Abstract
Twenty endophytic bacteria were isolated from surface-sterilized stems and roots of cucumber plants. After removal of potential siblings and human pathogens, the remaining seven strains were identified based on their 16S rDNA as Pseudomonas fluorescens (2 strains) and P. putida (5 strains). Three strains, namely P. fluorescens CS1, P. fluorescens CR2 and P. putida CR3, were able to suppress tomato foot and root rot (TFRR). Special attention was paid to the characterization of the BIOLOG carbon oxidation profiles of the isolated pseudomonads in order to identify nutrients which might be important for their endophytic lifestyle. Comparative analysis of the profiles of these seven strains with those of seven rhizospheric Pseudomonas spp. revealed that endophytes were able to oxidize l-arabinose and 2,3-butanediol significantly more often than the rhizospheric group. An independent growth experiment performed in tubes using l-arabinose and 2,3-butanediol as sole carbon sources showed the same results as seen using BIOLOG for l-arabinose, but not for 2,3-butanediol. Since l-arabinose is one of the most abundant sugars in xylem of cucumber plants and was not detected in their rhizosphere, our data suggest that utilization of l-arabinose might be a trait contributing to the endophytic lifestyle of the isolated Pseudomonas endophytes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants live in association with many bacteria which can be classified as rhizobacteria, epiphytic bacteria and endophytic bacteria. Endophytic bacteria are referred to as those which are able to colonize plants internally without causing any apparent harm. Due to their endophytic lifestyle, bacterial endophytes establish a more stable and long-lasting relationship with a plant than other plant-associated bacteria do (Hardoim et al. 2008). In addition, some endophytic bacteria have beneficial effects on plants. Therefore, bacterial endophytes with plant-beneficial traits are considered to be promising bio-inoculants for agricultural application (Strobel 2006).
Once endophytes establish themselves inside a plant, some of them can stimulate plant growth and/or protect plants against phytopathogens. Endophytic bacteria are able to promote plant growth directly by the secretion of phytohormones (Spaepen et al. 2008; Sgroy et al. 2009), by nitrogen fixation (You et al. 2005; Pedraza 2008) and by phosphate solubilization (Taurian et al. 2009; Lopez et al. 2011). In addition, several beneficial bacteria contain the enzyme 1-aminocyclopropane-1-carboxylate (ACC) deaminase, which hydrolyzes ACC, the precursor of the plant hormone ethylene, to NH3 and α-ketobutyrate (Glick 2005). The bacteria utilize NH3 as a source of N and thereby decrease the ACC and ethylene levels within the plant and, as a result, can stimulate plant growth (Sun et al. 2009). The ACC deaminase activity of endophytic bacteria and its responsibility for growth promotion of Solanum nigrum was reported for various strains of Pseudomonas (Long et al. 2008).
Endophytic bacteria can also promote plant growth indirectly via biocontrol of phytopathogens. Known mechanisms of biocontrol mediated by endophytic bacteria include (1) antibiosis through the production of antibiotics (Cho et al. 2003) or exo-enzymes (Downing and Thomson 2000) and (2) induction of systemic resistance (Van Wees et al. 2008; Yasuda et al. 2009).
Endophytic bacteria are able to colonize intercellular spaces of the cell walls and xylem vessels which together form the plant apoplast (Compant et al. 2010). Biochemical studies of nutrients which are present in the apoplast indicate that this niche contains sugars, alcohols, amino acids, organic acids, growth factors and mineral elements (Bacon and Hinton 2006). The question remains open of which carbon sources are utilized by endophytic bacteria in the plant apoplast.
The main aims of the present study were (1) to isolate and identify the major culturable heterotrophic bacterial endophytes from stems and roots of cucumber plants grown in a greenhouse, (2) to select novel beneficial strains based on their abilities (a) to promote growth of radish and/or (b) to control TFRR caused by the fungus Fusarium oxysporum f.sp. radicis-lycopersici (Forl) and (3) to characterize and compare oxidation profiles of carbon sources of endophytic and rhizospheric Pseudomonas spp. in an attempt to identify compounds which might be involved in the endophytic lifestyle.
