical regions. Therefore, our results provide insights into the response of soil microorganisms to long-term fertilizer use in subtropical regions.
REFERENCES
D. F. Acton and L. J. Gregorich, Health of Our Soils: Toward Sustainable Agriculture in Canada (Minister of Supply and Services Canada, Ottawa, 1995).
R. S. Ali, E. Kandeler, S. Marhan, M. S. Demyan, J. Ingwersen, R. Mirzaeitalarposhti, F. Rasche, G. Cadisch, and C. Poll, “Controls on microbially regulated soil organic carbon decomposition at the regional scale,” Soil Biol. Biochem. 118, 59–68 (2018). https://doi.org/10.1016/j.soilbio.2017.12.007
A. Belay, A. Claassens, and F. Wehner, “Effect of direct nitrogen and potassium and residual phosphorus fertilizers on soil chemical properties, microbial components and maize yield under long-term crop rotation,” Biol. Fertil. Soil 35 (6), 420–427 (2002). https://doi.org/10.1007/s00374-002-0489-x
R. Bobbink, K. Hicks, J. Galloway, T. Spranger, R. Alkemade, M. Ashmore, et al., “Global assessment of nitrogen deposition effects on terrestrial plant diversity: a synthesis,” Ecol. Appl. 20, 30–59 (2010). https://doi.org/10.1890/08-1140.1
C. M. Clark, E. E. Cleland, S. L. Collins, J. E. Fargione, L. Gough, K. L. Gross, et al., “Environmental and plant community determinants of species loss following nitrogen enrichment,” Ecol. Lett. 10, 596–607 (2007). https://doi.org/10.1111/j.1461-0248.2007.01053.x
V. Chaudhry, A. Rehman, A. Mishra, P. S. Chauhan, and C. S. Nautiyal, “Changes in bacterial community structure of agricultural land due to long-term organic and chemical amendments,” Microb. Ecol. 64 (2), 450–460 (2012). https://doi.org/10.1007/s00248-012-0025-y
Y. L. Chen, Y. Cao, and S. Liu, “Effects of long-term mineral fertilizer application on soil nutrients, yield, and fungal community composition,” Eurasian Soil Sci. 54, 597–604 (2021). https://doi.org/10.1134/S1064229321040049
X. B. Dai, H. M. Wang, and X. L. Fu, “Soil microbial community composition and its role in carbon mineralization in long-term fertilization paddy soils,” Sci. Total Environ. 580 (15), 556–563 (2015). https://doi.org/10.1016/j.scitotenv.2016.11.212
W. Dan, Q. Yang, J. Z. Zhang, S. Wang, X. L. Chen, X. L. Zhang, and W. Q. Li, “Bacterial community structure and diversity in a black soil as affected by long-term fertilization,” Pedosphere 18 (5), 582–592 (2018).https://doi.org/10.1016/S1002-0160(08)60052-1
G. Dantas and M. A. Sommer, “How to fight back against antibiotic resistance,” Am. Sci. 102 (1), 42–51 (2014). https://doi.org/10.1511/2014.106.42
J. W. Doran, M. Sarrantonio, and M. A. Liebig, “Soil health and sustainability,” Adv. Agron. 56 (8), 1–54 (1996). https://doi.org/10.1016/S0065-2113(08)60178-9
P. G. Falkowski, T. Fenchel, and E. F. Delong, “The microbial engines that drive Earth’s biogeochemical cycles,” Science 20 (5879), 1034–1039 (2008). https://doi.org/10.1126/science.1153213
R. K. Gangwar, M. Makádi, I. Demeter, A. Táncsics, M. Cserháti, G. Várbíró, J. Singh, Á. Csorba, M. Fuchs, E. Michéli, and T. Szegi, “Soil chemical and biological properties of salt affected soils under different land use practices in Hungary and India,” Eurasian Soil Sci. 54, 1007–1018 (2021). https://doi.org/10.1134/S1064229321070048
