Abstract
Purpose
The cytotoxic drug cyclophosphamide (CP) is bioactivated into 4-hydroxy-cyclophosphamide (4-OH-CP) through cytochrome P450 enzymes and cleared through aldehyde dehydrogenase and glutathione S-transferase. This prospective study analyzes the influence of drug metabolizing enzyme genotype on (1) plasma 4-OH-CP:CP ratio and (2) myelotoxicity in breast cancer patients on 500 mg/m2 cyclophosphamide.
Methods
Sixty-eight female breast cancer patients on FAC (fluorouracil, adriamycin, cyclophosphamide) were included. Genotyping of cytochrome P450 enzymes CYP2B6, CYP2C9, CYP2C19, CYP3A5, aldehyde dehydrogenase (ALDH3A1), and glutathione S-transferase (GSTA1) was done either through RFLP or pyrosequencing. Plasma CP and 4-OH-CP were measured immediately and 1 and 2 h after the end of infusion through LC-MS. The leukocyte count was determined on day 10 and 20 after chemotherapy.
Results
At CP dose of 500 mg/m2, the 4-OH-CP:CP ratio was negatively affected by CYP2C19*2 genotype (p = 0.039) showing a gene-dose effect. Moreover ALDH3A1*2 genotype increased 4-OH-CP:CP ratio (p = 0.037). These effects did not remain significant in a univariate analysis of variance including all genotypes. GSTA1*B carriers were at increased risk of severe leucopenia (OR 6.94; 95% CI 1.75–27.6, p = 0.006).
Conclusion
The myelotoxicity in patients receiving FAC is related to the activity of the phase-II enzyme GSTA1 but is independent of the formation of 4-OH-CP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most prevalent malignancy among females around the world, with a recently reported rise [1]. There has been a tremendous surge in attempts to elucidate diagnostic and prognostic markers of breast cancer and in particular to optimize the outcome individually as well as to prevent severe side effects caused by the cytostatic therapies.
Cyclophosphamide (CP) is commonly applied in the treatment of breast cancer among other indications. It is used at a dose of 500 mg/m2 in conjunction with doxorubicin (Adriamycin® = A) and either fluorouracil (F) or a taxane (T) as FAC or TAC protocols, respectively, which are regarded as a cost effective first line chemotherapy regimen [2].
CP is an inactive prodrug that undergoes metabolic activation to 4-hydroxycyclophosphamide (4-OH-CP) mainly by hepatic cytochrome P450 (CYP) 2B6, 2 C9, 2 C19, and 3A4 [3, 4]. 4-OH-CP is transformed non-enzymatically to its isomer aldophosphamide, which is either inactivated to carboxyphosphamide through aldehyde dehydrogenase (ALDH) or disintegrated into the ultimate bivalent alkylating nucleophile phosphoramide mustard. The reactive nucleophiles are inactivated especially by glutathione S-transferase (GSTA1) [5]. For details of the activation and detoxification pathway, see [6]. GSTs also play an important role in disposition of doxorubicinol, the active metabolite of doxorubicin [7].
Previous studies indicated an impact of CYP2C19 genotype on the elimination constant of cyclophosphamide [4, 8], whereas others reported an impact of CYP2B6 variants [9, 10]. However, only CYP2C19 genotype has been found to have an impact on the elimination constant of cyclophosphamide at a dose <1,000 mg/m2 [4].
Data on the relationship between CYP2C19 genotype and efficacy or side effects are rare, e.g., the CYP2C19*2 variant allele was associated with premature ovarian failure in lupus erythematosus–related nephritis [11]. A case report suggested that a fully functional CYP2C19 and a compromised GST render the recipient excessively exposed to active metabolites of cyclophosphamide leading to severe leucopenia even at small doses of cyclophosphamide [12]. On the other hand, functional ALDH3A1 has been associated with conferring resistance to cyclophosphamide [13].
This study aims to analyze the influence of genotypes of CYP2B6, CYP2C9, CYP2C19, CYP3A5, ALDH31, and GSTA1 on plasma 4-OH-CP as a ratio to CP and iatrogenic leucopenia in a sample of 68 Pakistani breast cancer patients receiving the FAC regimen.
