Abstract
Purpose
Natural language processing (NLP) can be used for automatic flagging of radiology reports. We assessed deep learning models for classifying non-English head CT reports.
Methods
We retrospectively collected head CT reports (2011–2018). Reports were signed in Hebrew. Emergency department (ED) reports of adult patients from January to February for each year (2013–2018) were manually labeled. All other reports were used to pre-train an embedding layer. We explored two use cases: (1) general labeling use case, in which reports were labeled as normal vs. pathological; (2) specific labeling use case, in which reports were labeled as with and without intra-cranial hemorrhage. We tested long short-term memory (LSTM) and LSTM-attention (LSTM-ATN) networks for classifying reports. We also evaluated the improvement of adding Word2Vec word embedding. Deep learning models were compared with a bag-of-words (BOW) model.
Results
We retrieved 176,988 head CT reports for pre-training. We manually labeled 7784 reports as normal (46.3%) or pathological (53.7%), and 7.1% with intra-cranial hemorrhage. For the general labeling, LSTM-ATN-Word2Vec showed the best results (AUC = 0.967 ± 0.006, accuracy 90.8% ± 0.01). For the specific labeling, all methods showed similar accuracies between 95.0 and 95.9%. Both LSTM-ATN-Word2Vec and BOW had the highest AUC (0.970).
Conclusion
For a general use case, word embedding using a large cohort of non-English head CT reports and ATN improves NLP performance. For a more specific task, BOW and deep learning showed similar results. Models should be explored and tailored to the NLP task.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Hospital emergency departments (ED) are increasingly being overwhelmed [1]. Non-contrast head computed tomography (CT) is the most frequently performed CT scan in the ED [2,3,4]. Flagging of reports could help prioritize patient care. Radiological reports are usually stored as unstructured free-text. This makes the extraction of data difficult [5,6,7,8,9,10]. NLP algorithms are designed to structure such free-text. The role of NLP in structuring electronic medical records (EMR) has been previously discussed in the medical literature [11,12,13,14,15]. In radiology, NLP has various applications: flagging and categorization of imaging findings, patient prioritization, generation of imaging protocols and research [9, 10].
Different algorithms have been developed for NLP tasks. Previous studies have reported very good results for rule-based NLP systems and, in this respect, they may be considered highly successful, but they are difficult to develop and maintain [16].
BOW is a known machine learning NLP model which has shown promising results within various radiological domains, including head CT [6].
In recent years, deep learning algorithms have made a large impact on industry and academia. These algorithms have presented tremendous abilities in image analysis [17]. The number of publications employing deep learning techniques for medical images is exponentially increasing [18,19,20,21,22,23]. These algorithms are already being used for commercial applications, such as automatic analysis of head CT scans [24]. It should be noted that computer vision deep learning models require large research cohorts for training. Flagging of radiology reports using NLP can help create large research cohorts for computer vision tasks.
Recently, deep learning methods have also shown promising results in performing various NLP tasks [25,26,27,28,29]. These methods include, among others, LSTM which is an algorithm designed to analyze sequential data such as sentences [30]; ATN algorithms which have recently shown state-of-the-art results for NLP tasks [31]; and word embedding which is a technique for representing words in a multi-dimensional space [32]. These technological innovations make deep learning feasible for medical tasks other than image analysis.
In this study, we aimed to assess the potential of using state-of-the-art deep learning models for classifying non-English head CT reports.
Materials and methods
Study design
This retrospective study was granted an institutional review board (IRB) approval.
We obtained head CT reports of all patients who underwent a head CT in our hospital. The reports were performed in the ED, inpatient, and outpatient settings between January 2011 and December 2018. All reports were signed by board-certified radiologists in a non-English language (Hebrew).
Reports of adult ED patients from January to February for each year between 2013 and 2018 were manually labeled. The rest of the reports were used to pre-train an embedding layer.
We evaluated deep learning models (LSTM, ATN) with and without pre-training a word embedding layer. We also compared deep learning models with a BOW model.
Data preprocessing
Reports were manually labeled by two residents (YB and SS) supervised by a senior radiologist (EK). Each report was labeled by one resident. The supervising radiologist adjusted the labeling in 341 reports.
We explored two use cases: (1) general labeling use case, in which reports were labeled as normal vs. pathological; (2) specific labeling use case, in which reports were labeled as with and without intra-cranial hemorrhage.
