Abstract
Acinetobacter species remain alive in hospitals on various surfaces, both dry and moist, forming an important source of hospital infections. These bacteria are naturally resistant to many antibiotic classes. Although the role of the quorum sensing system in regulating the virulence factors of Acinetobacter species has not been fully elucidated, it has been reported that they play a role in bacterial biofilm formation. The biofilm formation helps them to survive under unfavorable growth conditions and antimicrobial treatments. It is based on the accumulation of bacterial communication signal molecules in the area. In this study, we compared the bacterial signal molecules of 50 nosocomial Acinetobacter baumannii strain and 20 A. baumannii strain isolated from soil. The signal molecules were detected by the biosensor bacteria (Chromobacterium violaceum 026, Agrobacterium tumefaciens A136, and Agrobacterium tumefaciens NTL1) and their separation was determined by thin-layer chromatography. As a result, it has been found that soil-borne isolates can produce 3-oxo-C8-AHL and C8-AHL, whereas nosocomial-derived isolates can produce long-chain signals such as C10-AHL, C12-AHL, C14-AHL and C16-AHL. According to these results, it is possible to understand that these signal molecules are found in the infection caused by A. baumannii. The inhibition of this signaling molecules in a communication could use to prevent multiple antibiotic resistance of these bacteria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The antibiotics used in the treatment of infections have been considered as one of the most important discoveries of the century. In the past few years, extensive and improper use of antibiotics have caused the microorganisms and antimicrobials to become immune which has cast a cloud on the success achieved against the infectious agents and it has become an issue threatening the health of the people. Microorganisms become immune with different mechanisms toward the antibiotics, which they are sensitive to, and be effective in pathogenicity. An environmental sensing system, in other words, the bacterial communication system used among microorganisms called quorum sensing, promises hope in antimicrobial treatment and draws hope as a system where the damage to the host is at minimum levels. Briefly, the communication system known as “quorum sensing” can be defined as the population increase of species in the same environment in accordance with these species detecting this increase and, as a result, showing a change in behavior such as all individuals organizing their particular genes expressions in coordination (McClean et al. 1997; Shaw et al. 1997). Although the quorum sensing system was first determined in Gram-negative bacteria, it is also a microbial communication system used in Gram-positive bacteria and some fungus (Chen et al. 2004; de Kievit and Iglewski 2000). Enlightening this communication system and determining the active molecules in infection will make it easier for the treatment of an infection. According to the research conducted by World Health Organization (WHO), it is seen that Turkey takes place next to the top of the countries that have become immune to antibiotics observed in almost all kinds of bacteria (Bozkurt et al. 2014). The environments mostly causing the development of an immunity against antibiotics are hospitals. Hospital infections are stated as the infections that occur when a patient comes to the hospital with a health problem, ends up acquiring another infection. According to Ertek, hospital infections usually develop 48–72 h after hospitalization and within 10 days after being discharged from the hospital (Ertek 2008). They extend the duration of hospitalization, along with causing significant health problems, also having losses and the length of treatment extending with the cost of treatment rising, and sometimes causing the treatment to end in failure. Among the factors of the hospital infections, Gram-negative species such as Pseudomonas aeruginosa, Escherichia coli, Klebsiella spp., Enterobacter spp., Acinetobacter spp. and Serratia spp. are the utmost ones (Tariq 2014). The multi-drug resistance that is observed against the antibiotics in these microorganisms is confronted with a problem that is gradually increasing (Koul et al. 2016). According to the results of some studies conducted in our country that covers a wide range, some bacteria that are frequently isolated from intensive care units, such as Acinetobacter, show resistance against antibiotics and have very limited alternative treatments. Hence, it is observed that the Acinetobacter strains which are the ones isolated from clinic samples and the ones isolated from the environment differ in terms of antibiotic resistance. One of the reasons of this differentiation is bacterial communication system. In this context, there is an observational proof showing that different types of the bacterial communication system can take an antibacterial effect in different ways (Priya et al. 2013; Waheed et al. 2016; Zahin et al. 2010). In this study, the quorum sensing signal molecules used by Acinetobacter strains, which were isolated from patients with hospital infections in various clinics, were determined; concurrently, signal molecules produced by Acinetobacter strains isolated from nature were compared, and which signal molecules, which were active in infections, were researched.
