Abstract
Nucleic acid enzymes (NAzymes) are a class of nucleic acid molecules with catalytic activity, which can be modulated by the presence of different species such as metal ions, genetic biomarkers, small molecules or proteins, among others. NAzymes offer several important advantages for development of novel bioanalytical strategies, resulting from their functionality as specific recognition elements and as amplified analytical signal generators, making them ideal candidates for developing highly specific bioanalytical strategies for the detection of a wide variety of targets. When coupled with the exceptional features of inorganic nanoparticles (NPs), the sensitivity of the assays can be significantly improved, allowing the detection of targets using many different detection techniques including visual readout, spectrophotometry, fluorimetry, electrochemiluminescence, voltammetry, and single-particle inductively coupled plasma-mass spectrometry. Here we provide an overview of the fundamentals of novel strategies developed to achieve analytical signal amplification based on the use of NAzymes coupled with inorganic NPs. Some representative examples of such strategies for the highly sensitive detection of different targets will be presented, including metal ions, proteins, DNA- or RNA-based biomarkers, and small molecules or microorganisms. Furthermore, future prospective challenges will be discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Nucleic acid enzymes and their potential in bioanalytical applications
Genetic nanomaterials with intrinsic enzyme-like properties have attracted growing interest due to their striking advantages over traditional enzymes, such as low cost, easy surface modification, high stability and robustness, and tunable activity [1]. These features enable them to be considered as a potent substitute for natural enzymes to construct novel analytical platforms for detecting various analytes, from small molecules to proteins and cells. In this vein, researchers have recently been exploiting both natural and synthetic properties of nucleic acids, including catalysis by nucleic acid enzymes (NAzymes) for the development of novel and improved bioanalytical strategies and sensors [2]. NAzymes are composed of single-stranded sequences of either DNA (deoxyribozymes or DNAzymes) or RNA (ribozymes), organized into domains required for enzymatic activity (catalytic core domains) and substrate recognition (substrate binding domains) [3, 4]. These NAzymes are capable of catalyzing a broad range of chemical reactions, including cleavage, ligation, phosphorylation, or deglycosylation of DNA or RNA. Catalytic nucleic acids, sometimes in combination with aptamers, have been studied extensively for their capacity to provide the basis for biosensors.
The most widely studied NAzymes are those with cleaving capability of RNA substrates, producing the rupture between two RNA bases, and resulting in two shorter different sequences. The sequences of these DNAzymes were generated in vitro by prior selection from a library of DNA sequences. Their structure consists of a central sequence, which makes up the catalytic core, and a sequence that is complementary to the substrate containing the RNA bases to be cleaved (see Fig. 1). Using appropriate labels at both ends of the substrate, the scission produced can generate a change in the analytical signal [5, 6].
Scheme of RNA-cleaving NAzyme conformations. Left: A DNAzyme with cleaving capability is represented in green. When a substrate containing two RNA bases (in blue) hybridizes with the DNAzyme, the cleavage takes place, generating two shorter sequences that are easily displaced. Hence, a new substrate molecule can bind the DNAzyme to start the process again. P1 and P2 are two probes located at both ends (nanoparticles [NPs], dyes, fluorophores, etc.). Changes in the distance between them will serve to monitor the presence of substrate. Right: An MNAzyme is represented in green. The catalytic core has been divided into two parts, and another sequence (complementary to the target sequence) was introduced at the bottom. The behavior is similar to the DNAzyme on the left, but the presence of a target is needed to activate the catalytic activity of the MNAzyme
In addition, to increase the versatility and utility of NAzymes, Mokany et al. developed in 2010 multicomponent complexes, known as multicomponent nucleic acid enzymes (MNAzymes), which produce amplified “output” signals in response to specific “input” signals (a certain target sequence). MNAzymes were developed by breaking the catalytic core into two parts (partzyme A and partzyme B) and including new sequences in each partzyme [7]. Therefore, MNAzymes have two different specific recognition areas, the substrate-binding arm, which interacts with the substrate to be cleaved, and a second recognition area commonly called sensor arm, that is complementary to the target. The partzymes assemble into active MNAzymes only in the presence of a specific assembly species such as a target nucleic acid. Once formed, MNAzymes catalytically cut the substrate through the RNA bases, generating an amplified signal that heralds the presence of the target while leaving the target intact (Fig. 1) [8].
The catalytic core of the enzyme is made of about 13 or 15 nucleotides that require divalent metal cations for their operation. Depending on the nucleotide sequence of the catalytic core, specific cations are associated with the activity of the enzyme, such as Pb2+, Mg2+, Cu2+, Zn2+, and Hg2+, among others [9]. Hence, target-specific NAzymes are active under certain assay conditions that have to be customized for each NAzyme, such as different media and activation temperatures.
Besides the aforementioned and widely used DNAzymes with cleaving catalytic capabilities, DNAzymes made of guanine-rich oligonucleotides can form a four-stranded structure (known as G-quadruplex or G4) which exhibits a rather different catalytic activity. In these structures, G bases are hydrogen bonded to each other forming planar conformations known as the G-tetrad [10, 11]. Due to van der Waals interactions, ligands such as metalloporphyrins are able to stack on the terminal G-tetrads of the G-quadruplex. An iron porphyrin, typically hemin (the active center of heme-proteins as peroxidase), is the most studied metalloporphyrin in this field because of its ability to bind to the G-quadruplex with high affinity, and also due to its peroxidase-like activity as consequence of the reversible transformation of Fe(III)/Fe(II) [12]. Such hemin/G-quadruplex complexes with peroxidase-like activity are in fact a new kind of DNAzyme that, as will be summarized in the bioanalytical applications section, are mainly exploited for the detection of proteins [13].
Such special features offered by NAzymes, including high stability, extraordinary catalytic activity, and the possibility of being tailored with high specificity against a certain target, make them a promising tool to be exploited in different areas, including clinical biosensing, environmental monitoring, or food control.
Besides the inherent advantages of NAzymes, the synergistic use of inorganic NPs as labels for analytical detection can also add value to improve the sensitivity of the detection assay. Because of this, most of the applications combining NAzymes and NPs take advantage of the optoelectronic characteristics of gold NPs (AuNPs), but other types of NPs have been also evaluated, including platinum NPs (PtNPs), quantum dots (QDs), magnetic NPs (MNPs), or cerium oxide NPs, among others. The different types of NPs introduce different valuable optoelectronic and physical properties that significantly improve the sensitivity of the bioanalytical methods.
In the following sections, selected bioanalytical applications based on the use of DNAzymes and MNAzymes coupled to NPs will be presented. First, developments of DNAzymes coupled to NPs for sensitive detection of different biochemical targets will be summarized. Second, some bioanalytical applications of MNAzymes, using different types of NPs as labels, will be presented. The section devoted to bioanalytical applications of DNAzymes and NPs has been classified according to the type of analyte detected, because the number of publications in this topic is higher, and therefore, a more general information is provided. Nevertheless, Table 1, which includes the main figures of merit for each publication has been included to show more detail of each publication. Conversely, the section related to the bioanalytical applications of MNAzymes with NPs has been classified according to the type of NPs used, and provides more information about each publication because the number of publications is much lower when compared to those based on DNAzymes, and the novelty of the strategies is higher.