General labeling use case
Pathological reports were defined as those containing acute or chronic findings: brain infarction, dense artery sign, intra-cranial hemorrhage, brain or bone space-occupying lesion, brain edema, pneumocephalus, fractures, sinusitis or post-surgical findings, hydrocephalus.
The following findings were labeled as normal: vascular calcifications, old lacunar infarcts, chronic white-matter ischemic changes, and other incidental findings deemed as having no clinical significance.
Specific labeling use case
Reports were labeled as either with intra-cranial hemorrhage (intra- or extra-axial) or without intra-cranial hemorrhage.
Text cleaning included removing punctuations and low-frequency words (appearing in less than three reports). This was done separately for each training fold. We also limited texts to 1500 characters.
Data exploration—word importance
We evaluated the association of words with pathological labeling. We used the mutual information formula to measure the joint mutual information between the pathology class (C) and the word (W). Chi-square test evaluated the significance (p < 0.05) of the associations.
NLP models
Experiments were written in Python (version 3.7). The deep learning models were written using the Keras library (version 2.2.4) and TensorFlow module (version 1.13.1) as backend. The Word2Vec model was written using the Gensim library (version 3.8.1). The BOW model was written using the scikit-learn package (version 0.19.1). Computations were done on an Intel i7 CPU and two NVIDIA GeForce GTX 1080Ti GPUs.
Models were evaluated using tenfold cross-validation. In each experiment, nine folds were used for training and one held-out fold was used for testing. The results of the ten experiments were averaged.
For the specific use case, we have up-sampled the positive cases to a rate of 1:1. Up-sampling was done exclusively in the training folds.
We used the area under the curve (AUC), accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) metrics. A default probability of 0.5 was used to determine the measures other than AUC.
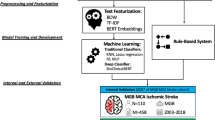
Student’s t test evaluated statistical differences between models’ metrics. Figure 1 shows a schematic representation of the deep learning models in the study.
BOW model
In the BOW model, reports are represented as an unordered collection (bag) of its words. Then, a classifier (such as logistic regression) is trained to classify the paragraphs based on the frequency of words in the bags.
We employed term frequency–inverse document frequency (tf-idf) approach on the BOW collections. tf-idf balances between how important a word is to a document (tf), to how common it is in the corpus (idf). The tf-idf formula for each word (w) in one document is:
The tf-idf has been computed separately for each training fold.
Word2Vec model
Word embedding represents words as multi-dimensional vectors. In the embedding process, the algorithm tries to map relations between words. By that, similar words will have similar vectors. For instance, for head CT reports, words such as hematoma and bleed will have similar vectors. The most common word embedding algorithms are Word2vec and Glove. We employed the Word2Vec model.
LSTM model
LSTM are networks that take chronological order into account [30]. This differs from BOW, in which the order of words is of no importance. Chronological awareness makes LSTM a good fit for NLP as the order of words in a sentence is meaningful.
Attention model
During LSTM encoding of a data sequence, intermediate calculations (states) are conducted. The ATN algorithm [33] utilizes these states to add context to the words in the sequence. The context of the word comes from the surrounding words in the sentence. Giving context to words augments the representation of the embedding layer.
Models’ hyper-parameters
BOW
For the logistic regression classifier, we have used l2 regularization with inverse of regularization strength = 1.0.
We have also conducted the following experiments for the BOW approach:
-
1)
We used gradient boosting (XGBoost) as a classifier. Default XGBoost hyper-parameters were used, with n_estimators = 1000. Tree-based methods are unaffected by normalization (in each node they cut above and below the desired value). This is why we used a CountVectorizer instead of tf-idf vectorizer for the XGBoost experiments.
-
2)
We have assessed the added value of using unigrams and bigrams (sequences of one and two words) as tokens.
Word2Vec
We used 200-dimensional vectors for the embedding layer. The model was trained using a continuous bag-of-words (CBOW) with a window size of 5.
LSTM
The bidirectional LSTM encoder consisted of 128 hidden units.
LSTM-ATN
On top of the LSTM layer, we stacked an attention layer and, on top of that, a 64-neuron fully connected layer.
Deep learning models
The LSTM and LSTM-ATN models were trained twice: first, by using a randomly assigned embedding layer and then by using a pre-trained embedding layer. The deep learning models were optimized using Adam optimizer [34]. We employed an early stopping criterion on the training set [35].
Results
We retrieved 176,988 head CT reports conducted in our hospital to pre-train an embedding layer. The embedding layer contained 30,002 vectors, corresponding to the number of unique words in all CT reports.
