Abstract
This review focuses on the possibilities and limits of nontarget screening of emerging contaminants, with emphasis on recent applications and developments in data evaluation and compound identification by liquid chromatography–high-resolution mass spectrometry (HRMS). The general workflow includes determination of the elemental composition from accurate mass, a further search for the molecular formula in compound libraries or general chemical databases, and a ranking of the proposed structures using further information, e.g., from mass spectrometry (MS) fragmentation and retention times. The success of nontarget screening is in some way limited to the preselection of relevant compounds from a large data set. Recently developed approaches show that statistical analysis in combination with suspect and nontarget screening are useful methods to preselect relevant compounds. Currently, the unequivocal identification of unknowns still requires information from an authentic standard which has to be measured or is already available in user-defined MS/MS reference databases or libraries containing HRMS spectral information and retention times. In this context, we discuss the advantages and future needs of publicly available MS and MS/MS reference databases and libraries which have mostly been created for the metabolomic field. A big step forward has been achieved with computer-based tools when no MS library or MS database entry is found for a compound. The numerous search results from a large chemical database can be condensed to only a few by in silico fragmentation. This has been demonstrated for selected compounds and metabolites in recent publications. Still, only very few compounds have been identified or tentatively identified in environmental samples by nontarget screening. The availability of comprehensive MS libraries with a focus on environmental contaminants would tremendously improve the situation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, the focus on organic trace pollutants in the aquatic environment has shifted to an increasing number of polar, highly water soluble compounds. The so-called emerging contaminants (ECs) are environmental pollutants that have not yet been considered in environmental screening programs. Most ECs are released by the discharge of municipal, industrial, and agricultural wastewater into surface waters, and afterwards to other environmental compartments such as soil, air, and groundwater. The importance of the issue of ECs has been shown by a survey of various pharmaceuticals in different surface waters in Europe and the USA, which revealed pharmaceuticals as ubiquitously occurring contaminants [1–8]. Recent reviews have provided an excellent overview of the multitude and variety of newly detected contaminants from domestic, commercial, and industrial use, e.g., artificial sweeteners, perfluorinated compounds, pharmaceuticals, hormones, disinfection by-products, UV filters, brominated flame retardants, benzotriazoles, naphthenic acids, siloxanes, musk fragrances, and transformation products (TPs) [9, 10]. The high number and the wide range of chemical structures which have to be considered pose a big challenge for analytical methods to monitor ECs.
In addition, TPs increase difficulty of the analytical work owing to their high numbers, their often unknown structures, and their unknown impact and fate in the environment [11–16]. TPs are metabolites from human and animal metabolism, and are also produced when anthropogenic pollutants undergo biological, chemical, and photochemical degradation in the environment or in different steps of water treatment, such as biological or chemical processes (e.g., chlorination, ozonation, and advanced oxidation).
To cope with those challenges, there is much interest in prioritizing ECs on the basis of their occurrence and toxicity data to preselect the most relevant compounds [17]. The use of multiresidue analysis methods by liquid chromatography (LC)–tandem mass spectrometry (MS/MS) is the measurement approach to obtain information on the occurrence and fate of ECs in the aquatic environment [18–26]. The increased availability and application of LC–high-resolution mass spectrometry (HRMS) is a considerable step forward and has been developed to a powerful tool for screening of environmentally relevant compounds over the last few years [10]. Liquid chromatographs coupled to quadrupole time-of-flight (Q-TOF) mass spectrometers and linear ion trap instruments with orbitrap technology [27] are mostly applied in environmental analysis [28–30]. High mass resolving power between 20,000 and 100,000 (at full width at half maximum, FWHM), good mass accuracy (often better than 1 ppm), good isotopic abundance accuracy (3–20 %), and high sensitivity in the picomolar to femtomolar range characterize the new generation of HRMS instruments [30, 31]. It has to be emphasized that these figures of merit are often not directly comparable since they depend on several operating parameters, such as the scan speed and mass range, and the properties of the analytes, such as the ionization efficiency and the molecular weight.
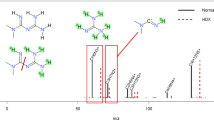
HRMS measurements increase the selectivity for screening of known micropollutants in complex matrices, but for unknown water contaminants more importantly they allow the deduction of the elemental composition from the accurate mass. The key to the deduction of elemental composition is that isotopes of all chemical elements have different mass defects, as exemplified for selected isotopes in Fig. 1. The mass defect is the difference between the nominal and the exact mass of a chemical element. This can lead to positive or negative mass defects, which theoretically allow one to determine a unique elemental composition assuming that the mass spectrometer has a mass resolving power and a mass accuracy in the range of 0.1 mDa even for very complex mixtures [32, 33]. Currently, this is generally not achieved in LC–mass spectrometry (MS) even with high-mass-resolving Fourier transform ion cyclotron resonance mass spectrometers. Current Fourier transform ion cyclotron resonance instruments achieve a mass accuracy of approximately 1 mDa or lower, whereas Q-TOF and orbitrap instruments achieve a range of 2–10 mDa depending on the mass [30, 31].
