Abstract
According to the EU and Swiss legislation for food, only approved traits of transgene plants are allowed to be imported and sold to the consumer. In order to control imports of rice and rice products from retailers, dealers and importing companies, efficient and reliable methods for the detection and quantification are a prerequisite. Therefore, a novel pentaplex real-time polymerase chain reaction system was developed and validated for the quantitative determination of three genetically modified rice lines at once. This system simultaneously determines DNA contents of the phospholipase-gen, a rice species specific gene, the 35S:BAR-construct, as the promotor of different transgene rice lines and the specific systems for LL62, LL601 and Bt-63-rice (Shanyou63). The test exhibits a good specificity and sensitivity for the transgenes in the range of 0.01–1%. It proved its efficiency and reliability in daily routine. Due to the lack of appropriate reference material for the Bt-63-rice, a reference oligonucleotide was artificially constructed. This oligonucleotide proved its applicability in diagnostic analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
On 31 August 2006, public laboratories found at Rotterdam in a cargo ship containing US long grain rice, traces of transgenic rice. Further analyses revealed that this rice corresponds with the not so far in US officially approved transgenic rice line LL601 from Bayer crop science. At that moment, the closely related line transgenic rice LL62 has been commercialised in the US and Canada already. The LL601 rice as well as the later in rice noodles detected transgenic Bt-63-rice (Shanyou63) from China was solely used for experimental field studies and not for human consumption. All of the transgenic rice lines are neither approved in the European community nor in Switzerland. Switzerland imports rice mainly from the European countries and from USA. But rice and rice products are also imported from eastern countries in high quantities. As a reaction, the European and Swiss authorities decided to implement a severe control of rice and rice products from the US and China in order to protect consumers from illegal transgenic rice and its products. As only an efficient diagnostic tool would enable laboratories to perform these investigations in a reasonable time and to reasonable costs, a novel pentaplex quantitative polymerase chain reaction (PCR) system called AllRice, to detect and quantify these three transgenic rice lines was developed.
Materials and methods
Plant material and DNA samples
Plant material from LL62-rice was obtained from American Oil chemists’ Society DNA samples. Plant material and DNA samples from LL601-transgene rice lines were kindly provided by the Federal Office of Public Health. Due to lack of standard material (Bt-63-rice) or insufficient amounts of reference material (LL601), an artificial standard was created.
Artificial standards
For the transgenic rice Bt63, no transgenic plant material was available. Therefore, the only solution for this lack was to produce an artificial reference material as reported in the past already [8–15, 17]. As the amplicon for the transgenic rice Bt63 is only 84 bp long, it was possible to synthesise two overlapping oligonucleotides and to amplify this fragment using the published primers (for details see Fig. 1).
This artificial DNA standard was calibrated against non-transgenic rice DNA using its additionally cloned part containing the gene for phospholipase enclosed in all rice genomes. Parallel amplification lead to the factor for the dilution of the artificial standard to assign the appropriate percentage. Dilutions of this calibrated artificial standard solutions were used for the quantification of the samples. It is not known how many insertions of the transgenic cassette, the transgenic line Bt63 comprises [1, 2]. Therefore, if the transgenic line Bt63 comprises more than one introduction site, the quantitative results presented here may overestimate the real percentage of the transgenic rice in the sample.
The equal procedure for the construction was used to create an artificial standard enclosing the target sequence for LL601-rice and the target sequence of the phospholipase-PCR system. The amount of reference material received for LL601 was sufficient to calibrate the artificial DNA standard. This was done by amplification of dilution rows of the genomic DNA of LL601 (100%) and the artificial standard for LL601. The factor gained was used to dilute the artificial standard to the appropriate percentage used for the validation and the routine diagnostic.
For routine diagnostic, these artificial standards were applied to generate quantitative results. The amounts obtained after construction is immense, posing also a high risk for contamination. To reduce this, risk-only dilutions of at least 1:1,000 were handled.
DNA extraction
DNA extraction from all sample matrices was performed using a Wizard Plus Minipreps DNA purification system (Promega, Madison, USA). Usually 200 mg of grinded sample material was extracted and DNA was eluted in 50 μl elution buffer according to the producers manual. The concentration was determined photometrically and adjusted by dilution to 20 ng/μl.
Primers and probes
The primers and probes were taken from published single PCR systems (details see Table 1). All primers and probes were synthesised by Microsynth AG, Balgach, Switzerland. Labelling of probes with fluorescent markers was done according to the recommendations of the Rotorgene 6000 manual and are listed in Table 1.
Real-time PCR procedure
5 μl DNA extracts were added to 20 μl of reaction mix containing QuantiTect multiplex PCR no ROX Master Mix (Qiagen AG, Germany), and all primers and probes listed in Table 1. For final concentrations see Table 1. PCR was performed on a Rotorgene 6000 real-time system (Corbett, Australia) according to the following cycling protocol: Initial step of 15 min at 95 °C; followed by 45 cycles of 10 s at 95 °C and 1 min at 60 °C (according to the recommendations for the QuantiTect multiplex PCR no ROX MasterMix).
