Abstract
Purpose
The aims of this study were to develop a population pharmacokinetic model for allopurinol and oxypurinol and to explore the influence of patient characteristics on allopurinol and oxypurinol pharmacokinetics.
Methods
Data from 92 patients with gout and 12 healthy volunteers were available for analysis. A parent–metabolite model with a two-compartment model for allopurinol and a one-compartment model for oxypurinol was fitted to the data using non-linear mixed effects modelling.
Results
Renal function, fat-free mass (FFM) and diuretic use were found to predict differences in the pharmacokinetics of oxypurinol. The population estimates for allopurinol clearance, inter-compartmental clearance, central and peripheral volume were 50, 142 L/h/70 kg FFM, 11.4, 91 L/70 kg FFM, respectively, with a between-subject variability of 33 % (coefficient of variance, CV) for allopurinol clearance. Oxypurinol clearance and volume of distribution were estimated to be 0.78 L/h per 6 L/h creatinine clearance/70 kg FFM and 41 L/70 kg FFM in the final model, with a between-subject variability of 28 and 15 % (CV), respectively.
Conclusions
The pharmacokinetic model provides a means of predicting the allopurinol dose required to achieve target oxypurinol plasma concentrations for patients with different magnitudes of renal function, different body mass and with or without concomitant diuretic use. The model provides a basis for the rational dosing of allopurinol in clinical practice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gout is a painful form of arthritis caused by the presence of uric acid crystals in joints. Uric acid is the ultimate breakdown product of purine nucleotide degradation in humans and is formed when hypoxanthine is first converted to xanthine and then further degraded to uric acid by xanthine oxidase. Frequent attacks of gout can cause permanent joint and bone damage. The successful long-term management of gout requires a sustained serum urate of <0.36 mmol/L [1, 2].
Allopurinol [4-hydroxy-[3,4-d)pyrazolopyrimidine] is the most commonly used urate-lowering therapy in the management of gout. It is rapidly and almost entirely metabolised to an active metabolite, oxypurinol [3]. Both parent and metabolite are structural analogues of hypoxanthine and xanthine (respectively) and reduce urate concentrations by competitively inhibiting xanthine oxidase, thereby preventing uric acid production [3]. Oxypurinol is primarily eliminated by the kidneys and has an elimination half-life of 18–30 h in patients with normal renal function [3]. Unlike allopurinol, oxypurinol accumulates extensively in plasma after repeated dosing and is believed to account for most of the urate-lowering effect [3].
There is little agreement in published sources about the optimal dosing of allopurinol, particularly for patients with renal impairment [1, 4–6]. The most commonly used guideline in clinical practice recommends a dose adjustment according to the patient’s renal function, estimated using creatinine clearance (CLcr) [5]. This dosing strategy was developed by Hande et al. [5] and is based on a postulated relationship between elevated oxypurinol plasma concentrations and the development of allopurinol-hypersensitivity syndrome (AHS), a rare but life-threatening adverse effect of allopurinol [5]. However, adherence to CLcr-based dosing guidelines has been found to result in inadequate reductions in serum urate and a failure to achieve target serum urate concentrations in the majority of patients [7–9]. In addition, there is little evidence to support an association between higher doses of allopurinol, elevated oxypurinol plasma concentrations and AHS [7, 8, 10, 11].
An individualised dosing strategy for allopurinol has recently been proposed where doses are cautiously escalated until a sustained serum urate of <0.36 mmol/L is attained, regardless of renal function [10, 11]. This approach was found to be successful in achieving target serum urate concentrations in 75 % of samples when oxypurinol plasma concentrations were >100 μmol/L [10]. On the basis of these findings, a revised therapeutic range for oxypurinol of 100–150 μmol/L was proposed [10]. However, the allopurinol dose required to achieve target serum uric acid concentrations was found to be highly variable between patients (ranging from 50 to 600 mg daily [10]), and no satisfactory means of predicting the maintenance dose a priori for an individual patient was obvious. This suggests that a better understanding of the relationship between allopurinol dose, oxypurinol plasma concentrations and serum uric acid reduction is required, particularly of the factors which determine the variability in allopurinol and oxypurinol pharmacokinetics between patients. This information can be used to predict the dose require to achieve target oxypurinol plasma concentrations and would provide the basis for the safe and effective prescribing of allopurinol in clinical practice
The aims of this study were (1) to develop a population pharmacokinetic model for allopurinol and oxypurinol and (2) to explore the influence of patient characteristics on allopurinol and oxypurinol pharmacokinetics.
Materials and methods
Data sources
Data were sourced from five studies [10–14]. Three studies were conducted in Christchurch, New Zealand and led by one of the co-authors (LS). Data from two studies were digitally extracted from the literature. All participants in the Christchurch cohorts gave written, informed consent. Demographic details are presented in Table 1. The study designs and the evaluation of the digitally extracted data are briefly summarised in Electronic Supplementary Material (ESM) 1.