Materials and methods
Bacterial strains and culture conditions
Fifty Acinetobacter baumannii isolates used in the study were isolated from different clinical samples and 20 A. baumannii isolates were isolated from soil samples. Clinical samples from hospitalized patients were inoculated on blood agar medium and incubated at 37 °C for 24 h. Preliminary identification was performed by Gram staining, colony morphology, culturing on MacConkey’s agar (oxoid), non-lactose fermenting bacteria, oxidase activity, catalase activity identified by the traditional biochemical test according to the additional data confirmed by Vitek 32 system. Acinetobacter baumannii (ATCC 19606) strain was used as a positive control when defining bacteria. Isolation of Acinetobacter species from soil was carried out according to Baumann (1968). Strains isolated were checked in CHROMagar Acinetobacter (CHROMagar, France). The AHL reporter strains Chromobacterium violaceum CV026, Chromobacterium violaceum 31532, Chromobacterium violaceum 12472, Agrobacterium tumefaciens NTL1 (pZLR4) and Agrobacterium tumefaciens A136 were kindly supplied by Prof. Dr. Robert J.C. McLean (Texas State University-San Marcos), Prof. Dr. Stephen K Farrand (University of Illinois at Urbana-Champaign), Prof. Dr. Stephen Winans (Cornell University) and Associate Professor Scott Rice (University of New South Wales), respectively. The bacteria were grown in Luria–Bertani medium (Merck) at 30–37 °C overnight, apart from A. tumefaciens A136 and A. tumefaciens NTL1 which was cultured on a supplemented minimal AB medium at 30 °C (Miller 1972). For determination of the presence of the signal molecules, the bacterial culture supernatant was collected, and it was filtered through a 0.22-m filter and stored at -20℃.
Screening of AHL by C. violaceum CV026 and A. tumefaciens A136 or A. tumefaciens NTL1 biosensor
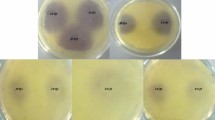
Fresh cultures of the A. baumannii strains were used with the strains isolated from soil incubated in Tryptic Soy Broth Medium in 30 °C for 6 h and ones from clinic samples in 37 °C for determining quorum sensing molecules. For determining the production of Acyl homoserine, Acinetobacter spp. and biosensor C. violaceum 026 strain were inoculated side by side in a way that they had a 1 cm gap between them (McClean et al. 1997) (Fig. 1). C. violaceum 026 was assessed positive or negative according to the color changes in biosensor strain.
Whether Acinetobacter species produce quorum sensing signal molecules or not was examined after being left for incubating in 30 °C for 24 h after inoculating process in Luria–Bertani Agar. This test conducted to determine the existence of C4–C6 AHL signal molecules was conducted in three parallels (McLean et al. 2004; Morohoshi et al. 2008). As suggested, A. tumefaciens A136 species were inoculated in Luria–Bertani medium with 50 mg/ml spectinomycin and 4.5 ug/ml tetracycline. A. tumefaciens NTL1 (pZLR4) was inoculated in Luria–Bertani medium with 30 mg/ml gentamicin and it was incubated at 28 °C for 48 h (Fuqua et al. 1994; Sio et al. 2006). P. aeruginosa PAO1 known to produce 3-oxo-C8 AHL was used as a positive control and it was incubated in LB agar medium at 37 °C for 24 h (Fig. 1). Acinetobacter species were produced in the ready 37 °C Tryptic Soy Broth medium for the detection of N-acyl homoserine lactone. One of the sensor strains of A. tumefaciens A136 or A. tumefaciens NTL1 (pZLR4) selected in a way that there was 1 cm between Acinetobacter species was inoculated next to each other. X-gal is disseminated over AB Mineral agar medium previously including 0.2% Mannitol in sterile conditions with the aid of glass beads (Fig. 2).
After inoculation process, whether Acinetobacter species left for incubation for 72 h at 30 °C produced quorum sensing signal molecules, was analyzed (González et al. 2001; Pina et al. 1998). The medium used for the detection of the quorum sensing is AB Mineral Medium. The prepared medium was sterilized at 121 °C for 25 min in an autoclave. 60 μg/ml X-gal was added to the sterilized surface of the medium and the glass beads were used to spread on the surface. The surface of the medium was dried on a fume hood without an exposure to sunlight. This test was repeated three times for the detection of the presence of C4–C12 AHL signal molecules. Acinetobacter species were inoculated as mentioned above and incubated at 28 °C for 48 h in AB mineral agar medium (Shaw et al. 1997).