We manually labeled 7784 ED CT reports. The number of unique words in the manually labeled group was 5141. There were no words that appeared in the manually labeled group but did not appear in the non-labeled cohort. Examples of low-frequency terms included words with typos, words that describe specific patients’ comorbidities such as “ovary,” in a woman with ovarian cancer, and unique anatomical terms such as “Galen.”
The reports in the manually labeled group were signed by 30 different board-certified radiologists (average reports per radiologist 259.5 ± 418.7). Of the 7784 reports, 3604 (46.3%) were labeled as normal and 4180 (53.7%) were labeled as pathological. 7.1% of the reports described intra-cranial hemorrhage.
The distribution of the number of characters in each document is presented in Fig. 2.
Data exploration—word importance
Tables 1 and 2 present data exploration results. Table 1 shows words (tokens) with a high affinity to the pathological report group. Table 2 shows words with high affinity to the intra-cranial hemorrhage group. This is reflected by the high mutual information score of these words.
For the general use case, the word “seen” is part of sentences describing lesions. The words “right” and “left” relate to the lesions’ location. The words “examination” and “compared” relate to comparison to previous examinations. The word “post” relates to previous surgeries.
For the specific use case, words with high affinity include words related to hemorrhage (“parenchymal”), and location (e.g., “lateral”).
Performance of model BOW approach
Table 3 presents the results of the experiments with BOW models for the general use case and the specific use case. Using both unigrams and bigrams showed a small improvement in accuracy both in the general use case and in the specific use case. This was true both for logistic regression and for XGBoost classifiers.
Deep learning general use case
The results of the models are presented in Table 4 which shows the means of the metrics in the study. The best performing model was LSTM-ATN with Word2Vec (AUC = 0.967 ± 0.006, accuracy 90.8% ± 0.01) (Fig. 3a).
Deep learning models were more accurate than the BOW model (for gradient boosting unigrams/bigrams BOW accuracy 88.9%). This was significant for LSTM-ATN (accuracy 90.2%, p < 0.01), LSTM-Word2Vec (accuracy 90.5%, p < 0.01), and LSTM-ATN-Word2Vec (accuracy 90.8%, p < 0.01) but not for LSTM alone (accuracy 89.0%, p = 0.879).
Adding an ATN layer to LSTM improved the accuracy (LSTM accuracy 89.0% vs. LSTM-ATN accuracy 90.2%, p = 0.02). This improvement was not significant after pre-training with Word2Vec (LSTM-Word2Vec accuracy 90.5% vs. LSTM-ATN-Word2Vec accuracy 90.8%, p = 0.54).
Finally, adding a pre-trained embedding layer significantly improved the accuracy of both LSTM (LSTM accuracy 89.0% vs. LSTM-Word2Vec accuracy 90.5%, p < 0.01) and LSTM-ATN (LSTM-ATN accuracy 90.2% vs. LSTM-ATN-Word2Vec accuracy 90.8%, p < 0.01).
Some examples of false negative cases include the following: a case of “nasal bones fracture,” a case of “bilateral hygromas,” and a wrongly positively labeled case.
Deep learning specific use case
The results of the specific use case (intra-cranial hemorrhage vs. no intra-cranial hemorrhage) are presented in Table 5. Unlike the general use case, in the specific use case, all models showed quite similar accuracies. The best AUCs were shown for the unigrams and the unigrams/bigrams logistic regression BOW models and the LSTM-ATN-Word2Vec model (for all these models, AUC of 0.970).
Discussion
In this work, we employed state-of-the-art neural networks for flagging ED head CT reports. For the general labeling use case, the best model was an LSTM-ATN with an embedding layer pre-trained on a large cohort. Learning the dictionary from a large cohort of similar documents improves NLP performance. The ATN layer adds context to the words in the sentence and thus further improves the LSTM accuracy. For the specific labeling use case, BOW and deep learning showed similar results.
The evolution of biomedical technology has increased the amount of healthcare data [36]. NLP research is needed for advancing the structuring of this accumulated data. In the ED setting, there is a need for optimized patient triage [1]. By classifying the reports according to the presence of findings, “red flags” can be raised in the EMR. This is like systems that are already implemented in the EMR that raise “red flags” for pathological blood tests, for instance, alerting on abnormal potassium levels and the like. Moreover, in systems that give the reports back as a list, sorting can be performed. “Normal” reports can be pushed down, and reports with specific findings (such as intra-cranial hemorrhage) can be pushed up.