Experimental procedures
Isolation of endophytic bacteria
Endophytic bacteria were isolated from stems and roots of cucumber (Cucumis sativus L.) plants collected from greenhouses in the Tashkent area, Uzbekistan. Plant samples were treated with 70 % ethanol for 3 min, followed by 4 % sodium hypochlorite for 5 min and several rinses with sterile water. To verify adequate surface sterilization, aliquots of water from the last rinsing were plated on 1/20 strength TSA control plates. Subsequently, the surface-sterilized plant samples were crushed under sterile conditions, and the resulting juices were plated on 1/20 strength tryptic soy agar (TSA, Difco Laboratories, MI, USA) plates. After incubation at 28 °C for 3 days, colonies originating from plant juice with empty control plates were used for further analysis.
Microbial strains and growth conditions
All isolated bacterial strains and eight Pseudomonas rhizospheric strains (P. fluorescens WCS365, PCL1444 and PCL1751 and P. putida PCL1760, PCL1759, PCL1758, PCL1603 and PCL1445), which belong to the collection of Institute of Biology Leiden, were grown and maintained on full strength TSA. The rhizospheric strains used for a comparative analysis originate from the following plants: avocado (P. putida 1603 and 1760), Barmultra grass (P. fluorescens 1444 and P. putida 1445), tomato (P.putida 1758 and 1759) and potato (P. fluorescens WCS365). P. fluorescens PCL1751, which is an excellent root colonizer (Kamilova et al. 2005) and is naturally resistant to kanamycin, was used as a reference strain for competitive cucumber root tip colonization experiments. Kanamycin was used at the final concentration of 50 μg ml−1.
The fungi Aspergillus niger, Forl, F. solani and the oomycete Pythium ultimum were routinely cultivated on potato dextrose agar (PDA, Difco Laboratories). To obtain spores for biocontrol experiments, Forl was routinely grown on Czapek-Dox liquid medium (Difco Laboratories) and incubated on a rotary shaker at 150 rpm at 28 °C.
Molecular characterization of endophytic strains
Amplified ribosomal restriction analysis (ARDRA) and identification of endophytic strains was performed as described previously (Malfanova et al. 2011). Briefly, 16S rRNA gene was amplified, cut with four different restriction enzymes and the resulting fragments were separated using a 2 % agarose gel. Those strains which gave a unique restriction pattern and appeared morphologically distinct were sent for sequencing to Service XS, Leiden, the Netherlands. Species they belonged to were identified as sharing at least 99 % homology with those of known species.
Characterization of potential plant-beneficial traits
Characterization of potential plant-beneficial traits such as the production of exo-enzymes (β- glucanase, cellulase, chitinase, lipase and protease), antifungal metabolites (AFM) and auxins was performed as described previously (Malfanova et al. 2011). The presence of ACC deaminase was judged by growth on 1-aminocyclopropane-1-carboxylate (ACC) as the sole N-source according to Belimov et al. (2005).
Plant growth promotion
Endophytic bacteria were tested for their ability to promote the growth of radish plants. This was done by soaking seeds of radish in a bacterial cell suspension (adjusted to 108 cfu/ml) in phosphate buffered saline solution (PBS) for 15 min. As a negative control, seeds were treated with sterile PBS. The treated seeds were subsequently planted in non-sterile potting soil and grown under greenhouse conditions at 80 % humidity and 16 h of daylight. Each variant consisted of four replicates with five seeds each. After 2 weeks of growth, the fresh weight of the roots was determined. All experiments were performed at least twice.
Biocontrol of tomato foot and root rot
Biocontrol of TFRR in soil was performed according to Kamilova et al. (2005) with small modifications. Briefly, tomato seeds of cultivar Carmello (Syngenta, Enkhuizen, the Netherlands) were coated with bacteria by dipping the seeds in a suspension of 1 % (w/v) methylcellulose (Sigma, St Louis, MO, USA) in PBS containing 108 cfu/ml. The treated seeds were then planted in non-sterile potting soil supplemented with 107 Forl spores per kg. For each treatment, 96 plants were tested in eight trays of 12 plants each. Plants were grown in a greenhouse at 21–24 °C, 70 % relative humidity and 16-h light. After 3 weeks of growth, plants were removed from soil and examined for symptoms of foot and root rot, such as brown spots, lesions, wilting or even death. Only roots without any of these symptoms were referred to as healthy and all others were scored as diseased. All experiments were performed at least twice.