Quorum sensing standard signal molecules
The quorum sensing signal molecules determined to have existence in Acinetobacter species have been evaluated with the signal molecule standards such as N-butyryl-l-homoserine lactone (BHL, C4-HSL, C8H13NO3), N-hexanoyl-l-homoserine lactone (HHL, C6-HSL, C10H17NO3), N-octanoyl-l-homoserine lactone (OHL, C8-HSL, C12H21NO3), N-decanoyl-l-homoserine lactone (DHL, C10-HSL, C14H25NO3), N-dodecanoyl-l-homoserine lactone (dDHL, C12-HSL, C16H29NO3), N-tridecanoyl-l-homoserine lactone (triDHL, C13-HSL, C17H31NO3), N-tetradecanoyl-l-homoserine lactone (tDHL, C14-HSL, C18H33NO3), N-hexadecanoyl-l-homoserine lactone (hDHL, C16-HSL, C20H35NO3), N-octadecanoyl-l-homoserine lactone (oDHL, C18-HSL, C22H39NO3). These standard signal molecules have been obtained from The School of Molecular Medical Sciences, Nottingham University.
AHL extraction and analytical thin-layer chromatography
A loopful of Acinetobacter strains known to produce N-acyl homoserine lactone was taken from the culture and produced in 25 ml tryptic soy broth medium at 37 °C for 24 h. The obtained culture was centrifuged at 4000 rpm for 10 min at 4 °C. The supernatant part was transferred to a new tube and the ethyl acetate solution containing an equal volume of 0.1% acetic acid was added to it. The organic phase transferred to a new tube was filtered through a sterile syringe tip with the pore size of 0.2 μm (non-pyrogenic, Sartorius). Then, it was kept in the fume hood at room temperature for one week for the removal of ethyl acetate (González et al. 2009; Stacy et al. 2012). The residue was dissolved in methanol after the organic phase in tubes evaporated completely organic. Then, the residue was applied to reversed phase TLC plates (RP-18 F254, Merck). Chloroform–methanol (95: 5, v/v) was prepared as a mobile phase. The mobile phase was allowed to proceed to 2 cm below the top of the thin-layer plate. After the separation was completed, TLC plates were left to dry in the fume hood for 2 h. A. tumefaciens A136 or NTL1 was used as biosensor bacteria in the separation of the molecules. 50 ml AB mineral soft agar medium containing 0.9% agar (50 °C) + 10 ml of A. tumefaciens A136 (48 h, 100 ml) culture and X-gal (60 μg/ml) was added onto the dried TLC plate and allowed to spread. The TLC plate was incubated at 30° C for 24 h (González et al. 2001, 2009; Stacy et al. 2012). Methanol was used as a negative control during the test. Standard C4 AHL, C6-AHL, C8-AHL, AHL-C10, C12-AHL, AHL-C13, C14-AHL, AHL-C16, C18-AHL, and 20-AHL signal molecules were used. The regions where the AHL molecules were found were determined by the formation of a greenish spot on a white background.
Results and discussion
Gram feature of Acinetobacter strains used in our study, MacConkey agar reproductive status, oxidase activity, catalase activity, growth at 42–44 °C were controlled and identified. The number of the strains analyzed, the tests and the results of these tests are shown in Table 1. When the test results were examined, the samples taken from hospitals were defined as A. baumannii in accordance with the data of the automated Vitek-32 system. In this context, the results of the tests used for the identification of the soil and clinical isolates were found to be coherent.
As mentioned earlier, Chromobacterium violaceum 026 biosensor strain is used to detect the C4-C6 AHL molecules. By the presence of these molecules, production of the violacein pigment is triggered and CV026 produces purple pigments. According to the results of cross-validation tests with the Acinetobacter species, they are not producing homoserine lactones including the short chain of carbon. According to the data of some researchers, Acinetobacter species both produce the short-chain signal molecules in low rates and are able to degrade them (Chan et al. 2011). In this study, that the purple pigment is not seen can be explained by low rate of the production of homoserine lactone with short chains in the strains and fast degradation of molecules through the strains.