C. Hong, Y. Si, Y. Xing, and Y. Li, “Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas,” Environ. Sci. Pollut. Res 22 (14), 10788–10799 (2015). https://doi.org/10.1007/s11356-015-4186-3
A. L. Koch, “Oligotrophs versus copiotrophs,” BioEssays 23 (7), 657–661 (2001). https://doi.org/10.1002/bies.1091
Y. Liang, L. Wu, I. M. Clark, K. Xue, Y. F. Yang, and J. D. van Nostrand, “Over 150 years of long-term fertilization alters spatial scaling of microbial biodiversity,” mBio 6 (2), 15–22 (2015). https://doi.org/10.1128/mBio.00240-15
J. W. Leff, S. E. Jones, S. M. Prober, A. Barberán, E. T. Borer, J. L. Firn, et al., “Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe,” Proc. Natl. Acad. Sci. U.S.A. 112 (35), 10967–10972 (2015). https://doi.org/10.1073/pnas.1508382112
H. Liu, W. Xiong, R. Zhang, X. N. Hang, D. S. Wang, and R. Li, “Continuous application of different organic additives can suppress tomato disease by inducing the healthy rhizospheric microbiota through alterations to the bulk soil microflora,” Plant Soil 423 (2), 229–240 (2018). https://doi.org/10.1007/s11104-017-3504-6
L. L. Liu, X. Q. Huang, J. B. Zhang, Z. C. Cai, K. Jiang, and Y. Y. Chang, “Deciphering the combined effect and relative importance of soil and plant traits on the development of rhizosphere microbial communities,” Soil Biol. Biochem. 148, 107909 (2020). https://doi.org/10.1016/j.soilbio.2020.107909
W. Liu, L. Jiang, S Yang, Z. Wang, R. Tian, Z. Y. Peng, et al., “Critical transition of soil bacterial diversity and composition triggered by nitrogen enrichment,” Ecology 101 (8), e03053 (2020). https://doi.org/10.1002/ecy.3053
Q. Ma, Y. Wen, D. Wang, X. Sun, P. W. Hill, A. Macdonald, et al., “Farmyard manure applications stimulate soil carbon and nitrogen cycling by boosting microbial biomass rather than changing its community composition,” Soil Biol. Biochem. 144, 107760 (2020). https://doi.org/10.1016/j.soilbio.2020.107760
J. B. Martiny, B. J. Bohannan, J. H. Brown, R. K. Colwell, J. A. Fuhrman, and J. L. Green, “Microbial biogeography: putting microorganisms on the map,” Nat. Rev. Microbiol. 4 (2), 102–112 (2006). https://doi.org/10.1038/nrmicro1341
P. C. Nel, R. O. Barnard, R. E. Steynberg, J. M. De Beer, and H. T. Groeneveld, “Trends in maize grain yields in a long-term fertilizer trial,” Field Crops Res. 47 (1), 53–64 (1996). https://doi.org/10.1016/0378-4290(96)00006-8
A. Orgiazzi, E. Lumini, R. H. R. Nilsson, H. Nilsson, M. Girlanda, A. Vizzini, P. Bonfante, and V. Bianciotto, “Unravelling soil fungal communities from different Mediterranean land-use backgrounds,” PLoS One 7 (4), e34847 (2012). https://doi.org/10.1371/journal.pone.0034847
C. J. Stevens, P. Manning, L. J. L. van den Berg, M. C. C. de Graaf, G. W. W. Wamelink, A. W. Boxman, et al., “Ecosystem responses to reduced and oxidised nitrogen inputs in European terrestrial habitats,” Environ. Pollut. 159, 665–676 (2011). https://doi.org/10.1016/j.envpol.2010.12.008
P. O. Sorensen, A. C. Finzi, M. A. Giasson, A. B. Reimann, R. S. Demott, and P. H. Templer, “Winter soil freeze-thaw cycles lead to reductions in soil microbial biomass and activity not compensated for by soil warming,” Soil Biol. Biochem. 116, 39–47 (2018). https://doi.org/10.1016/j.soilbio.2017.09.026