Data analysis
Details of the high-performance liquid chromatography assay used to measure allopurinol and oxypurinol plasma concentrations in all three Christchurch studies have been published previously [10]. The intra- and inter-day coefficient of variation (CV) was <10.4 % and the lower limit of quantification (LLOQ) was 0.7 μmol/L for both allopurinol and oxypurinol [10].
Population pharmacokinetic analysis
The population analysis was conducted using the nonlinear mixed effects modelling software NONMEM (v7.2) [15]. NONMEM used an AMD-CPU Opteron 12-core processor and a GNU Fortran 95 compiler (GCC 4.6.0). The convergence criterion was to 3 significant digits. The Laplacian option was chosen so that a likelihood-based method for handling concentrations below the LLOQ could be used (see below).
The final model was developed in a step-wise fashion. First, a model was fitted to the extracted intravenous allopurinol data from the published studies. One- and two-compartment structural models were explored, as well as zero order elimination and covariance between the clearance and volume parameters. Models to describe residual error were also explored, including additive, proportional and combined (i.e. both additive and proportional). The parameter variability between individuals was assumed to follow a log-normal distribution, taking the generic form
where θ ip is the estimate of the p th parameter θ for the i th individual, \( {{\widehat{\mu}}_p} \) is the population mean value of the p th parameter, and η ip is the deviation from the mean of the p th parameter for the i th individual. η was assumed to be normally distributed with a mean of zero and a variance of ω 2.
In the second step, allopurinol data from all of the studies (oral and intravenous) were included in the model using the final parameter estimates from the first step as initial values. Structural and error models were explored as above, including correlation between the clearance and volume parameters. In the third step, the parameter estimates from step 2 were used as the initial values for a parent–metabolite model incorporating oxypurinol. Both a sequential modelling approach, where the allopurinol parameter estimates were fixed to the empirical Bayes estimates (IPP method [16]) so that only the oxypurinol pharmacokinetic parameters were estimated, and a simultaneous method, where both allopurinol and oxypurinol pharmacokinetic parameters were estimated simultaneously, were explored. Correlation between oxypurinol clearance and volume parameters was also assessed.
Covariate model
The choice of covariates to be tested in the model was based on biological plausibility and on prior knowledge of factors believed to influence allopurinol and oxypurinol pharmacokinetics. Non-genotypic covariates considered included total body weight (TBW), fat-free mass (FFM), normal fat-mass (NFM), CLcr, and drug interactions. FFM was calculated using the formula developed by Janmahasatian et al. [17], while NFM was determined using the method proposed by Anderson and Holford [18]:
where NFM = normal fat mass, FFM = fat-free mass, TBW = total body weight and Ffat is a parameter estimated during the modelling analysis and represents the fraction of fat mass that contributes to the estimation of the pharmacokinetic parameter (e.g. clearance). Clearance was allometrically scaled to an exponent of 0.75 and volume to an exponent of 1 [19]. CLcr was calculated according the Cockroft–Gault formula [20] standardised to 70 kg. Renal function (RF) was then normalised to a standard CLcr (\( C{L_{{c{r_{STD }}}}} \)) of 6 L/h/70 kg (100 mL/min/70 kg):
RF was included in the model using a linear independent combination of renal and non-renal clearance (CL) parameters, as follows:
where CLnon-renal is non-renal clearance and CLrenal is renal clearance.
Drugs known to alter the pharmacokinetics for allopurinol or oxypurinol (e.g. probenecid, furosemide [12, 21]) were considered as covariates. In addition, drugs which have been found to alter serum urate concentrations were tested as covariates on oxypurinol clearance under the (conservative) assumption that urate and oxypurinol are handled by the kidneys in a similar manner and by the same transporters. Drugs that have been found to lower serum urate and which were tested as covariates in the model included losartan [22, 23], statins [24] and calcium-channel blockers [23]. The number of patients taking each of these medicines was small so they were tested in the model as a single group called ‘drugs that lower urate’. Drugs associated with an increased risk of hyperuricaemia through a presumed or known alteration in the renal handing of urate were also considered, including angiotensin converting enzyme inhibitors, angiotension receptor blockers, beta-blockers and non-steroidal anti-inflammatory drugs (not including low-dose aspirin) [3, 22, 23].
The fractional influence of interacting drugs (and other discrete covariates) was estimated as a parameter in the model. Individuals with missing covariate information were assumed to have the standard value of the covariate.
Genetic variants of several apical and basolateral transporters expressed in the proximal renal tubules [25–31] were investigated as covariates on oxypurinol clearance, as well as genetic variants thought to influence the conversion of allopurinol to oxypurinol [32, 33]. Details of the genotypic covariates are summarised in ESM 2 (Table S1).