Materials and methods
Patients, treatment regimen, blood sampling, and toxicity assessment
The patients were recruited from two public-sector facilities, covering the majority of the population in Karachi. Ethics committee and institutional review board at Ziauddin University, Karachi, approved the study. After written informed consent, 68 chemotherapy naïve females suffering from infiltrating ductal carcinoma of the breast under care of a consultant oncologist were consecutively included in this study from April to September 2008. All patients received six cycles of FAC chemotherapy as fluorouracil 500 mg/m2, doxorubicin 50 mg/m2, and cyclophosphamide 500 mg/m2 administered on a single day after every 21 days. The chemotherapy was administered only if their baseline (20th postchemotherapy day for cycles 2–6) vital signs, blood glucose, blood counts, liver function, renal function, and cardiac output were in the normal range (data not shown).
Chemotherapy was administered over a total time of approximately 3 h. Doxorubicin and cyclophosphamide were given as “infusion-1” over 30 min followed by fluorouracil as “infusion-2” over 2 h. Peripheral venous blood was collected serially through venipuncture at a suitable site (a) before infusion, (b) immediately after infusion-1, (c) 60 min after infusion-1, and (d) 120 min after infusion-1. Fourteen patients refused to give a sample at 120 min, hence that particular data point is not available for analysis. For each patient, sample (a) served as the control as well as the source for genotyping. An aliquot of 500 μL plasma from samples b, c, and d was immediately added to 50 μL of 2 M solution of semicarbazide hydrochloride to derivatize 4-OH-CP to the stable 4-OH-CP-semicarbazide as described by Huitema et al. [14].
Patient characteristics included age, ethnicity, clinical staging, and histological grading. Patients were followed up on the 10th day and 20th day after receiving chemotherapy. A total leukocyte count (measured on the 10th day postchemotherapy) below 2,500/mm3 was considered severe leucopenia and warranted start of G-CSF or blood transfusion to prevent infections.
Chemicals and reagents
All the chemicals and reagents were of MS grade. Acetonitrile and water were from Merck, Darmstadt, Germany; cyclophosphamide and semicarbazide hydrochloride were from Sigma-Aldrich, Steinheim, Germany; ifosfamide was from Baxter Oncology, Halle, Germany. Since 4-hydroxycyclophosphamide is very unstable, 4-hydroperoxycyclophosphamide (4-OOH-CP), which spontaneously converts into 4-hydroxycyclophosphamide upon dissolving in water, was purchased from IIT GmbH/NIOMECH, Bielefeld, Germany. 4-Hydroxycyclophosphamide semicarbazide derivatization was accomplished by dissolving 4-hydroperoxycyclophosphamide directly in 2 M solution of semicarbazide hydrochloride to yield 1 mg/ml stock solution [14]. This work-up procedure was applied in an identical way to each patient sample.
Pharmacokinetic analysis
CP and 4-OH-CP (as semicarbazide) plasma concentrations were quantified by HPLC-ESI-MS at mass-to-charge ratios (m/z) of 283 and 356, respectively. Ifosfamide was used as internal standard (m/z 283, eluted earlier than CP). The LC-MS platform LCMS-2010EV (Shimadzu, Japan) was used equipped with a Pursuit XRs C18 column (150 × 2.0 mm, 3 micron) and pre-column (Varian, CA, USA). Gradient elution at a total flow rate of 0.2 ml/min was applied [acetonitrile/water: 15% (0–7 min), 30% (7–8 min), 75% (8–9 min), 15% (9–10 min), 15% (10–16 min)].
Plasma samples were purified by solid-phase extraction [15] using Bond Elut C18 cartridges, 1 ml (Varian). After conditioning with 1 ml acetonitrile and 1 ml water, plasma samples were loaded followed by washing with 2 ml water. Finally, samples were eluted with 100% acetonitrile, evaporated to dryness, and reconstituted in 1 ml water.