(Bio)analytical applications based on DNAzymes and NPs
Detection of metals
Interestingly, Pb2+ has played an important role in nucleic acids chemistry. This explains that, since 2000, DNA has been extensively used for selective detection of Pb2+. Such an ion can serve as the most active cofactor for RNA-cleaving DNAzymes. In addition, Pb2+ can tightly bind to various G-quadruplex sequences, inducing their unique folding and binding to other molecules.
Therefore, the detection of Pb2+ ions is undoubtedly one of the most investigated analytical applications based on the combination of DNAzymes and NPs, and it constitutes nearly 30% of the research articles published on this topic. In addition, among those publications, most of them employ AuNPs, and are based on the cleavage catalytic activity that takes place in the presence of Pb2+ ions, where a substrate strand can be cleaved at its ribonucleotide adenosine (rA) site. The strategies evaluated so far for the detection of Pb2+ are diverse, and it is not the purpose of this review to examine all of them, but to provide an overview of the main approaches developed based on different detection techniques. In this sense, surface plasmon resonance (SPR) sensors have been described using AuNPs, where the DNAzyme and substrate assembly are immobilized onto a gold solid film, containing a AuNP at the other end. The presence of Pb2+ activates the catalytic activity of the DNAzyme, and cleavage of the substrate takes place. This is the basis of a pioneer work where the cleavage of the substrate gives rise to a conformational change, and the AuNPs fall closer to the gold film, increasing the measured SPR signal [14]. In an alternative approach, the AuNPs are removed from the surface of the sensor, decreasing the localized surface plasmon (LSP) and the surface plasmon polariton (SPP) coupling [15].
There are also several publications that describe the detection of Pb2+ ions using electrochemical techniques. These methods are mainly based on the immobilization of the DNAzyme and substrate on a surface, such as graphene oxide, and the presence of Pb2+ ions produces a conformational change that can be followed through by differential pulse voltammetry (DPV) assisted by ferricyanide ([Fe(CN)6]3−/4−) [33]. Another example makes use of a PtNPs/TiO2/α-Fe2O3 composite and the DNAzyme immobilized onto a gold electrode. The presence of Pb2+ ions activates the cleavage of the DNAzyme, and the nanocomposite is deposited onto the electrode surface, increasing the signal measured by DPV [34]. This strategy was successfully applied to the detection of Pb2+ ions in samples of lake water and human serum.
Development of lateral flow tests (LFTs) to carry out the visual detection of lead ions has been also described, achieving a low detection limit of just 1 nM with the naked eye after a 1-h incubation step for the sample containing Pb2+ with the DNAzyme, and a second step of 15 min after the addition of the mixture on the pad of the strip for developing the assay [16].
The aforementioned applications for Pb2+ detection, which are based on ultraviolet–visible (UV-Vis) spectrophotometry, fluorimetry, or electrochemical techniques, achieve low detection limits on the nanomolar to picomolar order. When compared to the most commonly employed elemental techniques, including inductively coupled plasma mass spectrometry (ICP-MS), atomic absorption spectrometry (AAS), and inductively coupled plasma atomic emission spectrometry (ICP-AES), the limits of detection achieved are in the nanomolar range [55,56,57]. Hence, coupling DNAzymes with NPs has allowed for development of analytical methods with similar or lower detection limits without requiring the expensive and complex equipment used in the abovementioned elemental detection techniques.
DNAzyme-based sensors have been developed to detect other metal ions besides Pb2+, including Mg2+, Hg2+, Cu2+, Zn2+, Cd2+, and UO22+. Due to the extremely low detection limits achieved, the detection of uranyl ions based on the use of DNAzymes merits special attention, being highly competitive over other conventional detection systems (e.g., atomic mass spectrometry). In a similar manner to Pb2+ detection, studies in which DNAzymes were used for the detection of uranyl ions are mostly based on the use of AuNPs. A very simple approach is the use of AuNPs decorated with a uranyl-specific DNAzyme. The presence of UO22+ produces the cleavage of the DNA on the surface of the NPs, decreasing the stability of the AuNPs. The aggregation of the NPs is measured through resonance light scattering, achieving a limit of detection on the order of 4 nM [17]. A wireless magnetoelastic sensor with a similar detection limit was also developed for highly selective detection of uranyl ions. In this work, the presence of UO22+ produces the cleavage of the substrate strand of the DNAzyme to generate a single-stranded DNA (ssDNA) that is adsorbed onto the graphene oxide film, giving rise to a decrease in the resonance frequency of the magnetoelastic sensor [18]. Finally, a similar approach based on the lower stability of AuNPs after cleavage of the surface DNA but using colorimetric detection was also published, with a limit of detection around 50 nM [19].
Hg2+ sensing based on DNAzymes has also been proposed based on a highly effective strategy that merits a special highlight. The approach is based on the well-known thymine-Hg(II)-thymine (T-Hg2+-T) interaction, that generates the binding of two DNA strands to form the enzyme strand DNA (E-DNA). A fluorophore-labeled hairpin is located on the surface of the AuNPs. Hence, in the absence of Hg2+ a low fluorescence signal is observed due to a Förster resonance energy transfer (FRET) phenomenon between the fluorophore and the AuNPs. The presence of Hg(II) produces the E-DNA which binds to the hairpin to form a Mg(II)-dependent DNAzyme structure. Because the release of the fluorophore gives rise to a recovery of the fluorescence, the circular cleavage of the hairpins results in amplification, achieving a detection limit of 80 pM [20].
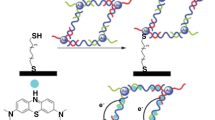
Electrochemical detection of Hg2+ was also carried out using Au/Ag core/shell NPs and T-Hg2+-T interactions. In this study, the NPs were surface-modified with different DNA sequences: one generated the G-quadruplex DNAzyme, and another complementary to the DNA sequence immobilized onto the gold electrode surface. In the presence of Hg2+, a double-stranded DNA structure is generated between the DNA immobilized onto the electrode and the DNA decorating the NPs as a consequence of the T-Hg2+-T interactions. Hence, DNAzymes are close to the surface of the electrode, and oxidation of H2O2 can be easily measured (see Fig. 2). The excellent sensitivity achieved in this work, with a limit of detection of 6 ng L−1, is due to several factors: (i) the bimetallic Au/Ag core/shell NPs can enhance the electrocatalytic oxidation of H2O2; (ii) the G-quadruplex DNAzyme employed also catalyzes the oxidation of H2O2; and (iii) the large surface area allows each NP to contain multiple DNAzymes [39].