Model discrimination and evaluation
A likelihood ratio test was used to discriminate between candidate nested models. The preferred model was chosen based on a decrease in the objective function value (OFV) of 3.84 units (χ2, p < 0.05) with 1 df. The model selection process consisted of initially developing a base model without any covariates (see above). Each covariate was sequentially added to the model in descending order of largest objective function change, and each successive model fitted to the data. At each forward addition of a covariate a likelihood ratio test was performed. Only those covariates that resulted in statistically significant reduction in OFV were retained. The final covariate model was further subjected to a backward elimination procedure. Each covariate was sequentially removed from the model and only retained if the objective function value increased by >6.6 units (χ2, p < 0.01) with 1 df.
Models were evaluated using visual predictive checks (VPCs) and non-parametric bootstrap statistics of the parameter estimate distribution. One hundred data sets were simulated under each evaluated model and the 5th, 50th and 95th percentiles, including the 95 % confidence interval (CI), were plotted against the same percentiles in the original data set. All VPC plots were created using R (v.2.15.0; R foundation for Statistical Computing, Vienna, Austria). A prediction-corrected design was used where the median prediction in each bin was used to normalise the observed and simulated observations in that bin [34]. The final pharmacokinetic model was also evaluated using a nonparametric bootstrap. The median parameter values and 95 % CI were determined from 1,000 non-parametric bootstrap runs and compared to the final parameter estimates.
Data below the limit of quantification
The oral data included 269 allopurinol concentrations that were reported as being below the LLOQ of 0.7 μmol/L [10]. No oxypurinol plasma concentrations below the LLOQ were reported. A likelihood based method (M3) [35] was used to analyse the allopurinol data below the LLOQ.
Genotyping
DNA was extracted and SLC2A9 (rs11942223), ABCG2 (rs2231142) and NPT1/SLC17A1 (rs1183201) were genotyped as previously described [29–31]. Single nucleotide polymorphisms (SNPs) rs3825018 (URAT1), rs17300741 (OAT4) and rs12129861 (PDZK1) were genotyped using TaqMan® assays (Applied Biosystems, Foster City, CA) in a Lightcycler® 480 Real-Time PCR System (Roche, Indianapolis, IN): rs17300741 (C_3234214_10), rs3825018 (C_27162391_10) and rs12129861 (C_26662675_10).
Model simulations
The final pharmacokinetic model for allopurinol and oxypurinol was implemented in MATLAB (v. 2012a). A series of deterministic simulations were conducted under the following conditions: (1) doses of 100, 200, 300, 400 and 600 mg daily for individuals with a total body weight of 70 kg and CLcr of 6 L/h (100 mL/min); (2) a dose of 300 mg daily for individuals with a total body weight of 70 kg and CLcr values of 1.2, 2.4, 3.6, 4.8 and 6 L/h; (3) a dose of 300 mg daily with and without concomitant diuretic therapy for individuals with a total body weight of 70 kg and CLcr of 6 L/h; (4) a dose of 300 mg for 14 days for individuals with a CLcr of 6 L/h and a total body weight of either 70 kg, 90 kg or 110 kg. For the purposes of these simulations, FFM was derived from the total body weight values stated above by calculating the FFM to TBW ratio in the observed data and using this value to predict FFM.
Results
Allopurinol and oxypurinol pharmacokinetic models
A sequential parent–metabolite model with first-order input, a two-compartment model for allopurinol (parent), and a one-compartment model for oxypurinol with a combined, additive and proportional residual variance provided the best fit to the data. The allopurinol model was developed first and the parameter estimates fixed in the sequential parent–metabolite model. The percentage of allopurinol converted to oxypurinol was fixed at 80 % based on values previously reported in the literature [3, 14]. Population-predicted and individual-predicted oxypurinol plasma concentrations versus the observed oxypurinol plasma concentrations are presented in Fig. 1. The ε-shrinkage was found to be 5.3 % (Table 2). There appears to be good agreement between predicted and observed values at lower concentrations (e.g. <200 μmol/L) while higher concentrations show a somewhat larger scatter around the line of identity (individual predictions) and a slight trend towards over-prediction (population predictions). Overall, nothing suggestive of model misspecification was evident from the goodness of fit plots.
The noteworthy covariate model building steps for the sequential model are presented in ESM 3 (Table S2). The influence of renal function on the clearance of oxypurinol caused the greatest reduction in the objective function value (64.7 points). Measures of body composition (weight, FFM and NFM) applied to both allopurinol and oxypurinol clearance and volume of distribution also reduced the objective function value compared to the base model. The three body composition measures did not differ appreciably in terms of model fit, nor were there any noticeable differences in VPCs generated under each model (not shown), so the simplest model (FFM) was retained.