The minimum limit of detection was 10 ng/ml for cyclophosphamide and 100 ng/ml for 4OH cyclophosphamide applying a signal-to-noise ratio of 3:1. The validation study involved concentrations of 0.125, 0.25, 0.5, and 1.0 μg/ml. The precision (coefficient of variance) was 7% at the lowest and 5% at the highest validation level; the mean accuracy for CP was 90%, whereas for 4-OH-CP, it was 103%. All determinations were performed six times. The analyte stability was confirmed at −80°C over 4 weeks.
Pharmacogenetic analysis
Genotyping for CYP2B6*4 (785A >G; rs2279343), *5 (1459 C >T; rs3211371), and *6 (516 G >T; rs3745274 and 785A >G); CYP2C9*2 (430 C >T; rs1799853) and *3 (1075A >C; rs1057910); CYP2C19*2 (681 G >A; rs4244285); CYP3A5*3 (6986A >G; rs776746); GSTA1 –69 C >T; rs3957357 and –52 G >A; rs3957356 (representing GSTA1*A and GSTA1*B haplotypes); and ALDH3A1*2 (985 C >G; rs2228100) was performed by PCR-RFLP or pyrosequencing as described previously [2, 6, 16].
Statistical analysis
The data were analyzed using SPSS® v.17.0. The sample size was calculated under the assumption that a change in metabolic ratios of 1.5 would be clinically significant. A post hoc analysis regarding genotype differences between the present study population and Caucasians revealed a statistical power of 89% using StatCalc® software. Student’s t-test was applied for continuous variables and χ2 (with Yates’ correction for continuity) or Fisher’s exact test for categorical variables. Mann-Whitney U test was applied followed by Jonckheere-Terpstra trend test where appropriate. Univariate/multivariate analysis was used to identify genotypes that significantly predict the outcome. Logistic regression analysis was applied to calculate the risk of severe leucopenia in relation to genotypes. All analyses were two-sided and only a p-value less than 0.05 was considered significant.
Results
The baseline characteristics of 68 consecutive breast cancer patients are presented in Table 1. The mean age (±SD; range) was 44 (±9.7; 25–70) years. All patients received six cycles of FAC chemotherapy protocol. Their hospital stay and pretreatment were similar. All major ethnic groups in Pakistan were represented in this study. There was a tendency to present with advanced disease at an early age. None of the patients presented at stage I, whereas 28% had a node negative disease. All investigated genotypes were in Hardy-Weinberg equilibrium. The genotype frequency data are presented elsewhere [6].
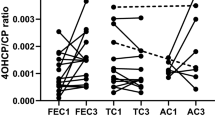
Plasma values of CP and 4-OH-CP are given in Table 2. On stratifying CP pharmacokinetics to single genotypes, neither the elimination of the parent compound CP nor of the metabolite 4-OH-CP was significantly influenced by any genotype of the investigated metabolic enzymes. Also application of the univariate analysis of variance revealed no further statistical significance. However, analysis of the ratio between 4-OH-CP and CP, which reflects the metabolic capacity of the involved enzymes, indicated that the 4-OH-CP:CP ratio after 120 min was significantly affected by CYP2C19*2 (p = 0.039) and ALDH3A1*2 (p = 0.037), and there was also a significant gene-dose effect for CYP2C19 (Jonkheere-Terpstra test, p = 0.045) but not for ALDH3A1 (p = 0.13). The carriers of the wild-type CYP2C19*1/*1 genotype exhibited a 1.19-fold higher ratio than heterozygote and a 1.3-fold higher ratio than homozygote CYP2C19*2 allele carriers (Fig. 1). In contrast, the carriers of the wild-type ALDH3A1*1/*1 genotype had lower 4-OH-CP:CP ratios after 2 h than heterozygotes (factor 0.72) or homozygotes (factor 0.66, Fig. 2). Although CYP2C9 low-activity genotypes also demonstrated lower formation of 4-OH-CP, this finding did not reach statistical significance (Table 3). These effects did not remain significant in a univariate analysis of variance including all genotypes.