The different mass defects of elemental isotopes enable a unique elemental composition for any molecule to be determined from a sufficiently accurate mass measurement. (Adapted from [32])
Therefore, high isotope accuracy is an important feature when using the isotope pattern as a further criterion to select the most probable molecular formula. One has to keep in mind that some chemical elements which may occur in ECs such as fluorine, phosphorus and iodine are monoisotopic and hence not suitable for this approach.
LC-HRMS has been increasingly used for nontarget screening of contaminants in environmental samples. Here we have to distinguish between different nontarget approaches depending on the information on the contaminants available from the sample and from spectral libraries or chemical compound databases. Recent reviews have discussed different analytical approaches for LC-HRMS screening of environmental micropollutants [30], structure elucidation of small molecules by MS in life sciences [31], processing and analysis of metabolomics data [34], and computer tools for structure elucidation in effect-directed analysis and metabolomics [35, 36]. The scope of this critical review is to discuss the possibilities and limits of nontarget screening of ECs, with a focus on recent applications and developments in data evaluation and identification used for nontarget screening. The applications have been chosen mainly from the area of water contaminants. Computer-based methods from the metabolomic field were selected only if they appear applicable in environmental analysis, and if they are not already included in most LC-MS workstations or software packages. The fundamental concept of this review is to demonstrate successful approaches and limits of nontarget screening for ECs. We do not consider recent improvements in instrumentation and ionization techniques or different screening approaches such as target, suspect, and nontarget screening. Recent developments in these fields can be found elsewhere [30, 31].
Applications of nontarget screening in water analysis
Suspect and nontarget screening approaches cannot be strictly distinguished from each other. Generally, in suspect screening, information on possibly occurring compounds is used for the evaluation of HRMS data, whereas real nontarget screening starts without any a priori information. Figure 2 illustrates the workflow for nontarget screening, and includes several or all of the steps that have been described in the recent literature [28–30, 37–39].
Nontarget screening typically starts with the accurate mass from LC-HRMS measurements followed by data processing steps to remove noise, blanks, or artifacts [40]. Next, automated deconvolution is performed to extract peaks of all possible compounds. The mass peaks of different ions of one compound are often merged to one feature (e.g., [M+H]+, [M+Na]+, [M+NH4]+). The resulting data set is then analyzed using statistical methods to evaluate the most relevant features by comparison of different samples and blanks. From the relevant features, the elemental composition is calculated and the most probable molecular formulae are evaluated by matching the isotope pattern. For identification, the molecular formulae are searched for in MS/MS databases or libraries. The retention time is often used as a further criterion to reduce the number of hits [29]. Identification is achieved when the MS fragmentation and retention time of the unknown compound fit to the library spectrum and the retention time of a reference compound.
If no match in an MS/MS database or library is available, searches in large chemical databases such as PubChem and ChemSpider are performed. This search generally results in several hundred to several thousand hits for a possible structure. MS fragmentation can be used as a criterion to select the most probable hits. Since the chemical databases generally do not contain any MS data, in silico fragmentation has to be used and then the fragments have to be matched against the measured MS fragments [41–43]. This results in a number of proposed compound structures. However, unequivocal identification still needs standards or complementary information from other analysis methods, such as NMR analysis in conjunction with MS fragmentation as was demonstrated for the structural identification of biotransformation products of iodinated X-ray contrast media [44, 45].