There was no identifiable influence of any individual genotype on either allopurinol or oxypurinol pharmacokinetics. Individuals with at least one minor allele for the PDZK1 rs12129861 SNP were found to have an oxypurinol clearance that was 12 % lower than homozygotes for the major allele; however, this effect became non-significant once differences in renal function and body mass were accounted for in the model. Similarly, individuals with at least one minor allele for the NPT1 SNP rs1183201 genotype showed a trend towards reduced oxypurinol clearance (34 % lower) compared to those with the major allele homozygote genotype but this did not reach statistical significance.
The renal clearance of oxypurinol was found to be significantly reduced in patients taking diuretics (median reduction 39 %) and beta-blockers (median reduction 29 %). These effects were retained once differences in renal function and body composition were included in the model, and the beta-blocker effect was retained after diuretic therapy was included in the model. Beta-blockers have been associated with increased serum urate [22], but an interaction with oxypurinol does not appear to have been documented. Note that beta-blocker therapy was not retained as a covariate in the final model as it did not meet the statistical criterion (see above) for inclusion during the backwards deletion process, nor were there any notable differences in VPCs generated from models with and without beta-blocker therapy. Thus, the clinical significance of this is uncertain.
The final model for oxypurinol clearance and volume were described by the equations
where CLoxy is oxypurinol clearance, CLcr is creatinine clearance, FFM is fat-free mass and Voxy is oxypurinol volume.
Parameter estimates for the base model and final covariate are presented in Table 2. The between-subject variance for oxypurinol clearance and volume of distribution decreased by about 50 % in the covariate model compared to the base model. Prediction-corrected VPCs showing the final covariate model predictions over the range of observed covariates are presented in Fig. 2.
Prediction-corrected visual predictive checks (VPCs) showing the 5th, 50th and 95th percentiles of the oxypurinol plasma concentrations predicted from the final model (solid line) and the observed oxypurinol data (dotted line) plotted against each covariate. a Not taking diuretics, b Taking diuretics, c over the range of creatinine clearance (CLcr), d over the range of fat-free mass (FFM). The grey-shaded area depicts the 95 % confidence interval
The median oxypurinol parameter estimates with the 95 % CI derived from 1,000 non-parametric, case-based bootstraps for the final sequential model are presented in Table 2. Note that all allopurinol pharmacokinetic parameters were fixed for these bootstraps. The mean bootstrap parameter values were comparable to the parameter estimates for the final model.
Prediction-corrected VPCs of the final model for allopurinol and oxypurinol are presented in Fig. 3. The median and 5th and 95th percentiles of the plasma concentrations predicted by the model follow the percentiles of the observed data, suggesting a good model fit in both cases.
VPC for the final allopurinol and oxypurinol pharmacokinetic models. a Observed allopurinol data with 5th, 50th and 95th percentiles. b Prediction-corrected VPC showing 5th, 50th and 95th percentiles of the observed allopurinol data (dotted red lines) and the allopurinol plasma concentrations predicted by the model (black solid (median) and dashed lines) with 95 % confidence interval (shaded area). c the observed oxypurinol data with 5th, 50th and 95th percentiles. d Prediction-corrected VPC showing 5th, 50th and 95th percentiles of the observed oxypurinol data (dotted red lines) and the oxypurinol plasma concentrations predicted by the model (black solid (median) and dashed lines) with 95 % confidence interval (solid line)
Model simulations
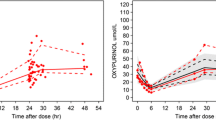
The predicted steady-state oxypurinol plasma concentrations for a typical individual in the population at different doses and at different values of CLcr, FFM and with or without concomitant diuretics are presented in Fig. 4. The model predicts that doses of 100 to 600 mg daily would be expected to produce oxypurinol plasma concentrations (measured at 6 h post-dose) ranging from about 20 to >180 μmol/L for a patient with normal renal function (6 L/h) and a total body weight of 70 kg (Fig. 4a). Oxypurinol plasma concentrations were predicted to increase in a proportional manner with declining renal function (Fig. 4b) and would be expected to be two to threefold higher in a patient with impairment renal function (e.g. CLcr of 1.2 L/h) compared to a patient with normal renal function (6 L/h). Diuretic therapy is predicted from the model to reduce oxypurinol clearance by 39 % (Fig. 4c). Simulations from the model also illustrated an important influence of FFM on oxypurinol plasma concentrations (Fig. 4d).