Agrobacterium tumefaciens NTL1 and A. tumefaciens A136 biosensor strains are used to determine C8-C14 AHL molecules. In the biosensor strains within the presence of these molecules, β-galactosidase enzyme production is triggered and a green color is observed due to the presence of X-gal in the environment. As a result of the cross-validation test with A. tumefaciens NTL1 and A. tumefaciens A136 biosensor strains, the Acinetobacter species which are included in our study, all strains are observed to produce signal molecules (Fig. 2).
Literature shows that co-authors for the detection of situated bacterial signaling molecules in Yersinia enterocolitica W828 and Serratia liquefaciens MG1 conducted cross-validation test with Chromobacterium violaceum 026 and when they were inoculated next to each other at the end of the study, it was observed that the violacein pigment production was triggered in Chromobacterium violaceum 026 strains and they detected the differences of these molecules through thin-layer chromatography (McClean et al. 1997). In another study, it was concluded that the bacterial communication molecule was widely produced in strains isolated from intensive care units such as Pseudomonas aeruginosa, A. baumannii, Escherichia coli and Klebsiella pneumoniae. Unlike other studies conducted, they showed that cross-validation tests of P. aeruginosa strain with C. violaceum 026 strains gave negative results. They suggested that a possible reason as such: P. aeruginosa isolates might have short-chain AHL or the level of the signal molecules might be too low (Boşgelmez-Tınaz et al. 2005).
As previously mentioned, A. tumefaciens NTL1 and A. tumefaciens A136 are used to determine the long-chain signal molecules. The mechanism causing A. baumannii to degrade the short-chain signal molecules is not effective on the long-chain molecules. In addition, biosensor bacteria have been shown to be able to detect these signal molecules when they can be produced at high rates. The use of biosensor bacteria for the detection of the signal molecule emerges as a common practice. Although the studies conducted with the biosensor bacteria focus more on pathogenic bacteria, nowadays the studies about bacterial communication from agriculture to health industry and food industry are being conducted.
The signal molecules of N-acyl homoserine lactone species were extracted from the Acinetobacter cultures, which were allowed to produce for 12 h. Then, they were separated from each other by applying a thin-layer chromatography method. The Rf values of the signal molecules which are used as a standard and the signal molecules produced by the species isolated from the hospitals and the soil are shown in Tables 2 and 3. As the short-chain C4-AHL and C6-AHL were not identified to be produced by the species via cross-validation, the thin-layer chromatography was not used as a standard. After the Rf values of the signal molecules isolated from the species were determined, a classification of the molecules took place by taking compliance of the Rf values to the standards into consideration. For example, in Table 3, the Rf1 value of the signal molecule produced by A1 isolate was 0.58 and the Rf range of 3-oxo-C8-AHL was known as 0.58 and 063; the molecule corresponding to this Rf range was accepted as 3-oxo-C8-AHL.
Acinetobacter baumannii isolated from the hospital show a multiple drug resistance. Considering the recent data from the hospitals, there is a wide range of a drug resistance as well as pan-drug resistance strains (Kostoulias et al. 2016; Modarresi et al. 2016). The antimicrobial resistance mechanism of A. baumannii strain has been described in many studies with the quorum sensing system (Bhargava et al. 2015; Kalia 2013). We believe that the identification of the signaling molecules produced by A. baumannii will form a background for the newly developed drugs. In our study, the species, which were nosocomial A. baumannii as a result of thin-layer chromatography, were determined to carry more than one signal molecules. When the Acinetobacter species isolated from the soil were examined, it was observed that they could produce a lower number of the signal molecules when compared to the Acinetobacter species. In particular, the molecules produced by the soil-based Acinetobacter including 3-oxo-C8-AHL and C8-AHL were observed to produce all of them or individually. Similar to the various studies summarized above, in our study, all strains isolated from the soil and the clinical samples were observed to produce the long-chain signal molecules. On the other hand, as mentioned above, the findings of not being able to identify the short-chain AHLs via the biosensor strains do not mean that these molecules are not certainly synthesized. Although it is possible that these molecules may be synthesized, it is also possible that a number of synthesized molecules are very little or it is degraded. This situation may be due to some enzymes that they have (Chow et al. 2014). A study has shown that the common signaling molecule C8-AHL produced by A. calcoaceticus–A. baumannii association can produce three different signal molecules (González et al. 2009). In A. baumannii as well as in other pathogenic bacteria, the biofilm formation and the movement are regulated by the QS system (Castillo-Juarez et al. 2017; González et al. 2009; Stacy et al. 2012). Similar to the former summarized studies with which the signal molecules of the nosocomial or Gram-negative bacteria isolated from nature, our study shows that A. baumannii can also produce different signal molecules (Chan et al. 2014; Chow et al. 2014). Upon analyzing the data, it is demonstrated that A. baumannii can produce all or a few of the 3-oxo-C8-AHL, C8-AHL, and C12-AHL molecule. In addition, with this study, it is determined that the nosocomial A. baumannii species could also produce C14-AHL and C16-AHL signal molecules. The long-chain signal molecules are produced for communication and pathogenicity.