Scheme of the Au/Ag core/shell NPs, surface-modified with the hemin/G-quadruplex DNAzyme and the DNA (green arrow) complementary to the sequence immobilized onto the electrode surface (purple arrow). The presence of Hg2+ generates the double-stranded DNA, bringing the NPs and the DNAzyme close to the surface of the electrode where the catalyzed oxidation of H2O2 is measured [39]. This figure is licensed under a CC BY 4.0 license and can be found at https://creativecommons.org/licenses/by/4.0/. It is attributed to Yanling Zhao and Xianmei Xie, and the original version can be found at https://doi.org/10.21577/0103-5053.20170133
The aforementioned examples demonstrated the utility of DNAzymes for metal sensing taking advantage of the metal-dependent activity of some DNAzymes. It is worth mentioning that the same principle has been exploited for intracellular imaging of metal ions. It is well known that simultaneous detection of Zn2+ and Cu2+ is attracting significant interest due to the important roles they play in regulation of biological systems. Hence, a two-color fluorescence nanoprobe was designed based on the use of AuNPs that were surface-modified with two DNAzymes specific to those ions, and their corresponding substrates were labeled with different fluorophores at the 5′ end (see Fig. 3; FAM for the Zn substrate, and Cy5 for the Cu substrate). In this nanoassembly, the fluorescence emission is quenched by the AuNPs and the quencher molecule. The presence of Zn2+ or Cu2+ causes the cleavage of the substrate. Thus, the distance between the fluorophore and both the quencher and the AuNP increases, producing an increase of the fluorescent signal that can be correlated with both the concentration and location of the intracellular Zn2+ and Cu2+. It should be noted that the use of AuNPs presents advantages beyond the obvious quenching efficiency. Because AuNPs present high stability in biological systems, they are capable of entering cells without the need for transfection agents, and are resistant to degradation [21].
Scheme of the simultaneous imaging of Zn2+ and Cu2+ in living cells based on DNAzyme-modified AuNPs [21]. Reprinted with permission from Analytical Chemistry 2015 87 (9), 4829–4835, Copyright © 2015 American Chemical Society
Recently, in an attempt to overcome the limitations of traditional one-photon fluorescent probes for intracellular applications, a two-photon fluorescent probe was designed to increase the light penetration depth and to minimize tissue autofluorescence and photodamage. The experimental design uses a two-photon fluorophore as a label, presenting a luminescence emission at 490 nm and a near-infrared (NIR) light as an excitation source (760 nm). With this strategy, the imaging of Zn2+ was successful, achieving tissue penetration up to 160 μm and excellent photostability (no significant light drift was observed even after 100 min of continuous irradiation). This method provides a superior strategy for designing other probes for metal ion imaging in living cells [23].
Detection of proteins
Many applications described in the literature using DNAzymes for the detection of proteins are based on the peroxidase mimic G-quadruplex DNAzyme. Among them, the detection of thrombin has been widely studied, using a specific aptamer to perform the recognition of the target in combination with different types of NPs, including AuPdNPs [31], AuNPs [25], or MNPs [41] to achieve the electrochemical or optical detection readouts. Among these works, the first one describes an interesting mode to obtain triple amplification, and consequently, high sensitivity for the voltammetric detection of thrombin. For this purpose, AuPdNPs surface-modified with horseradish peroxidase (HRP) and with a thrombin aptamer that can react with hemin to form a hemin/G-quadruplex DNAzyme were linked onto poly(o-phenylenediamine) microspheres (a redox-active material). AuNPs were bioconjugated to a thiolated thrombin aptamer and then electrodeposited onto a glassy carbon electrode (GCE). In a sandwich-type assay, the aptamer immobilized onto the GCE recognizes and captures the thrombin, and then, the solution containing the HRP/hemin/G-quadruplex/AuPdNPs is added, and specific recognition between the immobilized thrombin and the aptamer takes place. In this assay, HRP, the AuPdNPs, and the DNAzyme can catalyze the electrochemical reaction of poly(o-phenylendiamine) in the presence of H2O2, and the peak current, related to the amount of thrombin, is greatly amplified, achieving a limit of detection of 20 fM [31].
There are also few examples in the literature based on RNA-cleaving DNAzymes for the detection of proteins involving a thrombin aptamer as recognition element. For instance, ZrNPs [40] and CdSe@ZnS QDs [36] were combined with DNAzymes to perform electrochemical or electroluminescent detection, respectively. In this sense, a DNA hairpin composed of an 8-17 DNAzyme and a thrombin aptamer was built to carry out the thrombin recognition (see Fig. 4). Hence, the presence of the target opened the hairpin, and the DNAzyme started to cleave the substrate, constituting the first step of amplification. Those cleaved fragments were then available to open the capture DNA which was immobilized onto the gold electrode. Consequently, the capture DNA could also interact with biobar-coded AuNPs-CdSeTe@ZnS QDs to generate the electrochemiluminescence signal, achieving a second amplification step. This strategy resulted in a limit of detection for thrombin as low as 0.28 fM in serum samples, and obtaining ideal results in terms of selectivity as well.
Schematic illustration of the dual signal amplification strategy-based electro-chemiluminescence (ECL) aptasensor for detection of thrombin [36]. In the upper part of the figure, a hairpin DNA containing a thrombin aptamer and a DNAzyme is shown. The presence of thrombin opens the hairpin, so the substrate binds to the DNAzyme, that in the presence of Zn ions cleaves the substrate, which is liberated, and the DNAzyme is available to bind a new substrate sequence (first amplification step). In the lower part of the figure, a capture DNA probe is immobilized onto the surface of a gold electrode. The presence of the cleaved substrate opens the capture DNA that can now interact with the biobar-coded AuNPs-CdSeTe@ZnS QDs. Each AuNP contains many CdSeTe@ZnS QDs, which are the ECL signal probes that constitute the second amplification step. Reprinted with permission from ACS Appl. Mater. Interfaces 2015, 7, 1, 696–703, Copyright © 2014 American Chemical Society
Besides the most widely studied thrombin, some other proteins have been detected using a G-quadruplex DNAzyme. Most of the methods proposed exploit the peroxidase-mimic activity that in the presence of hemin generates H2O2. The resulting hydrogen peroxide is further employed to produce the oxidation of the colorless ABTS [2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate)], generating a blue-green product that will be employed for the colorimetric detection of tumor biomarker MUC1 [42], or the visual detection of myoglobin [26]. In other studies, the H2O2 is used to generate a chemiluminescent signal in the presence of luminol-p-iodophenol that is registered with a charge-coupled device (CCD) camera for the detection of cardiac troponin T [37]. Last but not least, G-quadruplex DNAzymes coupled with AuNPs have been also employed to electrochemically detect proteins such as carcinoembryonic antigen (CEA), where the generation of H2O2 has been exploited to perform voltammetric [35] or impedimetric [29] detection of CEA.
Detection of DNA/RNA biomarkers
The use of DNAzymes coupled with the detection of genetic targets using NPs has been also widely investigated. In a pioneer development, carboxylated nanotubes (CNTs) were modified with Pt via layer-by-layer assembly and further functionalized with the DNAzyme and the reporter probe through platinum–sulfur bonds. This sandwich-type detection is based on the use of a gold electrode modified with a capture probe that interacts with the target, while several DNAzyme/Pt NPs/CNTs are confined on the surface of the electrode. The addition of hemin activates the DNAzyme and catalyzes the reduction of the oxidized 3,3′,5,5′-tetramethylbenzidine (TMB), which was measured using chronoamperometry. The peak current of the DNA biosensor was logarithmically related to the target concentration across the range of 1.0 fM to 10 pM, with a limit of detection of 0.6 fM [38]. A similar approach was developed to detect the InvA gene of Salmonella. The target DNA was captured on a well surface, and subsequently, AuNPs modified with both the detection probe and the DNAzyme were added to the medium. Upon the addition of hemin, the oxidation of TMB occurs, and spectrophotometric detection is performed, achieving a limit of detection of 0.44 nM for the target sequence, corresponding to an interval of 3 × 103 to 3 × 106 CFU mL−1 for salmonella detection in water [27].