Oxypurinol plasma concentrations for a typical individual in the population simulated from the final pharmacokinetic model. a Allopurinol doses of 100, 200, 300, 400 and 600 mg daily, CLcr fixed at 6 L/h and total body weight fixed at 70 kg, b CLcr values of 1.2, 2.4, 3.6, 4.8 and 6 L/h, dose fixed at 300 mg daily and total body weight fixed at 70 kg, c dose of 300 mg daily with and without concomitant diuretic therapy, CLcr fixed at 6 L/h and total body weight fixed at 70 kg, d Fat-free mass of approximately 50 kg (corresponding to a total body weight of 70 kg in our data), 62 kg (corresponding to a total body weight of 90 kg) and 76 kg (corresponding to a total body weight of 110 kg), dose fixed at 300 mg and CLcr fixed at 6 L/h
Discussion
In this study we developed a population pharmacokinetic model for allopurinol and oxypurinol. To our knowledge, this is the first study to identify that body mass (FFM) is an important determinant of allopurinol and oxypurinol pharmacokinetics. Body mass is expected to predict clearance and volume parameters on strong biological grounds. The failure to observe this in other studies is likely a reflection of inadequate sample size and/or range of body sizes. We also found that renal function and the concomitant use of diuretics significantly influenced the pharmacokinetics of oxypurinol. By accounting for differences in these covariates between patients, the unexplained variability in oxypurinol clearance was reduced by 50 %. No detectable effect of renal transporter genotype on oxypurinol clearance was observed.
The pharmacokinetic model predicts that a typical patient given a daily allopurinol dose of 300 mg would produce oxypurinol plasma concentrations in the range of 60–90 μmol/L, if body mass were 70 kg (FFM of approximately 52 kg) and CLcr were 6 L/h. This is within the currently accepted therapeutic range for oxypurinol of 30–100 μmol/L but below the recently revised range of 100–150 μmol/L [10]. If we accept the new therapeutic range, then it is likely that many patients with normal kidney function will be under-treated if CLcr-based dosing guidelines are followed (e.g. Hande guidelines [5]). Predictions from the model also indicate that patients with normal renal function (CLcr 6 L/h) and a total body weight of 70 kg would require 450–500 mg daily on average to achieve oxypurinol plasma concentrations of 100–150 μmol/L (simulations not shown). The same patient with a CLcr of 3.6 L/h would require 300 mg daily, and if CLcr was 1.2 L/h a dose of 200 mg daily would be required. It is important to note that these predictions do not account for the impact of renal function on serum uric acid concentrations and, therefore, cannot be used to predict the probability of treatment success.
Diuretics were predicted to reduce oxypurinol clearance by an average 39 % by our model, an effect that may result in a clinically important increase in oxypurinol plasma concentrations. In addition, the model predicted a nearly twofold decrease in oxypurinol plasma concentrations for a patient at the upper end of the observed FFM in our data set (FFM of 99 kg, mean oxypurinol concentration 57 μmol/L) compared to a patient at the lower end of the range (FFM 35 kg, mean oxypurinol concentration 114 μmol/L) (simulations not shown). This suggests that the maintenance dose of allopurinol required to achieve a target oxypurinol plasma concentration could differ by as much as twofold based solely on differences in body mass.
There are few population analyses for allopurinol and, to our knowledge, only one published study involving patients with gout. Stocker et al. developed a population pharmacokinetic model for oxypurinol using data from 155 patients with gout [36]. The authors found that renal function, diuretic use and probenecid use influenced the pharmacokinetics of oxypurinol [36]. Unlike Stocker et al., we have developed a joint parent–metabolite model which accounts for the time course of allopurinol in plasma, as well as the active metabolite oxypurinol. Allopurinol is itself active but has a short half-life in the plasma compared to oxypurinol and, hence, its contribution to urate-lowering effect is poorly understood. Additionally, it is possible that the conversion of allopurinol to oxypurinol may be altered in some patients, and it was therefore a priority to include the parent drug in the pharmacokinetic model.
The findings of our covariate analysis were broadly similar to those of Stocker et al., although we additionally found that body mass had a significant impact on allopurinol and oxypurinol clearance and volume. Stocker reported that diuretic use led to a 41 % reduction in oxypurinol clearance on average [36], which is similar to the median value of 39 % predicted by our model. The genes studied overlapped between our studies with the SNPs examined being either identical or in strong linkage disequilibrium in European Caucasians.
The relationship between allopurinol dose, oxypurinol concentration and serum uric acid reduction is poorly understood. Indeed, data in the literature concerning this relationship are conflicting, with some studies suggesting a poor relationship between oxypurinol and uric acid concentrations [37, 38] and others showing a significant inverse correlation [10, 39]. A pharmacodynamic population analysis conducted using data from healthy volunteers found a shallow concentration–response relationship and an estimated C50 for oxypurinol (the plasma concentration at which drug effect is 50 % of the maximum) of about 35 μmol/L [39]. This C50 value is well below the steady-state oxypurinol concentrations generally achieved in clinical practice. From first principles, this suggests that there will be little benefit in terms of serum urate reduction from allopurinol doses above those that produce a steady-state plasma oxypurinol concentration of about 150 μmol/L on average (four times the reported C50).