Biocontrol of TFRR in stonewool substrate was conducted as described by Validov et al. (2007). Briefly, tomato seeds were placed in stonewool plugs which had been soaked in advance in plant nutrient solution (PNS) (Wageningen UR Greenhouse Horticulture, Bleiswijk, the Netherlands) supplemented with Forl spores (107 spores/L) and bacterial cells (106 cfu/ml). In the negative control, PNS was supplemented with spores only. Plants were grown for 14 days under greenhouse conditions at 80 % humidity and 16 h of daylight. The plants were then removed from the stonewool and examined for symptoms of foot and root rot. All experiments were performed twice. Homogeneity of variance and analysis of variance (ANOVA) at p = 0.05 were conducted with SPSS software (Chicago, IL, USA).
Carbon oxidation/utilization assay
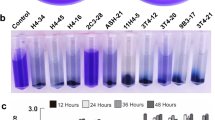
Seven Pseudomonas strains isolated from roots and stems of cucumber plants and seven rhizospheric strains of Pseudomonas spp. of different plant origin were tested for their ability to oxidize various carbon sources using BIOLOG GN2 Microplates (Biolog Inc., Hayward, CA, USA). Bacteria were grown overnight at 28 °C under aeration (150 rpm), harvested by centrifugation and subsequently resuspended in 0.85 % (w/v) NaCl solution to a final OD590 of 0.15. Aliquots of 150 μl were inoculated in each well of a 96-well microplate using a multichannel pipette. Plates were covered with a lid and incubated statically at 28 °C for 48 h. The appearance and intensity of a purple color, caused by the reduction of the tetrazolium salt, were read using a microplate reader at OD590 and by direct observation. Only results with values of OD590 > 0.3 and the visible presence of the purple color were scored as positive. In the case of border values between 0.29 and 0.3 and/or absence of purple color in the test wells, the results were scored as “±”. To check whether there is a significant difference in carbon oxidation abilities between the endophytic and rhizospheric group, the Chi-square test (p = 0.05) was performed using SPSS software.
To verify results obtained using the Biolog assay, we performed independent growth experiments. Endophytic and rhizospheric bacteria were grown overnight in LB broth and washed twice with 0.85 % NaCl to remove traces of extracellular carbon. Subsequently, bacteria were inoculated in M9 minimal medium containing (1) 0.2 % (w/v) l-arabinose or 2,3-butanediol (experiment), (2) 0.2 % (w/v) glucose (positive control), and (3) no added carbon source (negative control) to a final OD590 of 0.15. Falcon tubes containing 5 ml of each suspension were incubated for 24 h at 28 °C and 150 rpm. Subsequently, the bacterial growth was scored spectrophotometrically at OD590. All experiments were performed at least twice.
Competitive cucumber root tip colonization
In order to verify the ability of rhizospheric pseudomonads to efficiently colonize the rhizosphere of cucumber plants, competitive root tip colonization experiments were performed as described by Kamilova et al. (2005). Briefly, surface-sterilized cucumber seeds were inoculated with a 1:1 mixture of two bacterial strains (experiment vs. reference strain). The treated seeds were then planted in a gnotobiotic sand system (Simons et al. 1996) and grown for 7 days under greenhouse conditions. Subsequently, one cm of the root tip was cut off and vigorously shaken for 15 min to remove the adhered bacteria. Suspensions with bacterial cells were then diluted and plated on TSB with and without Km. The number of Km-sensitive (experiment) and Km-resistant (reference) colonies was determined, and the average of each group was calculated. All colonization experiments were performed in five replicates.
Results
Isolation and characterization of endophytic bacteria
Endophytic bacteria were isolated from stems and roots of greenhouse-grown cucumber plants. The controls showed that all living microorganisms on the plant surface were killed or became non-culturable.