To investigate the role of pharmacogenetics in detoxification, the side effect of acquired leucopenia was assessed in relationship to drug metabolizing enzyme genotypes (Table 4). Loss of at least one functional allele of GSTA1 predisposed to severe leucopenia (p = 0.004; OR 4.48; 95% CI 1.58–12.7). This observation remained valid even after binary logistic regression analysis correcting for age and all other dichotomous genotypes (OR 6.94; 95% CI 1.75–27.6, p = 0.006). Additional analyses did not reveal any significant correlation between any 4-OH-CP:CP ratios and presence of leucopenia (data not shown).
Discussion
In vitro studies have shown, though inconsistently, that several cytochrome P450 enzymes including CYP2B6 [3, 17], CYP2C9 [18, 19], CYP2C19 [18, 20], and CYP3A4/5 [19] are capable of 4-hydroxylation of cyclophosphamide.
In a study conducted on lupus nephritis patients who received pulsed low dose cyclophosphamide treatment [11], CYP2C19*2 carriers had a significantly lower risk of developing premature ovarian failure, indicating less bioactivation of cyclophosphamide. Timm et al. [4] demonstrated that metabolic clearance of cyclophosphamide is significantly dependent on CYP2C19 genotype at CP doses <1,000 mg/m2.
In contrast, Nakajima et al. [9] could not demonstrate any effect of CYP2C19 on CP pharmacokinetics in a larger but heterogeneous cohort. Some earlier studies had shown that CYP2C19 has low affinity (high Km) [18, 20], whereas CYP2C9 has high affinity (low Km) hydroxylase activity for cyclophosphamide [18, 19].
In the absence of the limitations encountered by Timm et al. [4] (no 4-OH-CP levels could be determined and no outcome data were available), our study confirms that CYP2C19 influences the bioactivation of cyclophosphamide and shows a gene-dose effect. All patients in our present study were female breast cancer patients who received low dose cyclophosphamide as a part of the FAC protocol. Their baseline hepatic and renal functions were within normal limits, the pretreatment was similar, and they were treated in a similar fashion. The in vivo involvement of CYP2C9 genotypes in cyclophosphamide metabolism seems to have a minor impact. In our study, CYP2C9 genotypes showed a trend similar to CYP2C19 but did not reach statistical significance, confirming observations by Timm et al. [4], Ekhart et al. [8], and Xie et al. [10].
Aldehyde dehydrogenases catalyze the formation of the inactive carboxyphosphamide from aldophosphamide thus contributing to detoxification. Therefore, it could be hypothesized that increased expression of ALDH3A1 may play a role in the development of tumor resistance. Indeed, transfection of ALDH3A1 expression vectors has been shown to decrease the sensitivity to cyclophosphamide [21], but Ekhart et al. [8] could not find an effect of ALDH3 genotypes on the pharmacokinetics of 4-OH-CP in a cohort of patients treated with high dose cyclophosphamide.
However, we could demonstrate that 4-OH-CP:CP ratio after 120 min was significantly influenced by ALDH3A1 in a gene-dose-related manner. After a univariate analysis of variance however, no genotype remained significant, possibly due to the limited sample size. The toxicity of the chemotherapy was analyzed with respect to the leukocyte count. Glutathione conjugation catalyzed by polymorphic GSTA1 contributes to the detoxification of reactive nucleophiles of cyclophosphamide [22]. This is also applicable to doxorubicin. However, there are not enough data that suggest a substantial preventive role of GSTA1 on the doxorubicin-induced cytotoxicity in vivo, especially in the lower doses commonly used in chemotherapy. There is some evidence in vitro for high doses of doxorubicin [23]. Therefore, GSTA1, the most abundant hepatic isoenzyme, was included to assess its effect on cyclophosphamide clearance in this study. In accordance with Ekhart et al. [8], Nakajima et al. [9], and Timm et al. [4], no effect of GSTA1 polymorphism could be demonstrated on plasma drug or metabolite levels. However, we could demonstrate that it strongly influences the myelotoxicity. Carriers of GSTA1*A/*A showed 5.7-fold less likelihood of experiencing a leukocyte drop below 2.5 × 109/L. As a consequence, they also required minimal correction of blood count and needed less vigorous antimicrobial therapy. This significant finding is in contrast to a recent paper by Yao et al. [24] showing no impact of GSTA1 but of GSTP1. In that study however, only one tagging SNP of GSTA1 was investigated. Since GSTs conjugate reactive nucleophiles, loss of function is reflected in terms of the susceptibility to toxicity rather than altering plasma levels of the parent drug. At present, these SNPs may be suitable to estimate or explain adverse effects. Further studies are needed to assign any role in a priori dose adapting.