To investigate the dose–response relationship of allopurinol further, we conducted an analysis of published data from four studies in gout patients [40–43] and from data available for patients initiating allopurinol in the vitamin C study described above. For each study, sequential serum uric acid concentrations for gout patients taking allopurinol therapy were available from baseline. Only serum urate values recorded after at least 7 days of allopurinol therapy at the same dose were included in the analysis. The percentage reduction in serum urate from baseline was calculated for each individual at each dose taken and presented in ESM 4 (Fig. S1 and Table S3). Data from 133 individuals were included in this analysis (note that some individuals provided data for more than one dose level). The dose–response of allopurinol was found to be nearly linear for doses from 50 to 500 mg daily. Daily doses of >500 mg produced diminishing reductions in serum urate, suggesting that these doses produce oxypurinol plasma concentrations above the C80 value (i.e. the plasma concentration at which the drug effect is 80 % of the maximum) and that the dose–response curve was approaching Emax (maximum possible effect). In addition, the results suggest that, at therapeutic doses, the maximum reduction in serum urate from baseline will be about 60 % on average.
Our analysis required that we pooled data from five different studies. The majority of the data (and all of the oxypurinol data) came from three studies in gout patients conducted in the same rheumatology clinic. We do not anticipate any bias in parameter estimates resulting from the pooling of these data. In addition, our goal was to understand the population pharmacokinetics of uncontrolled gout patients since this reflects the types of patients seen in clinical practice. The extraction of intravenous (IV) allopurinol data from studies in healthy volunteers was necessary to stabilise the allopurinol model. Estimates of allopurinol pharmacokinetic parameters from the IV data alone (e.g. clearance 48.8 L/h) were similar to those estimated using all five data sources (e.g. clearance 53.3 L/h). The 12 healthy volunteers represented in these studies were fairly homogenous in terms of age, kidney function and weight and so are not expected to contribute substantial information about the influence of covariates on the allopurinol pharmacokinetics.
While our pharmacokinetic model has provided an important step towards a better understanding of the variability in allopurinol dose–response between patients, our study was limited by the lack of a pharmacodynamic model to describe changes in serum urate concentrations during allopurinol therapy. For example, we were unable to determine if serum uric acid clearance was altered by diuretic therapy or renal impairment. Therefore, the impact of these covariates on allopurinol response cannot be determined without a fully integrated pharmacokinetic–pharmacodynamic model linking allopurinol dose, allopurinol and oxypurinol concentrations and changes in serum uric acid. Serum uric acid measurements were available for all patients in the Christchurch cohort, but the data was inadequate to identify and estimate parameters for a model for urate turnover.
Our results do not necessarily support the monitoring of oxypurinol plasma concentrations to guide allopurinol dosing. Given that serum urate concentrations provide a useful biomarker for clinical outcome in gout therapy, there is arguably little benefit to be gained from defining a therapeutic range for oxypurinol concentration. However, an understanding of the relationships between allopurinol dose, oxypurinol plasma concentration and serum urate reduction will provide a means of more accurately predicting the optimal dose of allopurinol to achieve treatment success, particularly in patients with renal impairment. This is particularly important because urate itself is retained in patients with impaired renal function, suggesting that oxypurinol plasma concentrations will need to be higher in these patients compared to patients with normal renal function to achieve the same effect on serum urate. Further research to clarify the time course and magnitude of allopurinol effect on serum urate and to enable the development of a population pharmacokinetic–pharmacodynamic model, including changes to urate turnover during allopurinol therapy, is therefore warranted.
Conclusions
We have developed a population pharmacokinetic model for allopurinol and oxypurinol. We have found that fat-free mass significantly influenced the pharmacokinetics of allopurinol while renal function, fat-free mass and the use of diuretics significantly influenced the pharmacokinetics of oxypurinol. By accounting for differences in these covariates, the unexplained variability in oxypurinol clearance and volume were both reduced by about 50 %. The pharmacokinetic model provides a means of predicting target oxypurinol plasma concentrations for individual patients. This research represents a step towards the rational dosing of allopurinol in clinical practice.