Immobilization of the DNAzyme on the electrode was also carried out for impedimetric detection of miRNA using AuNPs. In this case, in the presence of hemin, the DNAzyme catalyzes the oxidation of 4-chloro-1-napthol (CN) to form an insoluble product. The presence of the precipitate generates a barrier that increases the electron transfer resistance. With this scheme, and using electrochemical impedance spectroscopy and microgravimetric quartz crystal microbalance, the detection limit was 15 aM, achieving an excellent differentiation ability for single-base mismatch [30].
Coupling DNAzyme cleavage catalysis with the aggregation of AuNPs has been also used to detect an RNA sequence of dengue virus. In this approach, the target sequence interacts with the DNAzyme, and introduction of Mg2+ and a heating step produce the digestion of the viral RNA, deshielding the AuNPs and leading to their aggregation [24]. It is worth mentioning that the inclusion of sodium dodecyl sulfate (SDS) in the reaction mixture allows for detection directly on the supernatant of the cell culture, minimizing the sample treatment, and achieving a detection limit of 10 TCID50 units in a matter of 30 min.
AuNPs were also employed for miRNA-141 imaging in living cancer cells, since it is overexpressed in the cancer cell line LOVE-1, whereas the control cell line SMMC-7721 shows minimal levels of miRNA-141. In this study, the surface of the AuNPs was decorated with hairpin-locked DNAzyme strands with a fluorescent label. In absence of the target, the fluorescence of the dye is quenched by the AuNPs. The presence of the target provokes the opening of the hairpin, activating the DNAzyme, and cleavage of a DNA fragment containing the dye takes place. As consequence, the fluorophores are released from the AuNP surface, and a fluorescence enhancement takes place, while the target is also released to bind with another hairpin-locked DNAzyme, significantly improving the sensitivity. These results provide a new tool for understanding miRNA functionality in biological processes for fundamental research and clinical diagnostics [22].
Detection of small biomolecules and microorganisms
Sensitive detection of small biomolecules and microorganisms has also been achieved using DNAzymes. For instance, an efficient signal amplification strategy based on the use of an antibody-AuNP-DNAzyme assembly was designed for the detection and quantification of the carbohydrate antigen 19-9 (CA19-9), a well-known tumor marker, in human serum samples. For this purpose, multifunctional AuNPs were prepared by decorating their surface with both a specific antibody toward CA19-9 and a hemin/G-quadruplex DNAzyme. The target interacts specifically with the antibody, and the nanoassembly containing CA19-9 is separated from the one that did not interact with the target through capillary electrophoresis. The high sensitivity achieved is due to the amplification of the signal due to the hemin/G-quadruplex complex, which presents catalytic activity similar to that of peroxidase, and can be used to obtain a chemiluminescent signal in the presence of luminol [32].
A colorimetric strategy based on DNAzymes and MNPs (used as amplification and separation elements) was developed for the detection of cocaine. In this work, MNPs were decorated with an aptamer that specifically recognizes cocaine. Once again, using a hemin/G-quadruplex complex, the H2O2-mediated oxidation of TMB takes place, giving rise to a solution color change and providing a limit of detection of 50 nM [43].
Very few articles have described the use of DNAzymes for detecting genetic material specific to certain microorganisms to evidence the presence of such microorganisms in the media. In this sense, a specific aptamer for staphylococcal enterotoxin B (SEB) detection is immobilized on an enzyme-linked immunoassay (ELISA) plate with a probe “G.” In the absence of the target, the G-quadruplex sequences are not released, and no activity of the DNAzyme can be observed. Hence, the H2O2 reduces the gold ions, generating quasi-spherical and well-dispersed AuNPs in a red solution. Conversely, in the presence of a target, a conformational change occurs on the aptamer, liberating the G-quadruplex-forming sequences and converting them into active hemin/G-quadruplex peroxidase-mimicking DNAzymes. As a consequence, the H2O2 is consumed, and the kinetics of crystal growth slows, giving rise to the aggregation of the AuNPs as observed through a shift of the SPR peak toward higher wavelengths (changing from red to blue color) [28].
MNAzymes and NPs for bioanalytical applications
As previously described, MNAzymes have been developed to increase the versatility and utility of NAzymes. Synergistic combination of MNAzymes with inorganic NPs proved to be specially suited to achieve the extremely low detection limits often required for biomarker detection. Because there are still few publications related to this topic, the main approaches combining MNAzymes with inorganic NPs for bioanalytical purposes described in the literature are summarized in this section, and have been classified according to the type of nanomaterial employed.
AuNPs
MNAzymes can be used as tools for direct detection of nucleic acids in a protein-free, isothermal, optical format, when coupled with AuNPs as detection tags. The use of AuNPs coupled with MNAzymes has been exploited using two main approaches for the detection of long genetic sequences like DNA and RNA or shorter sequences such as miRNAs. The first approach, reported by Zagorovsky and Chan [44], takes advantage of the optoelectronic properties of AuNPs. In this study, the detection of a target is executed by evaluating the changes produced on the SPR peak when the AuNPs are aggregated or not aggregated in a colloidal suspension. For this purpose, AuNPs are surface-modified with short, thiolated DNA sequences, which are partially complementary to a substrate DNA sequence containing an rA base. This substrate is also complementary to the upper arms of the MNAzyme; thus, when the substrate is intact, the AuNPs are in close proximity. However, when the MNAzyme is activated in the presence of the target, cleavage of the substrate takes place in a multiple turnover, which is split into two shorter strands, increasing the distance between the AuNPs. Based on this strategy (see Fig. 5a), in the absence of the target, a dark purple color will be observed due to the aggregation of AuNPs, whereas the presence of the target will activate the MNAzyme, and the color will change to red/pink due to the good colloidal stability of AuNPs. Just by changing the sequence of the sensor arms, multianalyte detection is demonstrated. In fact, genetic targets corresponding to gonorrhea (Gon), syphilis bacteria (Syph), malaria parasite (Mal), and hepatitis B virus (HBV) have been detected in parallel with sensitivity on the order of 250 pM in 80 min. Following this strategy, Mohamed et al. [45] recently reported the implementation of this methodology for the detection of bacterial RNA from Staphylococcus aureus (S. aureus) and their genetic modifications that conferred them resistance to antibiotics such as penicillin, with clinical sensitivity of 90% in less than 3 h of assay. Hannah et al. [46] reported this strategy to differentiate COVID-19 (SARS-CoV-2) from influenza A (H3N2) virus with clinical sensitivity of 93% in 85 min. Finally, Zhu et al. [47] performed a similar aggregation assay for the detection of adenosine triphosphate (ATP) by modifying sensor arms of MNAzymes with longer sequences that specifically recognize ATP molecules, achieving a limit of detection of 5.3 pM in 60 min.