Results
Design of the multiplex real-time PCR Systems
For all applied PCR systems specificity is prerequisite. After checking primers for unspecific primer sites in the NCBI-databases by the Beacon-designer 5.1 software (Premier Biosoft, Palo Alto, CA, USA), they were first tested without probes in the single SYBR-green format. This approach is useful to detect amplification of unspecific products. After this, a first check of the specificity was performed. Therefore, DNA of several related plant species was isolated and used as template for the PCR reaction. In addition, a dilution row of each target revealed the amplification efficiency, which is crucial to reach maximal sensitivity for each target. If these first results were showed no cross-reactivity and amplification efficiencies close to 1, TaqMan-probes were ordered and labelled according to the channels of the Rotorgene 6000. We choose FAM, JOE, ROX, Cy5 and DY681 labelled TaqMan-probes (for details see Table 1).
Specificity
As one of the first steps establishing these PCR systems, all primers and probes were checked for relevant homologies by BLAST nr search within GenBank databases. To test the specificity extensively, DNA from a wide range of animals and other food ingredients was isolated. DNA of the following organisms were isolated and tested by this pentaplex PCR system: beef, trout, white beans, lentils, kidney beans, mung beans, runner beans, chickpeas, peas, beans, wheat, tomato, potato, rice, plum, apricots, peanut, hazelnut, almonds walnut, lupinus albus, lupinus augus, onion, garlic, carrot celery, chive, nutmeg, white pepper, cinnamon, aniseed, coconut and paprika. The cross-reactivity was measured as signal in % which a non-target DNA simulates (100% equal to 100 ng). Cross-reactivity above 1% emerged surprisingly for tomato. The cross-reactivity in the phospholipase-gen-system (rice) was 94% for tomato. However, it is unlikely to have tomato in raw rice but for products composed by many different ingredients this has to be considered. Additionally, several other transgene plants were tested for cross-reactivity: Corn (Bt176, Bt11, Mon810, GA21, T25, NK603, CBH-351, Mon863, TC1507, Mon810x683) and Rondup-Ready soy. Only CBH-351-corn lead to a 3.7% signal in the 35S:Bar-system. This must be considered when analysing mixed products of rice and maize.
Amplification characteristics
The amplification characteristics like amplification efficiency and correlation to the concentration of the target DNA was investigated using the multiplex row of standards (Table 2). This standard row simulates disproportional target concentrations leading to the numbers presented in Table 3. The system for LL62 shows an underperformance for the amplification efficiency which was also found in the single PCR modus (data not shown). We consider this to suboptimal primer sites. However, this system is an official method validated by the JRC [5] and therefore we decided to apply it.
Sensitivity, precision and uncertainty
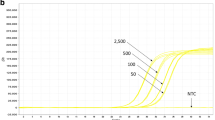
To evaluate the sensitivity, DNA extracts of each target species (DNA concentration 20 ng/μl) were diluted in half-logarithmic steps. Herring sperm DNA solution (20 ng/μl) was used as diluent. These single analyte dilution rows were mixed according to Table 3 to produce a multiplex standard, simulating calibration samples with signals for all five analytes. Each data-point was analysed six times (N = 6) during four runs. Data were collected and the relative standard deviation for the estimation of the precision and for the accuracy the mean deviation from the true value was taken (see compilation Table 4). The PCR systems detecting the transgene signals 35S:Bar, LL62 and Bt-63-rice 0.01% of target DNA was reliable detected, for LL601 even 0.0032% target DNA was detected reliable. None of the six negative controls showed a positive signal.
The quantification performance and range are shown in Table 4.
Due to the fact that the number of insertions is unknown, results for Bt-63-rice have to be interpreted carefully. However, artificial standards once created are stable and available in high amounts. They enable laboratories to solve principal problems due to the lack of reference material with reasonable input in a short time. Unfortunately, only one ringtrial was organised from Fapas (GeMD16) for LL62-rice including z scores. The assigned level was 1.17%. Applying the AllRice-system, we measured 1.5% leading to a satisfactory z score of 0.5.
Analysis of commercial samples
Fifteen rice-products were analysed. Three of them gave reproducible signals for traces of LL601-rice. Also signals for Bt-rice were detected, but due to the low concentration of target sequence, none of these signals were reproducible. In addition, processed products like rice-noodles were analysed for the presence of transgene rice. Of 15 samples most signals were below the quantification range for LL601. Signals for Bt-63-rice were detected but were not reproducible and/or below the quantification range. In total, only one product (Table 5: Parboiled Rice Liberty Gold, USA) gave reproducible signals for the LL601 and, as a confirmation, the 35S:Bar-construct was detected in this sample in addition. Therefore, this sample was considered to be GMO-positive according to the regulations [16]. The phospholipase-gene system that is enclosed in this pentaplex PCR system showed for all samples a positive signal. In addition, we diluted every sample to check if the results are proportional to the input-DNA. The column-based DNA cleanup in conjunction with these two tests confirm that no sample was inhibiting and therefore leading to false-negative results. In summary, this multiplex PCR system produced clear results without laborious repeated pipetting. Inconsistent results were produced in the range below the detection limit, which has to be expected. The accuracy of the here presented multiples PCR system has to be investigated further when certified reference material becomes available and in future ringtrials.