References
Zhang W, Doherty M, Bardin T, Pascual E, Barskova V, Conaghan P et al (2006) EULAR evidence based recommendations for gout. Part II: Management. Report of a task force of the EULAR Standing Committee for International Clinical Studies Including Therapeutics (ESCISIT). Ann Rheum Dis 65:1312–1324
Shoji A, Yamanaka H, Kamatani N (2004) A retrospective study of the relationship between serum urate level and recurrent attacks of gouty arthritis: evidence for reduction of recurrent gouty arthritis with antihyperuricemic therapy. Arthritis Rheum 51:321–325
Day RO, Graham GG, Hicks M, McLachlan AJ, Stocker SL, Williams KM (2007) Clinical pharmacokinetics and pharmacodynamics of allopurinol and oxypurinol. Clin Pharmacokinet 46:623–644
Jordan KM, Cameron JS, Snaith M, Zhang W, Doherty M, Seckl J et al (2007) British Society for Rheumatology and British Health Professionals in Rheumatology guideline for the management of gout. Rheumatology 46:1372–1374
Hande KR, Noone RM, Stone WJ (1984) Severe allopurinol toxicity. Description and guidelines for prevention in patients with renal insufficiency. Am J Med 76:47–56
US National Library of Medicine DailyMed: FDA information: allopurinol tablet (Zyloprim).Available at: http://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?id=13564. Accessed 15 Aug 2012
Dalbeth N, Kumar S, Stamp L, Gow P (2006) Dose adjustment of allopurinol according to creatinine clearance does not provide adequate control of hyperuricemia in patients with gout. J Rheumatol 33:1646–1650
Vazquez-Mellado J, Morales EM, Pacheco-Tena C, Burgos-Vargas R (2001) Relation between adverse events associated with allopurinol and renal function in patients with gout. Ann Rheum Dis 60:981–983
Stamp L, Gow P, Sharples K, Raill B (2000) The optimal use of allopurinol: an audit of allopurinol use in South Auckland. Aust NZ J Med 30:567–572
Stamp LK, Barclay ML, O’Donnell JL, Zhang M, Drake J, Frampton C, Chapman PT (2011) Relationship between serum urate and plasma oxypurinol in the management of gout: determination of minimum plasma oxypurinol concentration to achieve a target serum urate level. Clin Pharmacol Ther 90:392–398
Stamp LK, O’Donnell JL, Zhang M, James J, Frampton C, Barclay ML, Chapman PT (2011) Using allopurinol above the dose based on creatinine clearance is effective and safe in patients with chronic gout, including those with renal impairment. Arthritis Rheum 63:412–421
Stamp LK, Barclay ML, O’Donnell JL, Zhang M, Drake J, Frampton C, Chapman PT (2012) Furosemide increases plasma oxypurinol without lowering serum urate—a complex drug interaction: implications for clinical practice. Rheumatology 51:1670–1676
Appelbaum SJ, Mayersohn M, Dorr RT, Perrier D (1982) Allopurinol kinetics and bioavailability. Intravenous, oral and rectal administration. Cancer Chemother Pharmacol 8:93–98
Breithaupt H, Tittel M (1982) Kinetics of allopurinol after single intravenous and oral doses: noninteraction with benzobromarone and hydrochlorothiazide. Eur J Clin Pharmacol 22:77–84
Boeckmann AJ, Sheiner LB, Beal SL (2010) NONMEM users guide—part VIII. ICON Development Solutions, Ellicott City
Zhang L, Beal SL, Sheiner LB (2003) Simultaneous vs. sequential analysis for population PK/PD data I: best-case performance. J Pharmacokinet Pharmacodyn 30:387–404
Janmahasatian S, Duffull SB, Ash S, Ward LC, Byrne NM, Green B (2005) Quantification of lean bodyweight. Clin Pharmacokinet 44:1051–1065
Anderson BJ, Holford NHG (2009) Mechanistic basis of using body size and maturation to predict clearance in humans. Drug Metab Pharmacokinet 24:25–36
Anderson BJ, Holford NH (2008) Mechanism-based concepts of size and maturity in pharmacokinetics. Annu Rev Pharmacol Toxicol 48:303–332
Cockcroft DW, Gault MH (1976) Prediction of creatinine clearance from serum creatinine. Nephron 16:31–41
Stocker SL, Graham GG, McLachlan AJ, Williams KM, Day RO (2011) Pharmacokinetic and pharmacodynamic interaction between allopurinol and probenecid in patients with gout. J Rheumatol 38:904–910
Reyes AJ (2003) Cardiovascular drugs and serum uric acid. Cardiovasc Drugs Ther 17:397–414
Choi HK, Soriano LC, Zhang Y, Rodriguez LAG (2012) Antihypertensive drugs and risk of incident gout among patients with hypertension: population based case–control study. BMJ 344:d8190
Daskalopoulou SS, Tzovaras V, Mikhailidis DP, Elisaf M (2005) Effect on serum uric acid levels of drugs prescribed for indications other than treating hyperuricaemia. Curr Pharm Des 11:4161–4175