In conclusion, we could confirm the importance of CYP2C19 as a modulator of 4-OH-CP formation in a single approach. In addition, ALDH3A1 remained the only further significant factor contributing to 4-OH-CP clearance. The myelotoxicity of CP treatment, however, was associated with genotypes of the detoxifying phase-II enzyme GSTA1. Carriers of GSTA1*B were at significant increased risk of leucopenia compared to GSTA1*A/A subjects.
References
Althuis MD, Dozier JM, Anderson WF et al (2005) Global trends in breast cancer incidence and mortality 1973–1997. Int J Epidemiol 34:405–412
Ward S, Simpson E, Davis S et al (2007) Taxanes for the adjuvant treatment of early breast cancer: systematic review and economic evaluation. Health Technol Assess 11:1–144
Xie H, Yasar Ü, Lundgren S et al (2003) Role of polymorphic human CYP2B6 in cyclophosphamide bioactivation. Pharmacogenomics J 3:53–61
Timm R, Kaiser R, Lötsch J et al (2005) Association of cyclophosphamide pharmacokinetics to polymorphic cytochrome P450 2 C19. Pharmacogenomics J 5:365–373
Rowe JD, Nieves E, Listowsky I (1997) Subunit diversity and tissue distribution of human glutathione S-transferases: interpretations based on electrospray ionization-MS and peptide sequence-specific antiser. Biochem J 325(Pt 2):481–486
Afsar NA, Haenisch S, Mateen A et al (2010) Genotype frequencies of selected drug metabolizing enzymes and ABC drug transporters among breast cancer patients on FAC chemotherapy. Basic Clin Pharmacol Toxicol 107:570–576
Riddick DS, Lee C, Ramji S et al (2005) Cancer chemotherapy and drug metabolism. Drug Metab Dispos 33:1083–1096
Ekhart C, Doodeman VD, Rodenhuis S et al (2008) Influence of polymorphisms of drug metabolizing enzymes (CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP3A5, GSTA1, GSTP1, ALDH1A1 and ALDH3A1) on the pharmacokinetics of cyclophosphamide and 4-hydroxycyclophosphamide. Pharmacogenet Genomics 18:515–523
Nakajima M, Komagata S, Fujiki Y et al (2007) Genetic polymorphisms of CYP2B6 affect the pharmacokinetics/pharmacodynamics of cyclophosphamide in Japanese cancer patients. Pharmacogenet Genomics 17:431–445
Xie H, Griskevicius L, Ståhle L et al (2006) Pharmacogenetics of cyclophosphamide in patients with hematological malignancies. Eur J Pharm Sci 27:54–61
Takada K, Arefayene M, Desta Z et al (2004) Cytochrome P450 pharmacogenetics as a predictor of toxicity and clinical response to pulse cyclophosphamide in lupus nephritis. Arthritis Rheum 50:2202–2210
Tran A, Bournerias F, Le Beller C et al (2008) Serious haematological toxicity of cyclophosphamide in relation to CYP2B6, GSTA1 and GSTP1 polymorphisms. Br J Clin Pharmacol 65:279–280
Bunting KD, Townsend AJ (1996) Protection by transfected rat or human class 3 aldehyde dehydrogenase against the cytotoxic effects of oxazaphosphorine alkylating agents in hamster V79 cell lines. Demonstration of aldophosphamide metabolism by the human cytosolic class 3 isozyme. J Biol Chem 271:11891–11896
Huitema ADR, Tibben MM, Kerbusch T et al (2000) High performance liquid chromatographic determination of the stabilized cyclophosphamide metabolite 4-hydroxycyclophosphamide in plasma and red blood cells. J Liq Chrom Rel Technol 23:1725–1744