MNAzymes coupled with NPs as a signal amplification tool. a AuNPs as probes used in MNAzyme aggregation assays with visual/UV-Vis and sp-ICP-MS detection [44,45,46,47,48]. b AuNPs as probes used as quenchers of organic fluorophores (F) coupled to MNAzymes for fluorescence detection [49]. c MNPs as probes used in MNAzyme aggregation assays with optomagnetic detection [50]. d MNP probes used as carriers for multiple lanthanide (L) detection by ICP-MS [52]. e MNPs used as carriers for fluorescence assays with organic quencher (Q) and F as probes coupled with MNAzymes for fluorescence detection [51, 53]. f Catalytic NPs of CuMn-CeO2 modified with luminol as probes in an MNAzyme-mediated assay for ECL detection [54]
The colorimetric detection of AuNPs coupled with MNAzyme amplification is a simple, rapid, and cost-effective way to detect genetic targets or other types of small molecules with high sensitivity. However, alternative techniques have been described to detect AuNP aggregates formed during the assay, one being the elemental detection of gold atoms. For example, Yin et al. [48] ran the homogeneous assay based on MNAzymes and AuNPs, and the readout was performed using single-particle ICP-MS (sp-ICP-MS) capable of distinguishing between aggregated and non-aggregated AuNPs, achieving a sub-picomolar limit of detection. This work takes advantage of the differences between large aggregates of particles and small AuNPs formed by non-cleaved and cleaved substrate systems, respectively. Therefore, increasing the target concentration (i.e., more cleaved substrate) induces a decrease in the signal intensity of the aggregated particles, while at the same time, increasing the pulse frequency. Thus, size differences can be discriminated using sp-ICP-MS, enabling the application of this strategy to the detection of a long sequence from malaria DNA and a short sequence from miRNA-10 with a fitting range of 0.1–25 pM.
AuNPs can also be used as electron acceptors because they can absorb the fluorescence emission of fluorophores. This feature, coupled with the ability of MNAzymes to cleave a substrate sequence in the presence of the target, enables the development of methods based on FRET between AuNPs and a fluorophore with an emission band close to the absorption band of AuNPs (see Fig. 5b). Based on this approach, Wu et al. [49] developed an intracellular nanoprobe system for miRNA sensing in living cells. To achieve this, the substrate sequence to be cleaved by the MNAzyme in the presence of the miRNA is modified with AuNPs at one end and with the fluorophore FAM at the other end. Hence, the donor and acceptor are in close proximity in the absence of the target, and no fluorescent emission is observed. The rupture of the substrate upon activation of the MNAzyme in the presence of the target gives rise to a recovery of the fluorescence signal of FAM. It is noteworthy that AuNP-based probes can naturally permeate the cell membrane and that the intracellular target can activate MNAzymes under a physiological concentration of Mg2+, allowing for real-time fluorescence imaging of miR-21 in cancer cells with a detection limit of 10 pM.
MNPs and magnetic beads
Many variations of the basic MNAzyme design, some of which involve using MNPs, have provided further functionalities. The potential and versatility of MNPs arises from their paramagnetic and superparamagnetic properties, which make them suitable carrier probes because the MNPs can be guided via magnetic fields to a desired location. This fact, coupled with the ability of MNAzymes to cut a specific genetic substrate, guides the sequences (either the MNAzymes or the substrate) and sets them onto specific areas of recognition and introduces the possibility for separating the non-cleaved substrate from the rest of the components. Three strategies have been described using MNPs or magnetic beads (MBs) combined with MNAzymes.
A recent publication employs the disintegration of MNP assemblies for the detection of miRNA. MNP assemblies are tailored with the substrate sequences of the MNAzymes, which are activated in the presence of miRNA, leading to a rupture and disintegration of the MNP assemblies (see Fig. 5c). The concentration of released MNPs is proportional to the concentration of the target which is quantified by a 405-nm laser-based optomagnetic sensor, achieving a limit of detection on the order of a few pM [50].
The second detection approach using MBs is based on elemental detection by anchoring elemental tags or labels at one end of the substrate arm (see Fig. 5d). In this sense, Kang et al. [52] reported an assay employing lanthanide tags for the simultaneous detection of multiple miRNAs by ICP-MS. In this case, streptavidin-modified MNPs and three different substrate arms labeled with biotin at one end and lanthanide tags (Tb, Ho, Lu) at the other end were constructed. In the presence of the targets, three different pairs of MNAzymes were assembled with their corresponding substrate, releasing the fragment with the lanthanide tag that will be further monitored through ICP-MS. Using this approach, multiple-target recognition with a detection limit on the order of the picomolar was achieved. Finally, a third detection approach is based on the use of fluorophore–quencher interaction as assay probes. Both substrate ends are modified with a fluorophore and its respective quencher. As aforementioned, the presence of the target produces an increase of the fluorescence signal due to the cleavage of the substrate (see Fig. 5e). In this sense, Safdar et al. [53] reported a microwell-based bioassay for multiplex nucleic acid detection using MBs. Two sets of MBs were functionalized with two different MNAzyme sequences. To enable duplex detection, these sequences comprised the same core and included different target and substrate binding arms, with two different quenching and fluorophore probes, tailored for detection of either Streptococcus pneumoniae or human adenovirus targets. Then, both sets of MBs were mixed in a 1-to-1 ratio and positioned by permanent magnets on the microwell plate, with their respective substrate and target sequences. The presence of each target activated its corresponding MNAzyme, and cleavage of the specific substrate took place. Hence, a recovery of the fluorescence signal at 535 nm for Streptococcus pneumoniae and 567 nm for human adenovirus occurs. With this scheme, a detection limit on the sub-picomolar region was achieved. A similar approach based on fluorophore–quencher interaction was also reported by Bakshi et al. [51]. In this study, MNPs activated by a magnetic field act as carriers for the intracellular detection and imaging of a genetic biomarker for breast cancer.
Catalytic NPs (nanospheres of CuMn-CeO2)
It is well known that certain NPs present catalytic capacities that can be exploited in the development of improved electrochemical tests. The presence of metal oxides with special surface mixed-valence properties, such as cerium oxide (CeO2) NPs, which have a redox pair of Ce3+ / Ce4+, can switch oxidation states on the lattice surface. In addition to this redox feature, CeO2 nanocrystals can act as a host matrix for several doping elements, including Gd, Sm, Cu, or Mn, resulting in a significant increase of the catalytic performance of the NP. Inspired by the abovementioned principle, Ling et al. [54] reported the use of CuMn-CeO2 nanospheres with a large specific surface area that can act as nanocarrier for the development of an ECL biosensor for the detection of group B streptococci (GBS) with a detection limit in the attomolar region. In this pioneer work, a GCE with gold deposited on its surface was modified with a capture probe (CP), and CuMn-CeO2 nanospheres were modified with a signal probe (SP) and a chemiluminescence complex named polyethylenimine-luminol (PEI-luminol). In a previous step, the substrate hairpin sequence is cleaved by MNAzymes in the presence of the target analyte. The shorter sequence resulting from the cleavage of the substrate hairpin acts as linker and hybridizes with CP and SP, producing an approximation between the nanospheres modified with PEI-luminol and the gold electrode surface, stimulating oxidation of H2O2 test solution, and leading to an increase in the fluorescent signal of PEI-luminol (see Fig. 5f).
Summary and future trends
Clearly, the excellent properties exhibited by DNAzymes, including high stability, good catalytic activity that can be converted into a signal amplification step, and the possibility for introducing structural modifications to make them specific against a certain target, make such structures ideal tools to be exploited in the development of highly sensitive bioanalytical strategies. Many different chemical reactions, including RNA cleavage, oxidative or hydrolytic DNA cleavage, DNA or RNA ligation, or peroxidase reactions based on G-quadruplex, have now been carried out by DNAzymes. DNAzymes can catalyze their respective reactions in multiple turnovers, which allows for sensitive bioanalytical detection. Additionally, the synergistic combination of DNAzymes with the outstanding features offered by inorganic NPs allows for highly selective and ultrasensitive detection of a wide variety of targets, while also offering an easy integration into colorimetric, fluorometric, and electrochemical assays, making them well suited as chemosensors in simple and portable devices.