A total of 20 strains were randomly chosen from the colonies obtained after plating plant juices on 1/20 strength TSA. To eliminate potential siblings, these strains were compared for their colony morphology, motility, their ARDRA patterns and production of the exo-enzymes β-glucanase, cellulase, chitinase, lipase and protease. Eleven strains which were indistinguishable with respect to the mentioned traits were considered as likely siblings and not further studied.
The nine remaining strains were screened for their antagonistic activity toward four pathogens, their ability to produce auxin and their growth on ACC as the sole N-source (Table 1).
Strain CR3 is antagonistic toward the oomycete P. ultimum, but not A. niger, F. solanum or Forl and does not secrete any exo-enzymes. None of the other strains showed any antagonism against the phytopathogens tested or production of exo-enzymes except for CS1 that showed protease activity.
Five strains, namely CR3, CR6, CS4, CS5 and CS8, produce detectable amounts of auxins, but only in the presence of tryptophan in the growth medium. The level of auxin secreted by strain CS5 is more than 80 μg/ml. In the case of CS4 and CR3, the auxin level in the media with tryptophan is more than 15 μg/ml. Two other strains, namely CR6 and CS8 produce appr. 10 μg/ml of auxins.
The only strain able to utilize ACC as the sole nitrogen source is CS1.
Molecular identification of endophytic strains
The strains were identified based on comparison of their rRNA sequences with database sequences from correctly identified strains (Table 2). The following species were isolated from stems and roots of cucumber plants: one Bacillus cereus strain, two P. fluorescens strains, five P. putida strains and one Stenotrophomonas maltophilia. To see whether these strains are safe to be applied in the field as biocontrol and/or plant growth-promoting strains, we evaluated to which risk group (Anonymous 1998) they belong. Two strains, namely B. cereus and S. maltophilia belong to risk group 2, representing potential human pathogens. Therefore, they were excluded from further experiments. This left us with seven pseudomonads.
Plant growth promotion
Four auxin-producing strains, namely CR3, CR6, CS4 and CS5 and one ACC-utilizing strain, CS1, were tested for their ability to promote radish growth under greenhouse conditions. Radish was chosen as the model plant because its roots secrete a high level of tryptophan on filter paper (Kamilova et al. 2006), which can be used by many beneficial bacteria as the precursor of auxin. However, none of the tested strains showed plant growth promotion (results not shown).
Biocontrol of tomato foot and root rot
All seven Pseudomonas spp. were tested for their ability to suppress TFRR in soil and stonewool. Two strains, namely P. fluorescens CR2 and P. putida CR3, significantly decreased disease symptoms of tomato plants in soil from 38 % in the non-inoculated control to 22 and 24 %, respectively (Fig. 1a). Significant biocontrol activity of these strains was also found in the second soil experiment. In the stonewool biocontrol experiments strains CR2 and CR3 were not significantly active but another strain, P. fluorescens CS1, was able to suppress TFRR symptoms from 71 to 41 % in the first and from 58 to 36 % in the second experiment (Fig. 1b).
Carbon oxidation assay
Seven endophytic and rhizospheric Pseudomonas spp. were characterized and compared in respect to their carbon oxidation profiles in order to identify carbon sources, which might be important for their endophytic lifestyle (Table 3 and S1). Out of 95 different carbon sources, as many as 34 were oxidized by all tested strains. These include one sugar (α-d-glucose), 12 organic acids (cis-acetonic, citric, d-gluconic, β-hydroxybutyric, α-ketoglutaric, α-ketovaleric, d,l-lactic, propionic, quinic, d-saccharic, succinic and bromosuccinic acid), 13 amino acids (d-alanine, l-alanine, l-asparagine, l-aspartic, l-glutamic, l-histidine, hydroxy-l-proline, l-leucine, l-ornithine, l-proline, l-pyroglutamic acid, l-serine and gamma-amino butyric acid) and eight other compounds. A total number of 21 carbon sources were not oxidized by any of the strains. These comprise 11 sugars (d-cellobiose, l-fucose, gentiobiose, α-d-lactose, lactulose, maltose, d-melibiose, β-methyl-d-glucoside, d-raffinose, l-rhamnose and turanose), three sugar alcohols (adonitol, i-erythritol and xylitol), one amino acid (l-phenylalanine), one amino acid derivative (glycil-l-aspartic acid) and five other compounds.