Recent articles have illustrated the application of accurate mass measurements, the calculation of molecular formulae, and searches in user-defined or NIST libraries and the Merck index [28, 29, 46, 47]. With use of Q-TOF measurements and a user-defined database with 2,500 water pollutants plus 100 mass spectra, the structures of the three compounds N,N-dicyclohexyl-N-methylamine, carbamazepine, and triphenylphosphine oxide were proposed, but they have not been confirmed with standards [47]. The limited mass resolving power of 5,000 (FWHM) of a Q-TOF instrument resulted in a considerably high number of proposed molecular formulae for unknowns, which could be reduced by use of isotope patterns and a database search [46]. Four unknowns were identified and confirmed by matching mass fragments and retention times: the fungicide enilconazole and the herbicides prometryn, terbutryn, and diuron. The proposed structures of the molecular formulae of a further three unknowns (C10H9O2F2S2Cl3, C14H26O4, C13H12N2O2) could not be explained. In wastewater effluent, 463 features of Q-TOF measurements with a high mass accuracy of less than 2 ppm were found and matched to 51 compounds by use of retention times and accurate masses, from which 26 compounds were identified on the basis of data from a user-defined library with accurate masses, isotope patterns, and in-source fragments of 300 pesticides and 80 pharmaceuticals [29]. Seventeen of the detected compounds had no data on in-source fragments and were further subjected to scheduled MS/MS measurements. The compounds included five pesticides, 16 pharmaceuticals, and five metabolites. The retention time was an important criterion in preselection of relevant features and in library matching. The 463 features would have matched 463 potential compounds in the user-defined library had the retention time not been considered. Furthermore, several isomer pairs in the database such as sulfathiazole–ketoralac, theophylline–paraxanthine–theobromine, fluoxitine–nadolol, and ranitidine–clomipramine could only be distinguished by their retention times. The main TPs acetaminophen and azithromycin were identified on the basis of the matching of similar structural moieties and hence fragments with library compounds. A self-created database with a user-defined retention index based on fenuron and chloroxuron as internal standards has been used for the identification of metolachlor oxanilic acid, and alachlor minus chloromethane [39]. However, several unknowns could not be explained because of the lack of reference compounds. So far, the application of different approaches of nontarget screening by LC-MS has revealed some promising results. However, it is common that only one or a few compounds of real unknowns could be identified [28, 29, 37–39, 41, 47–53].
In comparison, screening by gas chromatography (GC)–MS and LC–particle beam–MS often resulted in more proposed compounds owing to the availability of comprehensive MS libraries for electron ionization (EI) MS and the often more meaningful EI mass spectra [37, 50, 51, 54–59]. In this context, it is also worth mentioning that the development of direct-EI interfaces could be a complementary technique to obtain EI mass spectra in LC-MS [60, 61]. However, in comparison with EI, soft ionization with electrospray ionization (ESI) facilitates the finding of the molecular mass of the unknowns if there is no library spectrum available [40].
Qualified nontarget screening
Generally, data reduction, MS library search, and the use of further MS and chromatographic data (MS fragmentation, retention time) are most promising. Mueller et al. [62] applied statistical approaches and a search in a user-defined MS library to evaluate potential drinking water contaminants from a landfill leachate. In contrast to using the signal intensity, all features with a retention time and mass-to-charge (m/z) ratio obtained by a Q-TOF full scan were used. Pattern matching of samples with temporal, spatial, or process-based relationships was done by computing the operations union (A ∪ B), intersection (A ∩ B), and complement (A/B) of the different data sets using Venn diagrams. Venn diagrams are used to teach elementary set theory and show all logical relations between a given collection of sets. This helped to reduce the total number of features detected in groundwater affected by a landfill leachate from 1,729 to eight relevant compounds occurring in the raw water taken for drinking water treatment. Three contaminants were then identified as relevant for drinking water quality, since they still persisted after ozone treatment. A search in a user-defined database (DAIOS: Database Assisted Identification of Organic Substances) [63] resulted in 1-adamantylamine, crotamiton, and carbamazepine as the most probable candidates.
Another approach to evaluate relevant compounds was presented by Helbling et al. [15], who investigated TPs of different pharmaceuticals and pesticides in batch experiments for biodegradation. They used a combinatory approach of suspect and nontarget screening. Proposals for suspected TPs were generated by a metabolite prediction software tool (University of Minnesota Pathway Prediction System, UM-PPS) [64] and their accurate masses were matched against measured full-scan MS data. For nontarget screening, Helbling et al. first performed a pattern matching of MS data of original samples at time 0 and at certain degradation times. After further use of a series of mass filters, they were able to reduce the number of measured features to a list of candidate TPs that were formed during the biotransformation experiment. The filters included the m/z ratio, a retention time domain constraint, a background subtraction algorithm, a constrained molecular formula fit, and a plausibility check based on the presence of 13C monoisotopic masses. The resulting candidate TPs were then confirmed or rejected by visual inspection of the mass and tandem mass spectral data regarding the relative abundance of 13C and/or 37Cl monoisotopic masses and/or adduct masses and product ions of each parent compound and TP. As a result, the suspect screening successfully predicted 21 plausible TPs, whereas the nontarget screening resulted in the proposal of 26 TPs. The most probable structures of the five additional TPs were obtained by interpretation of mass spectral data.