To further increase the versatility and utility of NAzymes, MNAzyme-based assays have recently appeared as an alternative type of isothermal, signal-amplifying genetic detection method. Many variations of the basic MNAzyme scheme design have already been successfully demonstrated for a wide variety of target analytes. The modular nature of MNAzymes provides a great potential for their integration into diverse devices such as diagnostic biosensors, molecular computers, and/or nanoscale machines.
As a result, a wide variety of DNAzymes and MNAzymes have been already successfully applied in biomedical imaging of metal ions, proteins, bacteria, and other targets, as summarized in this review. Most of the applications of the NAzymes described thus far exploit the cleavage activity of DNAzymes or MNAzymes in a way that, in the presence of the target, a substrate is cleaved, and the distance between a NP and another NP, fluorophore, or lanthanide label increases. This change in distance can be monitored through optical, electrochemical, or elemental detection, and it has been mainly applied to the detection of metals and DNA or RNA fragments that act as biomarkers or are related to the presence of microorganisms. On the other hand, detection of protein targets is mainly performed using G-quadruplex DNAzymes together with hemin. In these types of applications, the NPs act mainly as catalysts to produce H2O2 that will be used to generate a colorimetric response or to be used as labels in electrochemical systems. With the increasing number of publications on this topic nowadays, it is obvious that DNAzymes constitute a novel tool to be exploited in biosensing applications. Although there are few products on the market based on DNAzymes, there are hundreds of patents using DNAzyme- or MNAzyme-based biosensors, which reveals the significant potential that they offer to go from the lab to the market.
Despite the enormous progress in this field, to advance the field further and to reach the full potential of the DNAzymes and MNAzymes, we still need to meet several challenges.
An important issue that still remains unclear is the understanding of the specific catalysis mechanism. Understanding how the DNAzymes are capable of catalyzing reactions will reveal the experimental conditions needed to obtain the best catalytic performance, for instance, choosing the best divalent metal needed to activate a certain DNAzyme, the optimal NAzyme, substrates, reaction buffers, and operating conditions. Therefore, further efforts to comprehend DNAzymes at a more fundamental level would also allow for the generation of new and improved DNAzymes.
Most importantly, it is critical to identify DNAzymes and MNAzymes that can work well in real sample matrices. Most DNAzymes only work in simple solutions, under well-controlled conditions of pH, ionic strength, buffering agent, and temperature. As a result, a small change in reaction conditions can result in large changes in activity, hindering their applicability in real samples. Rational design of DNAzymes and MNAzymes whose activities can tolerate fluctuations of conditions should be explored.
Moreover, most DNAzymes and MNAzymes reported to date exhibit response times that are on orders of magnitude slower than conventional protein enzymes, resulting in quite long analysis times. Improved selection methods using lower target concentrations and shorter reaction times may help such genetic structures match the performance of protein enzymes.
Although some publications have shown the great potential of DNAzymes coupled with NPs for in vitro cellular imaging, e.g., using two-photon NIR fluorescent NPs, we have not found literature on this topic for in vivo applications, so this is another area to explore.
In brief, new innovative methodologies based on the synergistic combination of inorganic NPs, DNAzymes, and MNAzymes have been developed during the last decade, showing the potential of such combinations for highly sensitive detection of biomolecules. Designing bioanalytical devices using DNAzymes and MNAzymes is a growing field today, which undoubtedly will benefit bioanalysis given the ability to utilize multiple assay formats, with the possibility of a further signal amplification in combination with inorganic NPs, and to integrate these assays into a range of different devices. We expect that an exponential number of research developments on this topic will emerge in the near future.
References
Feng C, Zhu X. Signal Amplification. In: Nano-Inspired Biosensors for Protein Assay with Clinical Applications. Elsevier;2019. pp 287–312
Safdar S, Lammertyn J, Spasic D. RNA-Cleaving NAzymes: The Next Big Thing in Biosensing? Trends Biotechnol. 2020;38:1343–59. https://doi.org/10.1016/j.tibtech.2020.04.012.
Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem Biol. 1994;1:223–9. https://doi.org/10.1016/1074-5521(94)90014-0.
Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell. 1982;31:147–57. https://doi.org/10.1016/0092-8674(82)90414-7.
Ven K, Safdar S, Dillen A, Lammertyn J, Spasic D. Re-engineering 10–23 core DNA- and MNAzymes for applications at standard room temperature. Anal Bioanal Chem. 2019;411:205–15. https://doi.org/10.1007/s00216-018-1429-4.
Gao J, Shimada N, Maruyama A. Enhancement of deoxyribozyme activity by cationic copolymers. Biomater Sci. 2015;3:308–16. https://doi.org/10.1039/C4BM00256C.
Ruble BK, Richards JL, Cheung-Lau JC, Dmochowski IJ. Mismatch discrimination and efficient photomodulation with split 10–23 DNAzymes. Inorganica Chim Acta. 2012;380:386–91. https://doi.org/10.1016/j.ica.2011.10.068.
Mokany E, Bone SM, Young PE, Doan TB, Todd AV. MNAzymes, a Versatile New Class of Nucleic Acid Enzymes That Can Function as Biosensors and Molecular Switches. J Am Chem Soc. 2010;132:1051–9. https://doi.org/10.1021/ja9076777.
Peng H, Newbigging AM, Wang Z, Tao J, Deng W, Le XC, Zhang H. DNAzyme-Mediated Assays for Amplified Detection of Nucleic Acids and Proteins. Anal Chem. 2018;90:190–207. https://doi.org/10.1021/acs.analchem.7b04926.
Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–6. https://doi.org/10.1038/334364a0.
Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–4. https://doi.org/10.1038/344410a0.
Li T, Dong S, Wang E. G-Quadruplex Aptamers with Peroxidase-Like DNAzyme Functions: Which Is the Best and How Does it Work? Chem - An Asian J. 2009;4:918–22. https://doi.org/10.1002/asia.200900019.
Travascio P, Li Y, Sen D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem Biol. 1998;5:505–17. https://doi.org/10.1016/S1074-5521(98)90006-0.
Wang F, Zhang Y, Lu M, Du Y, Chen M, Meng S, Ji W, Sun C, Peng W. Near-infrared band Gold nanoparticles-Au film “hot spot” model based label-free ultratrace lead (II) ions detection via fiber SPR DNAzyme biosensor. Sensors Actuators B Chem. 2021;337:129816. https://doi.org/10.1016/j.snb.2021.129816.
Wu H, Wang S, Li SFY, Bao Q, Xu Q. A label-free lead(II) ion sensor based on surface plasmon resonance and DNAzyme-gold nanoparticle conjugates. Anal Bioanal Chem. 2020;412:7525–33. https://doi.org/10.1007/s00216-020-02887-z.
Lai C, Zhang Y, Liu X, Liu S, Li B, Zhang M, Qin L, Yi H, Li M, Li L, Fu Y, He J, Chen L. Electrochemical biosensor for amplified detection of Pb2+ based on perfect match of reduced graphene oxide–gold nanoparticles and single-stranded DNAzyme. Anal Bioanal Chem. 2019;411:7499–509. https://doi.org/10.1007/s00216-019-02146-w.