K. N. Suding, S. L. Collins, L. Gough, C. Clark, E. E. Cleland, K. L. Gross, et al., “Functional- and abundance-based mechanisms explain diversity loss due to N fertilization,” Proc. Natl. Acad. Sci. U.S.A. 102, 4387–4392 (2005). https://doi.org/10.1073/pnas.0408648102
J. F. Toljander, C. Juan, T. A. Santos-González, A. Tehler, and R. D. Finlay, “Community analysis of arbuscular mycorrhizal fungi and bacteria in the maize mycorrhizosphere in a long-term fertilization trial,” FEMS Microbiol. Ecol. 65 (2), 323–338 (2010). https://doi.org/10.1111/j.1574-6941.2008.00512.x
J. C, Wang, G. Rhodes, Q. W. Huang, and Q. R. Shen, “Plant growth stages and fertilization regimes drive soil fungal community compositions in a wheat-rice rotation system,” Biol. Fertil. Soils 54, 731–742 (2018). https://doi.org/10.1007/s00374-018-1295-4
J. J. Wang, Y. Zhang, B. X. Wang, X. D. Yang, and J. Shen, “Patterns of elevational beta diversity in micro- and macroorganisms,” Global Ecol. Biogeogr. 21 (7), 743–750 (2012). https://doi.org/10.1111/j.1466-8238.2011.00718.x
C. Xiong, Y. G. Zhu, J. T. Wang, S. Brajesh, L.L. Han, and L. L. Shen, “Host selection shapes crop microbiome assembly and network complexity,” New Phytol. 229, 1091–1104 (2020). https://doi.org/10.1111/nph.16890
Y. M. Xiong, L. Ruan, Z. Q. Li, S. P. Dai, Y. J. Pan, Y. Qiao, Y. Q. Qi and L. Hu, “Changes in metabolic functions of the soil microbial community in Eucalyptus plantations along an urban-rural gradient,” Eurasian Soil Sci. 54, 1912–1920 (2021). https://doi.org/10.1134/S1064229321120152
W. B. Xun, J. Zhao, C. Xue, G. S. Zhang, W. Ran, B. Wang, et al., “Significant alteration of soil bacterial communities and organic carbon decomposition by different long-term fertilization management conditions of extremely low-productivity arable soil in South China,” Environ. Microbiol. 18 (6), 1907–1917 (2016). https://doi.org/10.1111/1462-2920.13098
Y. Yu, M. Wu, E. Petropoulos, J. Zhang, J. Nie, Y. Liao, Z. Li, X. Lin, and Y. Feng, “Responses of paddy soil bacterial community assembly to different long-term fertilizations in southeast China,” Sci. Total Environ. 656, 625–633 (2019) https://doi.org/10.1016/j.scitotenv.2018.11.359
Y. H. Yun, B. M. Wang, X. Xiang, J. Zhou, X. Qiu, Y. Duan, and A. S. Engel, “The relationship between pH and bacterial communities in a single karst ecosystem and its implication for soil acidification,” Front. Microbiol. 116, 39–47 (2016). https://doi.org/10.3389/fmicb.2016.01955
ACKNOWLEDGMENTS
We are grateful to the staff of the Jingdong Soil and Fertilizer Station in Yunnan Province. This study was funded by the Key Research and Development Program of Yunnan (2018BB015), the Basic Application Research Project of Yunnan Province (2019YD096), and Technology Research and Development Program of ZunYi (HZ[2021]324).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors state no conflict of interest with respect to the research, authorship, and/or publication of this article.
Supplementary Information
Rights and permissions
About this article
Cite this article
Zhao, G.R., Fan, Z.W., An, T.X. et al. Long-Term Fertilizer Use Altered Soil Microbial Community Structure but Not α-Diversity in Subtropical Southwestern China. Eurasian Soil Sc. 55, 1116–1125 (2022). https://doi.org/10.1134/S1064229322080178
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1064229322080178