A similar approach was used by Kern et al. [65], who looked for plausible microbial TPs of xenobiotics formed in the environment. The examples revealed that if no previous information is available, identification of compounds based only on accurate mass and mass fragmentation data will be very challenging. This was also confirmed for the identification of metabolites of organic water pollutants using LC-HRMS [66]. The successful identification of unknowns is in some way limited by the availability of chemical compound databases and mass spectral libraries with high-resolution information.
LC-HRMS libraries and databases for organic water pollutants
As already mentioned, a simple but reliable approach for the successful identification and confirmation of unknowns is the comparison of measured accurate product/fragment ion mass spectra with accurate mass spectra of authentic reference compounds, as it provides additional selectivity instead of using only the exact mass and the corresponding isotopic pattern. Most of the reported approaches for the identification of unknowns in forensic, metabolomics, and environmental research use user-defined low-resolution and accurate tandem mass spectral or in-source fragmentation libraries [39, 47, 67, 68]. A variety of commercially available and user-defined mass spectral libraries have been developed for certain MS instrument types and settings. Poor reproducibility of tandem mass spectra from different instrument types is found in comparison with EI mass spectra. Various collision energies are applied in tandem mass spectra, and therefore, the relative intensities of ions differ considerably. However, similar product ion patterns have been observed across a range of collision energies [69]. Consequently mass spectral matching fails if the signal intensity is a criterion, but it is successful if only the fragment pattern is considered. Several studies have shown the reproducibility and transferability of tandem mass spectra for use with multiple instrument types [31, 70–72] and that instrument-independent tandem mass spectra can be obtained by application of multiple collision energies for fragmentation [73–75]. In 80 % of all cases, the mass spectrum of an unknown compound could be assigned to a structure if it was compared with two or more reference mass spectra recorded with different instruments or with different collision energies. Hence, a considerable collection of tandem mass spectra obtained with different collision energies and with different instruments can improve the overall performance of a successful library search.
Several commercially and publicly available spectral libraries aim to identify compounds independently of the instrument type and settings. Most of the LC-MS libraries have been developed and published by researchers in life sciences (e.g., proteomics and metabolomics). A recent review [36] on computational MS for metabolomics summarized existing compound libraries containing ESI mass spectra, which include the commercially available NIST reference library and the freely accessible metabolite libraries METLIN, Human Metabolome Database (HMDB), and MassBank. Only the NIST reference library, METLIN, and MassBank contain unit-resolution mass spectra and accurate mass spectral data. The NIST reference library released in 2011 contains 85,344 high- and low-resolution tandem mass spectra of 7,172 different ions from 3,877 compounds, and also includes environmentally relevant compounds.
METLIN [73, 76, 77] is a metabolite database currently containing 29,500 accurate tandem mass spectra from 5,327 metabolites. The tandem mass spectra are recorded with one type of Q-TOF instrument in the positive and negative ESI modes using four different collision energies (0, 10, 20, and 40 eV).
MassBank [78] contains 3,357 entries with accurate mass data and tandem mass spectra obtained with different types of instruments, settings, and ionization modes. It has many options for searching for a mass spectrum, such as by peak, compound name, exact mass, molecular formula, substructure, instrument type, single or multiple fragmentation, and type and mode of ionization. One of the major advantages of MassBank is the free accessibility and the possibility to upload both nominal and accurate mass spectra in common and different data formats [36]. This allows the collection of a considerable amount of useful mass spectral information from a broad research community, which might help to improve the overall performance of successful nontarget analysis. Although some data entries on metabolites are certainly useful for research on ECs, the mass spectral information associated with environmental pollutants is rather scarce. Nevertheless, computational techniques and tools for a reliable library search are well developed, and the spectrum-matching tools and search functions are already optimized for ESI-MS/MS library search. Therefore, it might be possible to extend the database with information on environmental pollutants [35, 78]. MassBank is currently being expanded with environmental pollutants using accurate tandem mass spectral data collected by a network of reference laboratories (NORMAN network) [79].
A database for water pollutants with emphasis on nontarget screening is the DAIOS database [80], which contains numeric information on the nominal and accurate masses of precursor and product ions (e.g., from MS/MS, TOF-MS, and Fourier transform MS). The mass spectral data can be searched for precursor and product ions. Additional useful metadata such as information on sampling points, existing production plants, agricultural uses, special urban situations, molecular data, and chromatographic conditions are compiled to constrain the search or to check the plausibility of the compounds searched. DAIOS currently contains about 344 substances, which is a comparably low number, but is open extension by further users.
Generally, the amount of accurate mass spectral information for environmental contaminants in currently available accurate mass libraries and databases is far from comprehensive. The situation is quite different for EI mass spectra; the current NIST reference library [81, 82] contains more than 240,000 EI mass spectra from 212,961 compounds, whereas the Wiley Registry (ninth edition) [83] contains 662,000 spectra from 592,000 compounds.