Liao X, Luo J, Wu J, Fan T, Yao Y, Gao F, Qian Y. A sensitive DNAzyme-based electrochemical sensor for Pb2+ detection with platinum nanoparticles decorated TiO2/α-Fe2O3 nanocomposite as signal labels. J Electroanal Chem. 2018;829:129–37. https://doi.org/10.1016/j.jelechem.2018.10.009.
Wang H-B, Ma L-H, Fang B-Y, Zhao Y-D, Hu X-B. Graphene oxide-assisted Au nanoparticle strip biosensor based on GR-5 DNAzyme for rapid lead ion detection. Colloids Surfaces B Biointerfaces. 2018;169:305–12. https://doi.org/10.1016/j.colsurfb.2018.05.020.
Zhou B, Shi L-F, Wang Y-S, Yang H-X, Xue J-H, Liu L, Wang Y-S, Yin J-C, Wang J-C. Resonance light scattering determination of uranyl based on labeled DNAzyme–gold nanoparticle system. Spectrochim Acta Part A Mol Biomol Spectrosc. 2013;110:419–24. https://doi.org/10.1016/j.saa.2013.03.036.
Yin J-C, Wang Y-S, Zhou B, Xiao X-L, Xue J-H, Wang J-C, Wang Y-S, Qian Q-M. A wireless magnetoelastic sensor for uranyl using DNAzyme–graphene oxide and gold nanoparticles-based amplification. Sensors Actuators B Chem. 2013;188:147–55. https://doi.org/10.1016/j.snb.2013.06.100.
Lee JH, Wang Z, Liu J, Lu Y. Highly Sensitive and Selective Colorimetric Sensors for Uranyl (UO 2 2+ ): Development and Comparison of Labeled and Label-Free DNAzyme-Gold Nanoparticle Systems. J Am Chem Soc. 2008;130:14217–26. https://doi.org/10.1021/ja803607z.
Yun W, Li F, Liu X, Li N, Chen L, Yang L. A catalytic cleavage strategy for fluorometric determination of Hg(II) based on the use of a Mg(II)-dependent split DNAzyme and hairpins conjugated to gold nanoparticles. Microchim Acta. 2018;185:457. https://doi.org/10.1007/s00604-018-2990-4.
Zhao Y, Xie X. A Novel Electrochemical Aptamer Biosensor Based on DNAzyme Decorated Au@Ag Core-Shell Nanoparticles for Hg2+ Determination. J Braz Chem Soc. 2017. https://doi.org/10.21577/0103-5053.20170133.
Li L, Feng J, Fan Y, Tang B. Simultaneous Imaging of Zn 2+ and Cu 2+ in Living Cells Based on DNAzyme Modified Gold Nanoparticle. Anal Chem. 2015;87:4829–35. https://doi.org/10.1021/acs.analchem.5b00204.
Yang C, Yin X, Huan S-Y, Chen L, Hu X-X, Xiong M-Y, Chen K, Zhang X-B. Two-Photon DNAzyme–Gold Nanoparticle Probe for Imaging Intracellular Metal Ions. Anal Chem. 2018;90:3118–23. https://doi.org/10.1021/acs.analchem.7b04171.
Wang X, Sun D, Tong Y, Zhong Y, Chen Z. A voltammetric aptamer-based thrombin biosensor exploiting signal amplification via synergetic catalysis by DNAzyme and enzyme decorated AuPd nanoparticles on a poly(o-phenylenediamine) support. Microchim Acta. 2017;184:1791–9. https://doi.org/10.1007/s00604-017-2160-0.
Rao X, Zhang J, Cui J, Hu Y, Liu T, Chai J, Cheng G, He P, Fang Y. Au nanoparticle-DNAzyme dual catalyst system for sensitively colorimetric detection of thrombin. Chem Res Chinese Univ. 2013;29:868–73. https://doi.org/10.1007/s40242-013-3039-1.
Zhu D, Luo J, Rao X, Zhang J, Cheng G, He P, Fang Y. A novel optical thrombin aptasensor based on magnetic nanoparticles and split DNAzyme. Anal Chim Acta. 2012;711:91–6. https://doi.org/10.1016/j.aca.2011.10.053.
Li W, Li H, Wu S, Feng C, Li G. Highly sensitive protein detection based on DNAzyme cycling activated surface assembly of peptide decorated nanoparticles. Electrochem commun. 2016;71:84–8. https://doi.org/10.1016/j.elecom.2016.08.011.
Xia H, Li L, Yin Z, Hou X, Zhu J-J. Biobar-Coded Gold Nanoparticles and DNAzyme-Based Dual Signal Amplification Strategy for Ultrasensitive Detection of Protein by Electrochemiluminescence. ACS Appl Mater Interfaces. 2015;7:696–703. https://doi.org/10.1021/am506980d.
Liu S, Xu N, Tan C, Fang W, Tan Y, Jiang Y. A sensitive colorimetric aptasensor based on trivalent peroxidase-mimic DNAzyme and magnetic nanoparticles. Anal Chim Acta. 2018;1018:86–93. https://doi.org/10.1016/j.aca.2018.01.040.
Wang Q, Yang X, Yang X, Liu F, Wang K. Visual detection of myoglobin via G-quadruplex DNAzyme functionalized gold nanoparticles-based colorimetric biosensor. Sensors Actuators B Chem. 2015;212:440–5. https://doi.org/10.1016/j.snb.2015.02.040.
Zong C, Zhang D, Yang H, Wang S, Chu M, Li P. Chemiluminescence immunoassay for cardiac troponin T by using silver nanoparticles functionalized with hemin/G-quadruplex DNAzyme on a glass chip array. Microchim Acta. 2017;184:3197–204. https://doi.org/10.1007/s00604-017-2331-z.
Shekari Z, Zare HR, Falahati A. Electrochemical sandwich aptasensor for the carcinoembryonic antigen using graphene quantum dots, gold nanoparticles and nitrogen doped graphene modified electrode and exploiting the peroxidase-mimicking activity of a G-quadruplex DNAzyme. Microchim Acta. 2019;186:530. https://doi.org/10.1007/s00604-019-3572-9.
Hou L, Wu X, Chen G, Yang H, Lu M, Tang D. HCR-stimulated formation of DNAzyme concatamers on gold nanoparticle for ultrasensitive impedimetric immunoassay. Biosens Bioelectron. 2015;68:487–93. https://doi.org/10.1016/j.bios.2015.01.043.
Dong X-Y, Mi X-N, Zhang L, Liang T-M, Xu J-J, Chen H-Y. DNAzyme-functionalized Pt nanoparticles/carbon nanotubes for amplified sandwich electrochemical DNA analysis. Biosens Bioelectron. 2012;38:337–41. https://doi.org/10.1016/j.bios.2012.06.015.
Luo R, Li Y, Lin X, Dong F, Zhang W, Yan L, Cheng W, Ju H, Ding S. A colorimetric assay method for invA gene of Salmonella using DNAzyme probe self-assembled gold nanoparticles as single tag. Sensors Actuators B Chem. 2014;198:87–93. https://doi.org/10.1016/j.snb.2014.02.104.