Three out of the 40 remaining carbon sources gave significantly different oxidation profiles between the endophytic and rhizospheric group, namely l-arabinose, 2,3-butanediol and p-hydroxyphenylacetic acid (Table 3 and S1). l-arabinose was oxidized by all seven endophytes and only by two rhizospheric strains, namely P. fluorescens strains WCS365 and PCL1444 (p < 0.05). Six out of seven endophytic pseudomonads (P. fluorescens CS1 and CR2 and P. putida CS5,CR3, CR6 and CR9) and one out of seven rhizospheric ones (P. fluorescens WCS365) were able to oxidize 2,3-butanediol (p < 0.05). Para-hydroxyphenylacetic acid was oxidized by five rhizospheric and one endophytic strain (p < 0.05).
The results of an independent growth experiment, as judged by an increase in the optical density (590 nm) compared to M9 medium without added carbon source, using l-arabinose as the sole carbon source showed the same results as the BIOLOG experiments obtained for l-arabinose. However, for 2, 3-butanediol as the sole carbon source, growth was measured for all endophytic strains and for six out of the seven rhizosperic strains.
Competitive cucumber root tip colonization experiment
To check whether the tested rhizospheric strains which originate from different plant hosts are cucumber rhizosphere competent and therefore can serve as suitable controls for the cucumber endophytes, we evaluated these strains in a competitive root tip colonization experiment against P. fluorescens PCL1751 (Kamilova et al. 2005), an excellent root colonizer. It appeared that all strains colonized the cucumber root tip in competition with P. fluorescens PCL1751 and therefore are cucumber rhizosphere competent (Table 4).
Discussion
General remarks about the isolated endophytes
After developing the protocol for the isolation of endophytic bacteria from cucumber plants, we used a similar strategy as described by Validov et al. (2007) for the elimination of siblings and potential pathogens. The fact that 11 out of the 20 strains are siblings indicates that the diversity among the isolated endophytes is low. Out of the nine remaining strains, two strains belong to risk group 2 (Table 2), indicating the presence of potential human pathogens among these endophytes. This phenomenon has been reported previously for both rhizospheric (Berg et al. 2005; Egamberdiyeva et al. 2008) and endophytic bacteria (Malfanova et al. 2011).
Seven remaining bacteria were identified as Pseudomonas spp. of which members have been found as endophytes of different plants (Mercado-Blanco and Bakker 2007; Ramesh et al. 2008). Several representatives of this group, namely P. fluorescens and P. putida, are widely known for their various plant-beneficial traits, which include production of antifungal metabolites and exo-enzymes, ACC deaminase activity and secretion of phytohormones (Mercado-Blanco and Bakker 2007). In our study, only a few identified pseudomonads displayed these characteristics (Table 1). The most common beneficial trait was the production of auxin in the presence of its precursor l-tryptophan.
Plant growth promotion
Auxin-producing strains were further tested under greenhouse conditions for their ability to promote the growth of radish roots. Auxins are known to be essential for plant physiology because they affect the root and shoot architecture (Spaepen et al. 2009). To our surprise, none of the tested pseudomonads had a significant effect on the root biomass of radish plants. The same results were obtained on tomato and cucumber plants (data not shown). The amount of l-tryptophan secreted by cucumber and tomato plants is low (1.8 and 7.4 ng per seedling, respectively), while the amount secreted by radish plants exceeds 0.29 μg per seedling (Kamilova et al. 2006) and is sufficient to stimulated microbial IAA production in nutrient-rich medium (Kravchenko et al. 2004). However, in nutrient poor soil, addition of high amounts of either l-tryptophan or IAA (up to 3 mg per kg soil) does not lead to a significant increase in radish root weight (Frankenberger et al. 1990). This may explain the absence of plant growth promotion in our experiments and highlights the importance of the substrate used during such investigations.