Therefore, an important approach of nontarget screening has to rely on general chemical databases and has to deal with the lack of MS library information.
Nontarget approaches based on comprehensive chemical databases and computer-based fragmentation
The nontarget approach is a rather challenging task if no compound databases or library information is available, and the proposal of compound structures is thus based only on HRMS data. The result of this approach is clearly limited to some extent. A novel approach to query chemical databases for structural interpretation in the metabolomics field was reported by Hill et al. [43]. They used measured monoisotopic molecular weights for 102 test compounds to retrieve candidates from a comprehensive chemical database (PubChem). On average, 272 candidates were proposed for each test compound. With the rule-based software program MassFrontier [84], fragmentation spectra were generated for all candidates and then compared with the experimental collision-induced-dissociation spectra of Q-TOF-MS measurements of the unknown structures. As a result, for 65 of 102 test compounds, the highest-ranking candidate matched the correct structure, for 87 compounds the right structure was within the first 20 candidates, and for 98 of 102 compounds the correct elemental formula was ranked first.
This reveals that matching experimental with computational fragment spectra is a promising approach to rapidly discriminate among compounds with the same molecular formula. This approach was refined and customized by Wolf et al. [42] in the form of the open source license software tool MetFrag. Direct database queries in PubChem, ChemSpider, and KEGG [36, 85–87] can be performed on a Web-based platform. From possible candidates, a rather fast algorithm for in silico fragmentation is performed using the bond disconnection approach and a small set of rules to describe molecular rearrangements. The numbers of molecular fragments which match the measured peaks are then scored on the basis of the number of fragments explaining the measured peaks and the bond dissociation energy. The higher the bond dissociation energy, the less likely the fragment is considered. Further details on MetFrag can be found in Wolf et al. [42], whereas details on the systematic bond disconnection approach can be found in Hill and Mortishire-Smith [84].
Other more specific approaches are promising to predict and explain some of the fragmentation mechanisms, but have much higher demands on computing time such as the application of ab initio calculations in density functional theory [88].
Other commercially available and freely accessible computational tools (e.g., ACD/Fragmenter, Mass Frontier, Sirius Starburst, and SmartFormula3D) are available to support the identification of unknown compounds. However, MetFrag revealed better performance than MassFrontier on the data set used by Hill et al. [43], with lower standard deviations of the correct ranks [36].
We are interested in how this approach is applicable to environmental contaminants and their metabolites. For this purpose, we selected mass spectral data of 21 ECs and metabolites from contaminated sites [62, 89] and from the compound classes pharmaceuticals [15], diagnostics [45, 90], and pesticides [62, 65]. We used the exact mass of the compounds as input for an upstream search in PubChem with mass windows of 2, 5, and 10 ppm using MetFrag. Between 28 and 2,420 candidate structures were retrieved for each compound (Table 1). By matching one or more exact mass fragments with in silico generated fragments of the first 100 hits, we considerably reduced the number of possible candidates to between 2 and 36, depending on the number of isomers which cannot be distinguished by mass spectral fragmentation. The correct structure was ranked first for eight compounds, and was among the first ten ranks in 18 cases. In four cases the correct compound could not be found in the chemical database (no. 21, the TP of iomeprol; nos. 1, 13, and 14, succinic acid derivatives of benzofuran and methylnaphthalene; see Table 1). Hence, this approach works only for compounds which are listed in a chemical database.
Further information on the sample and its contamination, and on the separation and analysis such as chromatographic retention times and ionization efficiencies would be necessary to further reduce the number of most likely candidates. This has been demonstrated for low-resolution GC-EI-MS and LC-EI-MS data [91]. The retention index and boiling point correlation, octanol–water partition coefficients, steric energies, and the linear solvation energy relationships approach have been further used to limit the number of candidate structures [92]. A combination of computer-based structure generation and mass spectral classifiers has been applied for low-resolution GC-MS data in effect-directed analysis [93]. This approach can be used as an alternative to library search or if a match in the database is not available.
Conclusions
The recent literature on different nontarget screening approaches using LC-HRMS reveals some promising results. In most cases only one or a few compounds could be identified by the nontarget approach, which requires several steps from measurement of data to compound identification. Generally, data reduction, MS library search, and the use of further MS and chromatographic data (MS fragmentation, retention time) are required for successful nontarget analysis. Unequivocal identification of unknowns still requires mass spectral information from authentic reference standards using user-defined, public, or commercially available ESI-MS/MS databases or libraries with high-resolution information.