While radiologists should communicate directly the referring physicians to convey critical imaging findings, and some PACS systems have an option for manual flagging of reports, an automatic system can be used as a backup.
In recent years, deep learning has made an impact on the way free-text can be processed. Several previous studies employed LSTM for radiology NLP tasks [37]. Carrodeguas et al. compared LSTM with support vector machine, random forest, and logistic regression for assessing follow-up recommendations in radiology reports. Their dataset consisted of 1000 randomly chosen reports. In their study, support vector machines, random forest, and logistic regression outperformed LSTM [38]. Yuan et al. studied models for detection and classification of changes in the description of pulmonary nodules in reports. They compared machine learning, convolutional neural networks (CNN), and LSTM for this task. CNN and LSTM showed similar results and outperformed the machine learning methods. In their work, they used word embedding with Word2Vec trained on a large cohort of approximately 1.5 million reports [39].
Zech et al. evaluated different classic machine learning models for classifying head CT reports. They used BOW with averaged word embedding vectors trained on 100,000 reports. This model showed a 0.966 AUC across all head CT findings, which is comparable with the results of our study [6].
ATN models have recently shown state-of-the-art results in different NLP tasks [31]. Recently, Zhang et al. described using the ATN-based pre-trained BERT model for extracting clinical information from clinical and radiological notes of breast cancer patients [40]. We evaluated LSTM-ATN algorithms’ ability to flag head CT reports with pathological findings. We have shown that, for a general labeling task, deep learning methods outperformed the machine learning BOW method. We have also demonstrated that pre-training using a large cohort and the ATN layer improves the accuracy of LSTM for this task. For a specific use case, deep learning and BOW showed similar results. This signifies a common saying in the data science world, “there is no such thing as a free lunch”—NLP models should be explored and specified depending on the task.
We conducted our research with reports written in a non-English (Hebrew) language. Although pre-trained language models exist, they are usually more attuned to English texts. Moreover, radiology reports have many domain specific words, which may be further specific for the originating institution. Our results suggest that other non-English datasets may benefit from a similar design and usage of a local large cohort of texts.
It should be noted that the BOW model showed high performance, especially for the specific task. BOW is a simpler and faster model and easier to implement. This should be taken into consideration for deployment decisions.
Our study has several limitations. It is a retrospective single-center study performed on a large cohort of digitally stored data. Second, neural networks can have a complex structure. We attempted to limit the complexity to one LSTM layer, one ATN layer, and one fully connected layer. Some decisions on hyper-parameter selection can be further explored. For example, we have limited the length of reports to 1500 characters. Although arbitrary, only 1.2% of the cohort had more than 1500 characters. In this study, we have explored one general labeling use case (with vs. without pathology) and one specific labeling use case (with vs. without intra-cranial hemorrhage). Other use cases can be explored, acute vs. non-acute, with vs. without ischemic infarct, etc. Moreover, hyper-parameters were optimized using a random search. Although the dataset was randomized between tuning of hyper-parameters and training, this can still cause over-fitting. Finally, accuracies around 90% may not be enough when considering medico-legal implications. Thus, for clinical implementation, further studies must be performed to augment on these proof-of-concept results.
Conclusion
For a general use case, word embedding using a large cohort of non-English head CT reports and ATN improves NLP performance. For a more specific task, deep learning and BOW showed similar results. Models should be explored and tailored to the NLP task.