Biocontrol of TFRR
Biocontrol results (Fig. 1) indicated that P. fluorescens strains CS1 and CR2 and P. putida strain CR3 have a strong ability to control TFRR. However, the biocontrol effect of these strains was substrate-dependent. Apparently, the biocontrol mechanisms used by the different strains do not necessarily function well in both substrates. A similar effect was reported previously by Validov et al. (2009) who found that flagellar motility, which is a key trait for the biocontrol ability of PCL1751in potting soil, is not important during colonization of stonewool by P. putida PCL1760.
Despite the fact that the selected biocontrol strains do not produce exo-enzymes and antifungal metabolites against Forl in vitro, they were able to suppress the disease caused by the fungus in vivo. This observation is in agreement with other reports (Berg and Hallmann 2006; Malfanova et al. 2011), indicating that in vitro and in vivo beneficial activity of some bacteria is not necessarily correlated. Possible mechanisms of biocontrol include induction of systemic resistance (ISR) and competition for niches and nutrient (CNN) (Lugtenberg and Kamilova 2009). It is tempting to speculate that CNN is likely to be involved in biocontrol mediated by the selected endophytic pseudomonads. This mechanism has been proven for P. fluorescens PCL1751 and P. putida PCL1760 which also do not inhibit Forl in the plate assay but show significant biocontrol of TFRR in stonewool (Kamilova et al. 2005; Validov et al. 2007, 2009). Since the three strains which showed biocontrol did only so on one of the two substrates, we did not study the mechanism(s) of action.
Utilization of carbon sources by endophytic and rhizospheric pseudomonads
Out of the 95 different carbon sources tested, three were significantly differentially oxidized between the rhizospheric and the endophytic groups (p < 0.05). An independent growth experiment using M9 medium confirmed the BIOLOG result in the sense that l-arabinose is utilized by all endophytic pseudomonads while only two out of the seven tested rhizospheric strains could use it as their sole carbon source. Interestingly, l-arabinose is one of the major sugars present in the apoplast of different plants, including cucumbers (Iwai et al. 2003). Therefore, our results suggest that utilization of l-arabinose might be a trait contributing to the endophytic lifestyle of the Pseudomonas endophytes isolated from cucumber plants.
Our suggestion about the role of l-arabinose may be extended to other endophytes and plants because Prakamhang et al. (2009) found that all 51 different endophytes isolated from rice were able to use l-arabinose as well as glucose as their sole carbon sources. l-arabinose was not detected in the root exudate of cucumber plants (Kamilova et al. 2006). This fact together with the results from the competitive cucumber colonization experiment, which showed that all tested rhizospheric strains are able to reach the tip of the root at high density (see Table 4), suggests that the root tip colonization ability of the rhizospheric strains is not dependent on utilization of l-arabinose. The results also suggest that those rhizosperic strains which are able to utilize l-arabinose have an enhanced possibility of becoming endophytes.
Further analysis of the BIOLOG results showed that 2, 3-butanediol was differentially oxidized. However, this could not be fully verified in an independent growth experiment although the endophytic strains were able to reach a higher cell density level than the rhizospheric strains (data not shown). It is interesting to note that 2, 3-butanediol is a volatile signaling molecule involved in plant growth regulation and triggering of ISR (Ryu et al. 2003). As can be expected from signal molecules, they can be degraded and several pseudomonads apparently are able do so.
In conclusion, the results of the present study show that among the seven isolated Pseudomonas cucumber endophytes three strains have biocontrol activity. Moreover, we also found that, in contrast to most rhizospheric Pseudomonas spp., endophytic pseudomonads were able to utilize l-arabinose, one of the most abundant sugars in the xylem fluid of various plants.
We interpret our results as strongly suggesting that utilization of l-arabinose is an important factor which contributes to the endophytic lifestyle of Pseudomonas spp. in cucumber. It is unlikely that l-arabinose utilization is the only trait relevant for this complex lifestyle. Moreover, other major xylem nutrients could play a role, certainly in crops with a different xylem composition as well as for other endophytic bacterial species. Attempts to test the role suggested for l-arabinose more thoroughly, by using an l-arabinose utilization-negative mutant of one of the isolated pseudomonads and comparing the abilities of wild type and mutant to live endophytically, failed because our many attempts to make wild-type cells endophytic were not successful (results not shown).