However, high-resolution mass spectral information associated with environmental pollutants is still scarce. The currently available ESI-MS/MS databases and libraries are still unsuitable for a comprehensive library search, and hence a comprehensive identification of nontargeted analytes is not possible. Freely accessible and publicly available MS libraries with the possibility to upload accurate mass spectra in common and different data formats allow a considerable amount of useful mass spectral information to be collected from a broad research community, which might help to improve the overall performance of successful nontarget analysis. Such libraries with well-developed computational search options already exist for use in metabolomics research.
A big step forward has been achieved with computer-based tools if no MS library or MS database entry is found for a compound. Our examples of selected compounds and metabolites from recent publications have demonstrated that numerous search results from a large chemical database can be limited to only a few by in silico fragmentation. Still, only very few compounds could be identified or tentatively identified in environmental samples by nontarget screening. In most cases the availability of comprehensive MS libraries with a focus on environmental contaminants is the limiting factor. Further information on the analyte characteristics in chromatography and ionization will gain increasing importance in nontarget screening.
References
Ferrer I, Thurman EM (2003) Trends Anal Chem 22(10):750–756
Loos R, Gawlik BM, Locoro G, Rimaviciute E, Contini S, Bidoglio G (2009) Environ Pollut 157(2):561–568
Schultz MM, Furlong ET, Kolpin DW, Werner SL, Schoenfuss HL, Barber LB, Blazer VS, Norris DO, Vajda AM (2010) Environ Sci Technol 44(6):1918–1925
Valcárcel Y, Alonso SG, Rodríguez-Gil JL, Maroto RR, Gil A, Catalá M (2011) Chemosphere 82(7):1062–1071
Verlicchi P, Galletti A, Petrovic M, Barceló D (2010) J Hydrol 389(3–4):416–428
Wang C, Shi H, Adams CD, Gamagedara S, Stayton I, Timmons T, Ma Y (2011) Water Res 45(4):1818–1828
Kolpin DW, Furlong ET, Meyer MT, Thurman EM, Zaugg SD, Barber LB, Buxton HT (2002) Environ Sci Technol 36(6):1202–1211
Ternes TA, Joss A, Siegrist H (2004) Environ Sci Technol 38(20):392A–399A
Richardson SD, Ternes TA (2011) Anal Chem 83(12):4614–4648
Richardson SD (2010) Anal Chem 82(12):4742–4774
Perez S, Eichhorn P, Celiz MD, Aga DS (2006) Anal Chem 78(6):1866–1874
Escher BI, Fenner K (2011) Environ Sci Technol 45(9):3835–3847
Kosjek T, Heath E (2008) Trends Anal Chem 27(10):807–820
Zwiener C (2007) Anal Bioanal Chem 387(4):1159–1162
Helbling DE, Hollender J, Kohler H-PE, Singer H, Fenner K (2010) Environ Sci Technol 44(17):6621–6627
Matamoros V, Jover E, Bayona J (2009) Anal Bioanal Chem 393(3):847–860
von der Ohe PC, Dulio V, Slobotnik J, De Deckere E, Kuehne R, Ebert R-U, Ginebreda A, De Cooman W, Schueuermann G, Brack (2011) Sci Total Environ 409(11):2064–2077
Gros M, Petrovic M, Barcelo D (2006) Anal Bioanal Chem 386(4):941–952
Zwiener C, Frimmel FH (2004) Anal Bioanal Chem 378(4):851–861
Zwiener C, Frimmel FH (2004) Anal Bioanal Chem 378(4):862–874
Al-Odaini NA, Zakaria MP, Yaziz MI, Surif S (2010) J Chromatogr A 1217(44):6791–6806
Hao C, Zhao X, Tabe S, Yang P (2008) Environ Sci Technol 42(11):4068–4075
Noedler K, Licha T, Bester K, Sauter M (2010) J Chromatogr A 1217(42):6511–6521
Lara-Martin PA, Gonzalez-Mazo E, Brownawell BJ (2011) J Chromatogr 1218(30):4799–4807
Rodil R, Quintana JB, Lopez-Mahia P, Muniategui-Lorenzo S, Prada-Rodriguez D (2009) J Chromatogr 1216(14):2958–2969