Graessler J, Graessler A, Unger S, Kopprasch S, Tausche A-K, Kuhlisch E, Schroeder HE (2006) Association of the human urate transporter 1 with reduced renal uric acid excretion and hyperuricemia in a German Caucasian population. Arthritis Rheum 54:292–300
Stark K, Reinhard W, Grassl M, Erdmann J, Schunker H, Illig T, Hengstenberg C (2009) Common polymorphisms influencing serum uric acid levels contribute to susceptibility to gout, but not to coronary artery disease. PLoS One 4:e7729
van der Harst P, Bakker SJL, de Boer RA, Wolffenbuttel BHR, Johnson T, Caulfield MJ, Navis G (2010) Replication of the five novel loci for uric acid concentrations and potential mediating mechanisms. Hum Mol Genet 19:387–395
Kolz M, Johnson T, Sanna S, Teumer A, Vitart V, Perola M et al (2009) Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet 5:e1000504
Hollis-Moffatt JE, Xu X, Dalbeth N, Merriman ME, Topless R, Waddell C, Gow PJ, Harrison AA, Highton J, Jones PBB, Stamp LK, Merriman TR (2009) Role of the urate transporter SLC2A9 gene in susceptibility to gout in New Zealand Maori, Pacific Island, and Caucasian case–control sample sets. Arthritis Rheum 60:3485–3492
Hollis-Moffatt JE, Phipps-Green AJ, Chapman B, Jones GT, van Rij A, Gow PJ, Harrison AA, Highton J, Jones PB, Montgomery GW, Stamp LK, Dalbeth N, Merriman TR (2012) The renal urate transporter SLC17A1 locus: confirmation of association with gout. Arthritis Res 14:R92
Phipps-Green AJ, Hollis-Moffatt JE, Dalbeth N, Merriman ME, Topless R, Gow PJ et al (2010) A strong role for the ABCG2 gene in susceptibility to gout in New Zealand Pacific Island and Caucasian, but not Maori, case and control sample sets. Hum Mol Genet 19:4813–4819
Roberts RL, Zhang M, Marinaki AM, Stamp LK (2010) Does genetic variability in aldehyde oxidase and molybdenum cofactor sulfurase predict nonresponse to allopurinol? Aliment Pharmacol Ther 32:310–311
Smith MA, Marinaki AM, Arenas M, Shobowale-Bakre M, Lewis CM, Ansari A, Duley J, Sanderson D (2009) Novel pharmacogenetic markers for treatment outcome in azathioprine-treated inflammatory bowel disease. Aliment Pharmacol Ther 30:375–384
Bergstrand M, Hooker AC, Wallin JE, Karlsson MO (2011) Prediction-corrected visual predictive checks for diagnosing nonlinear mixed-effects models. AAPS J 13:143–151
Ahn JE, Karlsson MO, Dunne A, Ludden TM (2008) Likelihood based approaches to handling data below the quantification limit using NONMEM VI. J Pharmacokinet Pharmacodyn 35:401–421
Stocker SL, McLachlan AJ, Savic RM, Kirkpatrick CM, Graham GG, Williams KM, Day RO (2012) The pharmacokinetics of oxypurinol in people with gout. Br J Clin Pharmacol 74:477–489
Day RO, Miners JO, Birkett DJ, Whitehead A, Naidoo D, Hayes J, Savdie E (1988) Allopurinol dose selection: relationships between dose and plasma oxypuirnol and serum urate concentrations and urinary rate excretion. Br J Clin Pharmacol 26:423–428
Peterson GM, Boyle RR, Francis HW, Oliver NWJ, Paterson J, von Witt RJ, Taylor GR (1990) Dosage prescribing and plasma oxypurinol levels in patients receiving allopurinol therapy. Eur J Clin Pharmacol 39:419–421
Graham S, Day RO, Wong H, McLachlan AJ, Bergendal L, Minors JO, Birkett DJ (1996) Pharmacodynamics of oxypurinol after administration of allopurinol to healthy subjects. Br J Clin Pharmacol 41:299–304
Delbarre F, Amor B, Auscher C, de Gery A (1966) Treatment of gout with allopurinol. A study of 106 cases. Ann Rheum Dis 25[6 Suppl]:627–633
Ta-F Y, Gutman AB (1964) Effect of allopurinol (4-Hydroxypyrazolo-(3,4-d)pyrimidine) on serum and urinary uric acid in primary and secondary gout. Am J Med 37:885–898
Klinenberg JR, Goldfinger SE, Seegmiller JE (1965) The effectiveness of the xanthine oxidase inhibitor allopurinol in the treatment of gout. Ann Intern Med 62:639
Rundles RW, Metz EN, Silberman HR (1966) Allopurinol in the treatment of gout. Ann Intern Med 64:229
Acknowledgments
At the time of writing, Dan Wright was the recipient of a University of Otago PhD scholarship. We wish to thank Dr. Rebecca Roberts, Department of Surgical Sciences, University of Otago, Dunedin for generous assistance with the AOX1 and MOCOS genotypes. We are also grateful to Jill Drake, Department of Rheumatology, Immunology and Allergy, Christchurch Hospital, for valuable assistance with the Christchurch data.
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wright, D.F.B., Stamp, L.K., Merriman, T.R. et al. The population pharmacokinetics of allopurinol and oxypurinol in patients with gout. Eur J Clin Pharmacol 69, 1411–1421 (2013). https://doi.org/10.1007/s00228-013-1478-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00228-013-1478-8