Conclusion
We state, that the pentaplex PCR system presented here, gives quantitative results with high sensitivity showing that multiplex quantitative results can be generated for five target sequences at once. Additionally, it was demonstrated that artificial standards as amplicons can be created rapidly and make analysis possible in cases where reference material is limited or not available at all. This multiplex quantitative real-time PCR system enables laboratories to process many sample in a reliable and cost-effective manner.
References
Yang L et al (2005) Estimating the copy number of transgenes in transformed rice by real-time quantitative PCR. Plant Cell Rep 23:10–11
Prior FA, Tackaberry ES, Aubin RA, Casley WL (2006) Accurate determination of zygosity in transgenic rice by real-time PCR does not require standard curves or efficiency correction. Transgenic Res 15:261–265
Grain testing method for the detection of rice GM event LLRice601 using RT-PCR protocols PGS0505 and PGS0476 (2006) Molecular and Biochemical Analytical Services, Bayer BioScience N.V., Belgium
Report on the Verification of a Construct-Specific Detection Method for Identification of GM-Events Containing P35SBAR using a Real-time PCR Assay, European Commission JRC
Event-specific method for the quantification of rice line LLRice62 using real-time PCR, European Commission JRC. http://gmo-crl.jrc.ec.europa.eu/statusofdoss.htm
Report on the Verification of an Event-specific Detection Method for the Identification of Rice GM-event LLRice601 Using a Real-time PCR Assay, European Commission JRC. http://gmo-crl.jrc.ec.europa.eu/statusofdoss.htm
Mäde D, Degner C, Grohmann L Detection of genetically modified rice: a construct-specific real-time PCR method based on DNA sequences from transgenic Bt rice, Landesamt für Verbraucherschutz Sachsen-Anhalt Dezernat 33, Freiimfelderstr. 66/68, 06112 Halle, Deutschland
Legoux P, Minty C, Delpech B, Minty AJ, Shire D (1992) Simultaneous quantitation of cytokine mRNAs in interleukin-1 beta stimulated U373 human astrocytoma cells by a polymerisation chain reaction method involving co-amplification with an internal multi-specific control. Eur Cytokine Netw 3(6):553–563
Biosmart GmbH (1999) Swiss Patent 698 698, 21 Dec 1999
Burns M, Corbisier P, Wiseman G, Valdivian H, McDonald P, Bowler P, Ohara K, Schimmel H, Charels D, Damant A, Harris N (2006) Comparison of plasmid and genomic DNA calibrants for the quantification of genetically modified ingredients. Eur Food Res Technol 224:249–258
Kuribara H, Shindo Y, Matsuoka T, Takubo K, Futo S, Aoki N, Hirao T, Akiyama H, Goda Y, Toyoda M, Hino A (2002) Novel reference molecules for quantitation of genetically modified maize an soybean. J AOAC Int 85(5):1177–1189
Block A, Schwarz G (2003) Validation of different genomic and cloned DNA calibration standards for construct-specific quantification of LibertyLink in rapeseed by real-time PCR. Eur Food Res Technol 216:421–427
Pardigol A, Guillet S, Pöpping B (2003) A simple procedure for quantification of genetically modified organisms using hybrid Amplicon standards. Eur Food Res Technol 216:412–420
Tavernier I, van Bockstaele E, de Loose M (2004) Cloned plasmid DNA fragments as calibrators for controlling GMO’s: different real-time duplex quantitative PCR methods. Anal Biochem 378:107–1198
Mattarucchi E, Weighardt F, Barbati C, Querci M, Van den Eede G (2005) Development and applications of real-time PCR standards for GMO quantification based on tandem-marker plasmids. Eur Food Res Technol 221:511–519
Directive 11 from the Swiss Federal Office of Public Health, 15 Dec 2006
Babekova R, Funk T, Pecoraro S, Engel K-H, Busch U (2008) Development of an event-specific real-time PCR detection method for transgenic Bt rice line KMD1. Eur Food Res Technol 228:707–716
Acknowledgments
We thank the Federal Office of Public Health for providing of reference material and the Cantonal Laboratory of Zürich for providing the resources for this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Köppel, R., Zimmerli, F. & Breitenmoser, A. Multiplex real-time PCR for the simultaneous detection and quantification of DNA from three transgenic rice species and construction and application of an artificial oligonucleotide as reference molecule. Eur Food Res Technol 230, 731–736 (2010). https://doi.org/10.1007/s00217-010-1213-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-010-1213-y