References
Anonymous (1998) Sichere Biotechnologie. Eingruppierung biologischer Agenzien: Bacterien, BG Chemie, Merkblatt B 006 8/98 ZH 1/346. Heidelberg, Germany: Jedermann-Verlag Dr Otto Pfeffer oHG
Bacon CW, Hinton DM (2006) Bacterial endophytes: the endophytic niche, its occupants, and its utility. In: Gnanamanickam SS (ed) Plant-associated bacteria. Springer, The Netherlands, pp 155–194
Belimov AA, Hontzeas N, Safronova VI, Demchinskaya SV, Piluzza G, Bullitta S, Glick BR (2005) Cadmium-tolerant plant growth-promoting bacteria associated with the roots of Indian mustard (Brassica juncea L. Czern.). Soil Biol Biochem 37:241–250
Berg G, Hallmann J (2006) Control of plant pathogenic fungi with bacterial endophytes. In: Schulz B, Boyle C, Sieber T (eds) Microbial root endophytes. Springer, Berlin, pp 53–69
Berg G, Krechel A, Ditz M, Sikora RA, Ulrich A, Hallmann J (2005) Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol Ecol 51:215–229
Cho S, Lim W, Hong S et al (2003) Endophytic colonization of balloon flower by antifungal strain Bacillus sp. CY22. Biosci Biotech Biochem 67:2132–2138
Compant S, Clément C, Sessitsch A (2010) Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678
Downing K, Thomson J (2000) Introduction of the Serratia marcescens chiA gene into an endophytic Pseudomonas fluorescens for the biocontrol of phytopathogenic fungi. Can J Microbiol 46:363–369
Egamberdiyeva D, Kamilova F, Validov S, Gafurova L, Kucharova Z, Lugtenberg B (2008) High incidence of plant growth-stimulating bacteria associated with the rhizosphere of wheat grown in salinated soil in Uzbekistan. Environ Microbiol 10:1–9
Frankenberger WT, Chang AC, Arshad M (1990) Response of Raphanus sativus to the auxin precursor, l-tryptophan applied to soil. Plant Soil 129:235–241
Glick BR (2005) Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol Lett 251:1–7
Hardoim PR, van Overbeek LS, Elsas JD (2008) Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16:463–471
Iwai H, Usui M, Hoshino H, Kamada H, Matsunaga T, Kakegawa K, Ishii T, Satoh S (2003) Analysis of sugars in squash xylem sap. Plant Cell Physiol 44:582–587
Kamilova F, Validov S, Azarova T, Mulders I, Lugtenberg B (2005) Enrichment for enhanced competitive root tip colonizers selects for a new class of biocontrol bacteria. Environ Microbiol 7:1809–1817
Kamilova F, Kravchenko LV, Shaposhnikov AI, Azarova T, Makarova N, Lugtenberg BJJ (2006) Organic acids, sugars, and l-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol Plant Microbe Interact 19:250–256
Kravchenko LV, Azarova TS, Makarova NM, Tikhonovich IA (2004) The Effect of tryptophan present in plant root exudates on the phytostimulating activity of rhizobacteria. Microbiology 73:156–158
Long HH, Schmidt DD, Baldwin IT (2008) Native bacterial endophytes promote host growth in a species-specific manner; phytohormone manipulations do not result in common growth responses. PLoS ONE 3:e2702
Lopez BR, Bashan Y, Bacilio M (2011) Endophytic bacteria of Mammillaria fraileana, an endemic rock-colonizing cactus of the southern Sonoran Desert. Arch Microbiol 193:527–541
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Ann Rev Microbiol 63:541–556
Malfanova N, Kamilova F, Validov S, Shcherbakov A, Chebotar V, Tikhonovich I, Lugtenberg B (2011) Characterization of Bacillus subtilis HC8, a novel plant-beneficial endophytic strain from giant hogweed. Microb Biotech 4:523–532
Mercado-Blanco J, Bakker PAHM (2007) Interactions between plants and beneficial Pseudomonas spp.: exploiting bacterial traits for crop protection. Antonie Van Leeuwenhoek 92:367–389
Pedraza RO (2008) Recent advances in nitrogen-fixing acetic acid bacteria. Int J Food Microbiol 125:25–35