Conclusion
Overall, our study shows that the nosocomial and soil-based Acinetobacter species use different signal molecules in bacterial communication. 3-oxo-C8-AHL and C8-AHL molecules can only be the ones used for Acinetobacters to recognize each other in bacterial communication. C10-AHL, C12-AHL, C14-AHL and C16-AHLs only seen in the nosocomial isolates are thought to be the active molecules in virulence. In addition, the study gives the idea that these molecules have roles in an easy adaptation of these bacteria, which have been isolated from various sources to various environments. In avoiding the development of antibiotic resistance, blocking this system will be a pre-stage for the future studies.
References
Baumann P (1968) Isolation of Acinetobacter from soil and water. J Bacteriol 96:39–42
Bhargava N, Sharma P, Capalash N (2015) Quorum sensing in Acinetobacter baumannii. Quorum sensing vs quorum quenching: a battle with no end in sight. Springer, India, pp 101–113
Boşgelmez-Tınaz G, Ulusoy S, Arıdoğan B, Eroğlu F, Kaya S (2005) N-butanoyl-l-homoserine lactone (BHL) deficient Pseudomonas aeruginosa isolates from an intensive care unit. Microbiol Res 160:399–403. doi:10.1016/j.micres.2005.03.005
Bozkurt F, Kaya S, Tekin R, Gulsun S, Deveci O, Dayan S, Hosoglu S (2014) Analysis of antimicrobial consumption and cost in a teaching hospital. J Infect Public Health 7:161–169. doi:10.1016/j.jiph.2013.09.007
Castillo-Juarez I et al. (2017) Exploiting quorum sensing inhibition for the control of Pseudomonas aeruginosa and Acinetobacter baumannii biofilms. Curr Top Med Chem 7. PMID:28056745
Chan K-GG et al (2011) Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol 11:51. doi:10.1186/1471-2180-11-51
Chan K-G, Cheng HJ, Chen JW, Yin W-F, Ngeow YF (2014) Tandem mass spectrometry detection of quorum sensing activity in multidrug resistant clinical isolate Acinetobacter baumannii. Sci World J. doi:10.1155/2014/891041
Chen H, Fujita M, Feng Q, Clardy J, Fink GR (2004) Tyrosol is a quorum-sensing molecule in Candida albicans. PNAS 101:5048–5052. doi:10.1073/pnas.0401416101
Chow JY, Yang Y, Tay SB, Chua KL, Yew WS (2014) Disruption of biofilm formation by the human pathogen Acinetobacter baumannii using engineered quorum-quenching lactonases. Antimicrob Agents Chemother 58:1802–1805. doi:10.1128/AAC.02410-13
de Kievit TR, Iglewski BH (2000) Bacterial quorum sensing in pathogenic relationships. Infect Immun 68:4839–4849. doi:10.1128/iai.68.9.4839-4849.2000
Ertek M (2008) Hastane Enfeksiyonları: Türkiye Verileri. İÜ Cerrahpaşa Tıp Fakültesi Sürekli Tıp Eğitimi Etkinlikleri 60:9–14
Fuqua WC, Winans SC, Greenberg EP (1994) Quorum sensing in bacteria: the LuxR–LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176:269–275 (PMC205046)
González RH, Nusblat A, Nudel BC (2001) Detection and characterization of quorum sensing signal molecules in Acinetobacter strains. Microbiol Res 155:271–277. doi:10.1016/S0944-5013(01)80004-5
González RH, Dijkshoorn L, Van den Barselaar M, Nudel C (2009) Quorum sensing signal profile of Acinetobacter strains from nosocomial and environmental sources. Rev Argent Microbiol 41:73–78 PMID:19623895
Kalia VC (2013) Quorum sensing inhibitors: an overview. Biotechnol Adv 31:224–245. doi:10.1016/j.biotechadv.2012.10.004