Liu FM, Bischoff G, Pestemer W, Xu W, Kofoet A (2006) Chromatographia 63(5–6):233–237
Makarov A, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S (2006) Anal Chem 78(7):2113–2120
Ibáñez M, Sancho JV, Hernández F, McMillan D, Rao R (2008) Trends Anal Chem 27(5):481–489
Gomez MJ, Gomez-Ramos MM, Malato O, Mezcua M, Fernandez-Alba AR (2010) J Chromatogr 1217(45):7038–7054
Krauss M (2010) Anal Bioanal Chem 397:943–951
Kind T, Fiehn O (2010) Bioanal Rev 2(1–4):23–60
Marshall AG, Rodgers RP (2008) Proc Natl Acad Sci USA 105(47):18090–18095
Kim S, Rodgers RP, Marshall AG (2006) Int J Mass Spectrom 251(2–3):260–265
Want E, Masson P (2011) In: Metz TO (ed) Metabolic profiling. Methods in molecular biology, vol 708. Springer, Berlin, pp 277–298
Schymanski EL, Schulze T, Hermans J, Brack W (2011) In: Brack W (ed) Handbook of environmental chemistry, vol.15. Springer, Berlin, pp 167–198
Neumann S, Böcker S (2010) Anal Bioanal Chem 398(7):2779–2788
Hernández F, Portolés T, Pitarch E, López FJ (2007) Anal Chem 79(24):9494–9504
García-Reyes JF, Hernando MD, Molina-Díaz A, Fernández-Alba AR (2007) Trends Anal Chem 26(8):828–841
Hogenboom AC, van Leerdam JA, de Voogt P (2009) J Chromatogr A 1216(3):510–519
Hernández F, Portolés T, Pitarch E, López FJ (2011) Trends Anal Chem 30(2):388–400
Pelander A, Tyrkko E, Ojanpera I (2009) Rapid Commun Mass Spectrom 23(4):506–514
Wolf S, Schmidt S, Muller-Hannemann M, Neumann S (2010) BMC Bioinform 11(1):148
Hill DW, Kertesz TM, Fontaine D, Friedman R, Grant DF (2008) Anal Chem 80(14):5574–5582
Kormos JL, Schulz M, Wagner M, Ternes TA (2009) Anal Chem 81(22):9216–9224
Kormos JL, Schulz M, Kohler H-PE, Ternes TA (2010) Environ Sci Technol 44(13):4998–5007
Ibáñez M, Sancho JV, Pozo ÓJ, Niessen W, Hernández F (2005) Rapid Commun Mass Spectrom 19(2):169–178
Bobeldijk I, Vissers JPC, Kearney G, Major H, van Leerdam JA (2001) J Chromatogr A 929(1–2):63–74
Hao H, Cui N, Wang G, Xiang B, Liang Y, Xu X, Zhang H, Yang J, Zheng C, Wu L, Gong P, Wang W (2008) Anal Chem 80(21):8187–8194
Meng C-K, Zweigenbaum JA (2010) J AOAC 93(2):703–711
Bester K, Huehnerfuss H, Lange W, Theobald N (1997) Sci Total Environ 207(2–3):111–118
Bester K, Theobald N (2000) Water Res 34(8):2277–2282
Mortishire-Smith RJ, O'Connor D, Castro-Perez JM, Kirby J (2005) Rapid Commun Mass Spectrom 19(18):2659–2670
Weigel S, Bester K, Huehnerfuss H (2001) J Chromatogr A 912(1):151–161
Petri M, Jiang JQ, Maier M (2010) Water Sci Technol Water Supply 10(5):806–814
Grigoriadou A, Schwarzbauer J (2011) Water Air Soil Pollut 214(1):623–643
Edler B, Zwiener C, Frimmel FH (1997) Fresenius J Anal Chem 359(3):288–292
Bester K, Huehnerfuss H, Lange W, Rimkus GG, Theobald N (1998) Water Res 32(6):1857–1863
Zwiener C, Frimmel FH (1998) Fresenius J Anal Chem 360(7–8):820–823
Gómez MJ, Gómez-Ramos MM, Agueera A, Mezcua M, Herrera S, Fernández-Alba AR (2009) J Chromatogr A 1216(18):4071–4082
Cappiello A, Famiglini G, Palma P, Pierini E, Termopoli V, Trufelli H (2011) Mass Spectrom Rev 30(6):1242–1255
Cappiello A, Famiglini G, Termopoli V, Trufelli H, Zazzeroni R, Jacquoilleot S, Radici L, Saib O (2011) Anal Chem 83(22):8537–8542
Mueller A, Schulz W, Ruck WK, Weber WH (2011) Chemosphere 85(8):1211–1219
Müller A, Schulz W, Weber W (2009) Paper presented at the 75. Jahrestagung der Wasserchemischen Gesellschaft, Stralsund, 18–20 May 2009
Ellis L, Wackett L, Liu Y, Turnbull M (2011) University of Minnesota. http://umbbd.msi.umn.edu/predict/. Accessed Dec 2011
Kern S, Fenner K, Singer HP, Schwarzenbach RP, Hollender J (2009) Environ Sci Technol 43(18):7039–7046
Hernandez F, Ibanez M, Sancho JV, McMillan D, Rao R (2008) Trends Anal Chem 27(5):481–489