Liu JJ, Kestell P, Findlay M et al (2004) Application of liquid chromatography-mass spectrometry to monitoring plasma cyclophosphamide levels in phase I trial cancer patients. Clin Exp Pharmacol Physiol 31:677–682
Ufer M, Dilger K, Leschhorn L et al (2008) Influence of CYP3A4, CYP3A5 and ABCB1 genotype and expression on budesonide pharmacokinetics: a possible role of intestinal CYP3A4 expression. Clin Pharm Ther 84:43–46
Roy P, Yu LJ, Crespi CL et al (1999) Development of a substrate-activity based approach to identify the major human liver P-450 catalysts of cyclophosphamide and ifosfamide activation based on cDNA-expressed activities and liver microsomal P-450 profiles. Drug Metab Dispos 27:655–666
Chang TK, Yu L, Goldstein JA et al (1997) Identification of the polymorphically expressed CYP2C19 and the wild-type CYP2C9-ILE359 allele as low-Km catalysts of cyclophosphamide and ifosfamide activation. Pharmacogenetics 7:211–221
Ren S, Yang J-S, Kalhorn TF et al (1997) Oxidation of cyclophosphamide to 4-hydroxycyclophosphamide and deschloroethylcyclophosphamide in human liver microsomes. Cancer Res 57:4229–4235
Griskevicius L, Yasar Ü, Sandberg M et al (2003) Bioactivation of cyclophosphamide: the role of polymorphic CYP2C enzymes. Eur J Clin Pharmacol 59:103–109
Sladek NE (1999) Aldehyde dehydrogenase-mediated cellular relative insensitivity to the oxazaphosphorines. Curr Pharm Des 5:607–625
Dirven HA, van Ommen B, Van Bladeren PJ (1994) Involvement of human glutathione S-transferase isoenzymes in the conjugation of cyclophosphamide metabolites with glutathione. Cancer Res 54:6215–6220
L’Ecuyer T, Allebban Z, Thomas R et al (2004) Glutathione S-transferase overexpression protects against anthracycline-induced H9C2 cell death. Am J Physiol Heart Circ Physiol 286:H2057–H2064
Yao S, Barlow WE, Albain KS et al (2010) Gene polymorphisms in cyclophosphamide metabolism pathway, treatment-related toxicity, and disease-free survival in SWOG 8897 clinical trial for breast cancer. Clin Cancer Res 16:6169–6176
Acknowledgments
The research grant was provided by Higher Education Commission Pakistan and Ziauddin University, Karachi, Pakistan. The authors are thankful to all the administration and staff of Karachi Institute of Radiotherapy and Nuclear Medicine (KIRAN) and Jinnah Postgraduate Medical Center (JPMC), Karachi, Pakistan, as well as Dr. Rubina Ghani, Molecular and Pathological Laboratories, Karachi; Prof. N.A. Jafarey, Ziauddin University, Karachi; Dr. Sandra Lächelt, Dr. Denisa May, Jutta Finger, and Marion Pauer of the Institute of Experimental and Clinical Pharmacology, CAU, Kiel, for their support.
The study is dedicated to Dr. Muhammad Noorul Afsar [1931–1991; MBBS (Dhaka, Bangladesh), DTM&H (Liverpool, UK)] for his philanthropic general practice and life-long support of rational drug use.
Conflict of interest
It is declared that there is no conflict of interest regarding this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Afsar, N.A., Ufer, M., Haenisch, S. et al. Relationship of drug metabolizing enzyme genotype to plasma levels as well as myelotoxicity of cyclophosphamide in breast cancer patients. Eur J Clin Pharmacol 68, 389–395 (2012). https://doi.org/10.1007/s00228-011-1134-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00228-011-1134-0