Kostoulias X et al (2016) Impact of a cross-kingdom signaling molecule of Candida albicans on Acinetobacter baumannii physiology. Antimicrob Agents Chemother 60:161–167. doi:10.1128/AAC.01540-15
Koul S, Prakash J, Mishra A, Kalia VC (2016) Potential emergence of multi-quorum sensing inhibitor resistant (MQSIR) bacteria. Indian J Microbiol 56:1–18. doi:10.1007/s12088-015-0558-0
McClean KH et al (1997) Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology (Reading, England) 143(Pt 12):3703–3711
McLean RJ, Pierson LS, Fuqua C (2004) A simple screening protocol for the identification of quorum signal antagonists. J Microbiol Methods 58:351–360. doi:10.1016/j.mimet.2004.04.016
Miller J (1972) Experiments in molecular genetics. Cold Springs Harbor University Press, Cold Spring Harbor
Modarresi F, Azizi O, Shakibaie MR, Motamedifar M, Mansouri S (2016) Cloning and expression of quorum sensing N-acyl-homoserine synthase (LuxI) gene detected in Acinetobacter baumannii. Iran J Microbiol 8:139 (PMC4906721)
Morohoshi T, Kato M, Fukamachi K, Kato N, Ikeda T (2008) N-acylhomoserine lactone regulates violacein production in Chromobacterium violaceum type strain ATCC 12472. FEMS Microbiol Lett 279:124–130. doi:10.1111/j.1574-6968.2007.01016.x
Pina PGP, Grosbuis S, Guyot L, Ghnassia JC, Allouch PY (1998) An Acinetobacter baumanii outbreak at the Versailles Hospital Center. Pathol Biol 46:385–394 PMID:9769866
Priya K, Yin W-FF, Chan K-GG (2013) Anti-quorum sensing activity of the traditional Chinese herb, Phyllanthus amarus. Sensors (Basel, Switzerland) 13:14558–14569. doi:10.3390/s131114558
Shaw PD, Ping G, Daly SL, Cha C, Cronan JE, Rinehart KL, Farrand SK (1997) Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. PNAS 94:6036–6041. doi:10.1073/pnas.94.12.6036
Sio CF et al (2006) Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun 74:1673–1682. doi:10.1128/IAI.74.3.1673-1682.2006
Stacy DM, Welsh MA, Rather PN, Blackwell HE (2012) Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native N-Acyl homoserine lactones. ACS Chem Biol 7:1719–1728. doi:10.1021/cb300351x
Tariq TM (2014) Bacteriologic profile and antibiogram of blood culture isolates from a Children’s Hospital in Kabul. J Coll Physicians Surg Pak 24:3396–3399 PMID:24953929
Waheed H, Hashmi I, Khan SJ, Kim SR, Arshad M, Nasir H (2016) Microbial population dynamics and profiling of quorum sensing agents in membrane bioreactor. Int Biodeterior Biodegrad 113:66–73. doi:10.1016/j.ibiod.2015.12.014
Zahin M, Hasan S, Aqil F, Khan MS, Husain FM, Ahmad I (2010) Screening of certain medicinal plants from India for their anti-quorum sensing activity. Indian J Exp Biol 48:1219–1224 PMID:21250604
Acknowledgements
The data of this study have been excerpted from the Ph.D. thesis of the author. This work was financed by Hacettepe University Scientific Research Project grant BAP 013BDYP604002.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All of the authors declare they have no competing interest.
Additional information
Communicated by Erko Stackebrandt.
Rights and permissions
About this article
Cite this article
Erdönmez, D., Rad, A.Y. & Aksöz, N. Quorum sensing molecules production by nosocomial and soil isolates Acinetobacter baumannii . Arch Microbiol 199, 1325–1334 (2017). https://doi.org/10.1007/s00203-017-1408-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-017-1408-8