References
Schuetz P, Hausfater P, Amin D, Haubitz S, Fassler L, Grolimund E, Kutz A, Schild U, Caldara Z, Regez K, Zhydkov A, Kahles T, Nedeltchev K, von Felten S, De Geest S, Conca A, Schafer-Keller P, Huber A, Bargetzi M, Buergi U, Sauvin G, Perrig-Chiello P, Reutlinger B, Mueller B (2013) Optimizing triage and hospitalization in adult general medical emergency patients: the triage project. BMC emergency medicine 13:12–11. https://doi.org/10.1186/1471-227x-13-12
Klang E, Barash Y, Soffer S (2019) Promoting head CT exams in the emergency department triage using a machine learning model. https://doi.org/10.1007/s00234-019-02293-y
Klang E, Beytelman A, Greenberg D, Or J, Guranda L, Konen E, Zimlichman E (2017) Overuse of head CT examinations for the investigation of minor head trauma: analysis of contributing factors. Journal of the American College of Radiology : JACR 14(2):171–176. https://doi.org/10.1016/j.jacr.2016.08.032
Ohana O, Soffer S, Zimlichman E, Klang E (2018) Overuse of CT and MRI in paediatric emergency departments. Br J Radiol 91(1085):20170434. https://doi.org/10.1259/bjr.20170434
Liew C (2018) The future of radiology augmented with artificial intelligence: a strategy for success. Eur J Radiol 102:152–156. https://doi.org/10.1016/j.ejrad.2018.03.019
Zech J, Pain M, Titano J, Badgeley M, Schefflein J, Su A, Costa A, Bederson J, Lehar J, Oermann EK (2018) Natural language-based machine learning models for the annotation of clinical radiology reports. Radiology 287(2):570–580. https://doi.org/10.1148/radiol.2018171093
Hassanpour S, Bay G, Langlotz CP (2017) Characterization of change and significance for clinical findings in radiology reports through natural language processing. J Digit Imaging 30(3):314–322. https://doi.org/10.1007/s10278-016-9931-8
Hassanpour S, Langlotz CP (2016) Information extraction from multi-institutional radiology reports. Artif Intell Med 66:29–39. https://doi.org/10.1016/j.artmed.2015.09.007
Cai T, Giannopoulos AA, Yu S, Kelil T, Ripley B, Kumamaru KK, Rybicki FJ, Mitsouras D (2016) Natural language processing technologies in radiology research and clinical applications. Radiographics: a review publication of the Radiological Society of North America, Inc 36(1):176–191. https://doi.org/10.1148/rg.2016150080
Pons E, Braun LM, Hunink MG, Kors JA (2016) Natural language processing in radiology: a systematic review. Radiology 279(2):329–343. https://doi.org/10.1148/radiol.16142770
Toyabe S (2012) Detecting inpatient falls by using natural language processing of electronic medical records. BMC Health Serv Res 12:448. https://doi.org/10.1186/1472-6963-12-448
Collier N, Nazarenko A, Baud R, Ruch P (2006) Recent advances in natural language processing for biomedical applications. Int J Med Inform 75(6):413–417. https://doi.org/10.1016/j.ijmedinf.2005.06.008
Byrd RJ, Steinhubl SR, Sun J, Ebadollahi S, Stewart WF (2014) Automatic identification of heart failure diagnostic criteria, using text analysis of clinical notes from electronic health records. Int J Med Inform 83(12):983–992. https://doi.org/10.1016/j.ijmedinf.2012.12.005
Carrell DS, Cronkite D, Palmer RE, Saunders K, Gross DE, Masters ET, Hylan TR, Von Korff M (2015) Using natural language processing to identify problem usage of prescription opioids. Int J Med Inform 84(12):1057–1064. https://doi.org/10.1016/j.ijmedinf.2015.09.002
Yim WW, Yetisgen M, Harris WP, Kwan SW (2016) Natural language processing in oncology: a review. JAMA oncology 2(6):797–804. https://doi.org/10.1001/jamaoncol.2016.0213
Lakhani P, Kim W, Langlotz CP (2012) Automated extraction of critical test values and communications from unstructured radiology reports: an analysis of 9.3 million reports from 1990 to 2011. Radiology 265(3):809–818. https://doi.org/10.1148/radiol.12112438
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. Paper presented at the Proceedings of the 25th International Conference on Neural Information Processing Systems - Volume 1, Lake Tahoe, Nevada,
Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, Kadoury S, Tang A (2017) Deep learning: a primer for radiologists. 37(7):2113–2131. https://doi.org/10.1148/rg.2017170077
Suzuki K (2017) Overview of deep learning in medical imaging. Radiol Phys Technol 10(3):257–273. https://doi.org/10.1007/s12194-017-0406-5
Soffer S, Ben-Cohen A, Shimon O, Amitai MM, Greenspan H, Klang E (2019) Convolutional neural networks for radiologic images: a radiologist’s guide. Radiology 290(3):590–606. https://doi.org/10.1148/radiol.2018180547
Klang E (2018) Deep learning and medical imaging. Journal of thoracic disease 10(3):1325–1328. https://doi.org/10.21037/jtd.2018.02.76