Prakamhang J, Minamisawa K, Teamtaisong K, Boonkerd N, Teaumroong N (2009) The communities of endophytic diazotrophic bacteria in cultivated rice (Oryza sativa L.). Appl Soil Ecol 42:141–149
Ramesh R, Joshi AA, Ghanekar MP (2008) Pseudomonads: major antagonistic endophytic bacteria to suppress bacterial wilt pathogen, Ralstonia solanacearum in the eggplant (Solanum melongena L.). World J Microbiol Biotechnol 25:47–55
Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Pare PW, Kloepper JW (2003) Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Science (USA) 100:4927–4932
Sgroy V, Cassán F, Masciarelli O, Del Papa MF, Lagares A, Luna V (2009) Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl Microbiol Biotech 85:371–381
Simons M, van der Bij AJ, Brand I, de Weger LA, Wijffelman CA, Lugtenberg BJJ (1996) Gnotobiotic system for studying rhizosphere colonization by plant growth-pro- moting Pseudomonas bacteria. Mol Plant Microbe Interact 9:600–607
Spaepen S, Dobbelaere S, Croonenborghs A, Vanderleyden J (2008) Effects of Azospirillum brasilense indole-3-acetic acid production on inoculated wheat plants. Plant Soil 312:15–23
Spaepen S, Vanderleyden J, Okon Y (2009) Plant Growth-promoting actions of rhizobacteria. In: van Loon LC (ed) Plant innate immunity. Elsevier, Amsterdam, pp 283–320
Strobel G (2006) Muscodor albus and its biological promise. J Indust Microbiol Biotech 33:514–522
Sun Y, Cheng Z, Glick BR (2009) The presence of a 1-aminocyclopropane-1-carboxylate (ACC) deaminase deletion mutation alters the physiology of the endophytic plant growth-promoting bacterium Burkholderia phytofirmans PsJN. FEMS Microbiol Lett 296:131–136
Taurian T, Anzuay MS, Angelini JG, Tonelli ML, Ludueña L, Pena D, Ibáñez F, Fabra A (2009) Phosphate-solubilizing peanut associated bacteria: screening for plant growth-promoting activities. Plant Soil 329:421–431
Validov S, Kamilova F, Qi S, Stephan D, Wang JJ, Makarova N, Lugtenberg B (2007) Selection of bacteria able to control Fusarium oxysporum f. sp. radicis lycopersici in stonewool substrate. J Appl Microbiol 102:461–471
Validov SZ, Kamilova F, Lugtenberg BJJ (2009) Pseudomonas putida strain PCL1760 controls tomato foot and root rot in stonewool under industrial conditions in a certified greenhouse. Biol Control 48:6–11
Van Wees SCM, Van der Ent S, Pieterse CMJ (2008) Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol 11:443–448
Yasuda M, Isawa T, Shinozaki S, Minamisawa K, Nakashita H (2009) Effects of colonization of a bacterial endophyte, Azospirillum sp. B510, on disease resistance in rice. Biosci Biotech Biochem 73:2595–2599
You M, Nishiguchi T, Saito A, Isawa T, Mitsui H, Minamisawa K (2005) Expression of the nifH gene of a Herbaspirillum endophyte in wild rice species: daily rhythm during the light–dark cycle. Appl Environ Microbiol 71:8183–8190
Acknowledgments
The research described here was supported by the Netherlands Organization for Scientific research (NWO, project number 047.018.001) and represents part of the Russian-Dutch collaboration project “Center of Excellence”. We thank Dr. Bernadette Kroon (Syngenta B.V., Enkhuizen, the Netherlands) for providing us with tomato and cucumber seeds. We thank Gerben Voshol for his help in some experiments and for critically reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Theo Hansen.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Malfanova, N., Kamilova, F., Validov, S. et al. Is l-arabinose important for the endophytic lifestyle of Pseudomonas spp.?. Arch Microbiol 195, 9–17 (2013). https://doi.org/10.1007/s00203-012-0842-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-012-0842-x