Wan J, Liu X, Zhang Y, Gao Q, Qi H, Zhang C. Sensitive impedimetric detection of microRNAs using a hairpin probe based on DNAzyme-functionalized gold nanoparticle tag-initiated deposition of an insulating film on gold electrode. Sensors Actuators B Chem. 2015;213:409–16. https://doi.org/10.1016/j.snb.2015.02.123.
Carter JR, Balaraman V, Kucharski CA, Fraser TS, Fraser MJ. A novel dengue virus detection method that couples DNAzyme and gold nanoparticle approaches. Virol J. 2013;10:201. https://doi.org/10.1186/1743-422X-10-201.
Yang Y, Huang J, Yang X, He X, Quan K, Xie N, Ou M, Wang K. Gold Nanoparticle Based Hairpin-Locked-DNAzyme Probe for Amplified miRNA Imaging in Living Cells. Anal Chem. 2017;89:5850–6. https://doi.org/10.1021/acs.analchem.7b00174.
Shi M, Zhao S, Huang Y, Zhao L, Liu Y-M. Signal amplification in capillary electrophoresis based chemiluminescent immunoassays by using an antibody–gold nanoparticle–DNAzyme assembly. Talanta. 2014;124:14–20. https://doi.org/10.1016/j.talanta.2014.02.032.
Du Y, Li B, Guo S, Zhou Z, Zhou M, Wang E, Dong S. G-Quadruplex-based DNAzyme for colorimetric detection ofcocaine: Using magnetic nanoparticles as the separation and amplification element. Analyst. 2011;136:493–7. https://doi.org/10.1039/C0AN00557F.
Zhou D, Xie G, Cao X, Chen X, Zhang X, Chen H. Colorimetric determination of staphylococcal enterotoxin B via DNAzyme-guided growth of gold nanoparticles. Microchim Acta. 2016;183:2753–60. https://doi.org/10.1007/s00604-016-1919-z.
Zagorovsky K, Chan WCW. A Plasmonic DNAzyme Strategy for Point-of-Care Genetic Detection of Infectious Pathogens. Angew Chemie Int Ed. 2013;52:3168–71. https://doi.org/10.1002/anie.201208715.
Abdou Mohamed MA, Kozlowski HN, Kim J, Zagorovsky K, Kantor M, Feld JJ, Mubareka S, Mazzulli T, Chan WCW. Diagnosing Antibiotic Resistance Using Nucleic Acid Enzymes and Gold Nanoparticles. ACS Nano. 2021;15:9379–90. https://doi.org/10.1021/acsnano.0c09902.
Kozlowski HN, Abdou Mohamed MA, Kim J, Bell NG, Zagorovsky K, Mubareka S, Chan WCW. A Colorimetric Test to Differentiate Patients Infected with Influenza from COVID-19. Small Struct. 2021;2:2100034. https://doi.org/10.1002/sstr.202100034.
Zhu S, Wang X, Jing C, Yin Y, Zhou N. A colorimetric ATP assay based on the use of a magnesium(II)-dependent DNAzyme. Microchim Acta. 2019;186:176. https://doi.org/10.1007/s00604-019-3244-9.
Yin X, Chen B, He M, Hu B. A Homogeneous Multicomponent Nucleic Acid Enzyme Assay for Universal Nucleic Acid Detection by Single-Particle Inductively Coupled Plasma Mass Spectrometry. Anal Chem. 2021;93:4952–9. https://doi.org/10.1021/acs.analchem.0c05444.
Wu Y, Huang J, Yang X, Yang Y, Quan K, Xie N, Li J, Ma C, Wang K. Gold Nanoparticle Loaded Split-DNAzyme Probe for Amplified miRNA Detection in Living Cells. Anal Chem. 2017;89:8377–83. https://doi.org/10.1021/acs.analchem.7b01632.
Tian B, Han Y, Wetterskog E, Donolato M, Hansen MF, Svedlindh P, Strömberg M. MicroRNA Detection through DNAzyme-Mediated Disintegration of Magnetic Nanoparticle Assemblies. ACS Sensors. 2018;3:1884–91. https://doi.org/10.1021/acssensors.8b00850.
Kang Q, He M, Chen B, Xiao G, Hu B (2021) MNAzyme-Catalyzed Amplification Assay with Lanthanide Tags for the Simultaneous Detection of Multiple microRNAs by Inductively Coupled Plasma–Mass Spectrometry. Anal Chem 93:737–744. https://doi.org/10.1021/acs.analchem.0c02455
Safdar S, Ven K, van Lent J, Pavie B, Rutten I, Dillen A, Munck S, Lammertyn J, Spasic D. DNA-only, microwell-based bioassay for multiplex nucleic acid detection with single base-pair resolution using MNAzymes. Biosens Bioelectron. 2020;152:112017. https://doi.org/10.1016/j.bios.2020.112017.
Bakshi SF, Guz N, Zakharchenko A, Deng H, Tumanov AV, Woodworth CD, Minko S, Kolpashchikov DM, Katz E. Nanoreactors based on DNAzyme-functionalized magnetic nanoparticles activated by magnetic field. Nanoscale. 2018;10:1356–65. https://doi.org/10.1039/C7NR08581H.
Ling J, Zhao M, Chen F, Zhou X, Li X, Ding S, Tang H. An enzyme-free electrochemiluminescence biosensor for ultrasensitive assay of Group B Streptococci based on self-enhanced luminol complex functionalized CuMn-CeO2 nanospheres. Biosens Bioelectron. 2019;127:167–73. https://doi.org/10.1016/j.bios.2018.12.012.
Cocherie A, Robert M. Direct measurement of lead isotope ratios in low concentration environmental samples by MC-ICP-MS and multi-ion counting. Chem Geol. 2007;243:90–104. https://doi.org/10.1016/j.chemgeo.2007.05.011.
Babu SH, Kumar KS, Suvardhan K, Kiran K, Rekha D, Krishnaiah L, Janardhanam K, Chiranjeevi P. Preconcentration technique for the determination of trace elements in natural water samples by ICP-AES. Environ Monit Assess. 2007;128:241–9. https://doi.org/10.1007/s10661-006-9309-3.
Mortada WI, Kenawy IMM, Abdelghany AM, Ismail AM, Donia AF, Nabieh KA. Determination of Cu2+, Zn2+ and Pb2+ in biological and food samples by FAAS after preconcentration with hydroxyapatite nanorods originated from eggshell. Mater Sci Eng C. 2015;52:288–96. https://doi.org/10.1016/j.msec.2015.03.061.
Funding
Financial support from the Ministry of Science and Innovation (Spain) through the projects MCI-20-PID2019-109698GB-I00 and PID2020-117282RB-I00, from the Fundación Española para la Ciencia y la Tecnología through the project MCIU-20-FCT-PRECIPITA, from Banco Santander and University of Oviedo through the project PAPI-19-PF-22, and from Principado de Asturias (Spain) through the project FC-GRUPIN-IDI/2021/000081 is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no competing interests.
Additional information
Published in the topical collection featuring Promising Early-Career (Bio)Analytical Researchers with guest editors Antje J. Baeumner, María C. Moreno-Bondi, Sabine Szunerits, and Qiuquan Wang.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sánchez-Visedo, A., Ferrero, F.J., Costa-Fernández, J.M. et al. Inorganic nanoparticles coupled to nucleic acid enzymes as analytical signal amplification tools. Anal Bioanal Chem 414, 5201–5215 (2022). https://doi.org/10.1007/s00216-022-03998-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-03998-5