Barash Y, Klang E (2019) Automated quantitative assessment of oncological disease progression using deep learning. Annals of Translational Medicine 7:S379-S379. https://doi.org/10.21037/atm.2019.12.101
Le Berre A, Kamagata K (2019) Convolutional neural network-based segmentation can help in assessing the substantia nigra in neuromelanin MRI. 61 (12):1387–1395. doi:https://doi.org/10.1007/s00234-019-02279-w
Ginat DT (2019) Analysis of head CT scans flagged by deep learning software for acute intracranial hemorrhage. Neuroradiology. 62:335–340. https://doi.org/10.1007/s00234-019-02330-w
Chen MC, Ball RL, Yang L, Moradzadeh N, Chapman BE, Larson DB, Langlotz CP, Amrhein TJ, Lungren MP (2018) Deep learning to classify radiology free-text reports. Radiology 286(3):845–852. https://doi.org/10.1148/radiol.2017171115
Hughes M, Li I, Kotoulas S, Suzumura T (2017) Medical text classification using convolutional neural networks. Studies in health technology and informatics. 235:246–250
Gehrmann S, Dernoncourt F, Li Y, Carlson ET, Wu JT, Welt J, Foote J Jr, Moseley ET, Grant DW, Tyler PD, Celi LA (2018) Comparing deep learning and concept extraction based methods for patient phenotyping from clinical narratives. PLoS One 13(2):e0192360. https://doi.org/10.1371/journal.pone.0192360
Lin C, Hsu CJ (2017) Artificial intelligence learning semantics via external resources for classifying diagnosis codes in discharge notes. 19(11):e380. https://doi.org/10.2196/jmir.8344
Luo Y, Cheng Y, Uzuner O, Szolovits P, Starren J (2018) Segment convolutional neural networks (Seg-CNNs) for classifying relations in clinical notes. Journal of the American Medical Informatics Association : JAMIA 25(1):93–98. https://doi.org/10.1093/jamia/ocx090
Hochreiter S, #252, Schmidhuber R (1997) Long short-term memory. Neural Comput 9 (8):1735–1780. doi:https://doi.org/10.1162/neco.1997.9.8.1735
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, Polosukhin I (2017) Attention is all you need. arXiv e-prints
Mikolov T, Sutskever I, Chen K, Corrado G, Dean J (2013) Distributed representations of words and phrases and their compositionality. arXiv e-prints
Bahdanau D, Cho K, Bengio Y (2014) Neural machine translation by jointly learning to align and translate. arXiv e-prints
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv e-prints
Mahsereci M, Balles L, Lassner C, Hennig P (2017) Early stopping without a validation set. arXiv e-prints
Cao C, Liu F, Tan H, Song D, Shu W, Li W, Zhou Y, Bo X, Xie Z (2018) Deep learning and its applications in biomedicine. Genomics, proteomics & bioinformatics 16(1):17–32. https://doi.org/10.1016/j.gpb.2017.07.003
Sorin V, Barash Y, Konen E, Klang E (2020) Deep learning for natural language processing in radiology-fundamentals and a systematic review. Journal of the American College of Radiology : JACR. https://doi.org/10.1016/j.jacr.2019.12.026
Carrodeguas E, Lacson R, Swanson W, Khorasani R (2019) Use of machine learning to identify follow-up recommendations in radiology reports. Journal of the American College of Radiology : JACR 16(3):336–343. https://doi.org/10.1016/j.jacr.2018.10.020
Yuan J, Zhu H, Tahmasebi A (2019) Classification of pulmonary nodular findings based on characterization of change using radiology reports. AMIA Joint Summits on Translational Science proceedings AMIA Joint Summits on Translational Science 2019:285–294
Zhang X, Zhang Y, Zhang Q, Ren Y, Qiu T, Ma J, Sun Q (2019) Extracting comprehensive clinical information for breast cancer using deep learning methods. Int J Med Inform 132:103985. https://doi.org/10.1016/j.ijmedinf.2019.103985
Funding
This study was conducted with the help of Accelerate, Redesign, Collaborate (ARC) - The Innovation Center at Sheba Medical Center.
Author information
Authors and Affiliations
Contributions
All authors made substantial contributions to the design and writing of the work. All authors have approved the final manuscript and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. A Sheba Medical Hospital institutional review board (IRB) approval was granted for this retrospective study (4025-17-MSC). The IRB committee waived informed consent.
Informed consent
For this retrospective study, formal consent is not required.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Barash, Y., Guralnik, G., Tau, N. et al. Comparison of deep learning models for natural language processing-based classification of non-English head CT reports. Neuroradiology 62, 1247–1256 (2020). https://doi.org/10.1007/s00234-020-02420-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00234-020-02420-0