Gomez C, Segura J, Monfort N, Suominen T, Leinonen A, Vahermo M, Yli-Kauhaluoma J, Ventura R (2010) Anal Bioanal Chem 397(7):2903–2916
Dresen S, Ferreiros N, Gnann H, Zimmermann R, Weinmann W (2010) Anal Bioanal Chem 396(7):2425–2434
Volna K, Holcapek M, Kolarova L, Lemr K, Caslavsky J, Kacer P, Poustka J, Hubalek M (2008) Rapid Commun Mass Spectrom 22(2):101–108
Bristow AWT, Webb KS, Lubben AT, Halket J (2004) Rapid Commun Mass Spectrom 18(13):1447–1454
Hopley C, Bristow T, Lubben A, Simpson A, Bul E, Klagkou K, Herniman J, Langley J (2008) Rapid Commun Mass Spectrom 22(12):1779–1786
Milman BL (2005) Rapid Commun Mass Spectrom 19(19):2833–2839
Pavlic M, Schubert B, Libiseller K, Oberacher H (2010) Forensic Sci Int 197(1–3):40–47
Oberacher H, Pavlic M, Libiseller K, Schubert B, Sulyok M, Schuhmacher R, Csaszar E, Kofeler HC (2009) J Mass Spectrom 44(4):485–493
Oberacher H, Pavlic M, Libiseller K, Schubert B, Sulyok M, Schuhmacher R, Csaszar E, Kofeler HC (2009) J Mass Spectrom 44(4):494–502
Sana TR, Roark JC, Li X, Waddell K, Fischer SM (2008) J Biomol Tech 19(4):258–266
Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G (2005) Ther Drug Monit 27(6):747–751
Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Tanaka K, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T (2010) J Mass Spectrom 45(7):703–714
Network of reference laboratories for monitoring of emerging environmental pollutants (Norman) (2011) http://www.norman-network.net/index_php.php. Accessed Dec 2011
Zweckverband Landeswasserversorgung (2010) http://www.daios-online.de/daios/. Accessed Dec 2011
Sparkman OD (2011) NIST 11: what's new and what value does it offer? Part I. Available via http://www.sepscience.com/mssolutions001
Sparkman OD (2011) NIST 11: what's new and what value does it offer? Part II. Available via http://www.sepscience.com/mssolutions001
Wiley Registry of Mass Spectral Data, 9th Edition (2011). John Wiley & Sons
Hill AW, Mortishire-Smith RJ (2005) Rapid Commun Mass Spectrom 19(21):3111–3118
National Center for Biotechnology Information (2011) http://pubchem.ncbi.nlm.nih.gov/. Accessed Dec 2011
Kanehisa Laboratories (2011) http://www.genome.jp/kegg/. Accessed Dec 2011
Royal Society of Chemistry (2011) http://www.chemspider.com/. Accessed Dec 2011
Levsen K, Schiebel H-M, Terlouw JK, Jobst KJ, Elend M, Preiß A, Thiele H, Ingendoh A (2007) J Mass Spectrom 42(8):1024–1044
Jobelius C, Ruth B, Griebler C, Meckenstock RU, Hollender J, Reineke A, Frimmel FH, Zwiener C (2010) Environ Sci Technol 45(2):474–481
Zwiener C, Glauner T, Sturm J, Woerner M, Frimmel FH (2009) Anal Bioanal Chem 395(6):1885–1892
Schymanski EL, Meringer M, Brack W (2011) Anal Chem 83(3):903–912
Ulrich N, Schuurmann G, Brack W (2011) J Chromatogr 1218(45):8192–8196
Schymanski EL, Meinert C, Meringer M, Brack W (2008) Anal Chim Acta 615(2):136–147
Prasse C, Schluesener MP, Schulz R, Ternes TA (2010) Environ Sci Technol 44(5):1728–1735
Acknowledgments
The authors would like to thank Karina Zedda for reviewing this manuscript and three anonymous reviewers for their helpful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in the topical collection Analytical Challenges in Environmental and Geosciences with guest editor Christian Zwiener.
Rights and permissions
About this article
Cite this article
Zedda, M., Zwiener, C. Is nontarget screening of emerging contaminants by LC-HRMS successful? A plea for compound libraries and computer tools. Anal Bioanal Chem 403, 2493–2502 (2012). https://doi.org/10.1007/s00216-012-5893-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-012-5893-y