Abstract
Loose cluster architecture is an important aim in grapevine breeding since it has high impact on the phytosanitary status of grapes. This investigation analyzed the contributions of individual cluster sub-traits to the overall trait of cluster architecture. Six sub-traits showed large impact on cluster architecture as major determinants. They explained 57% of the OIV204 descriptor for cluster compactness rating in a highly diverse cross-population of 149 genotypes. Genetic analysis revealed several genomic regions involved in the expression of this trait. Based on the linkage of phenotypic features to molecular markers, QTL calculations shed new light on the genetic determinants of cluster architecture. Eight QTL clusters harbor overlapping confidence intervals of up to four co-located QTLs. A physical projection of the QTL clusters by confidence interval-flanking markers onto the PN40024 reference genome sequence revealed genes enriched in these regions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grapevine (Vitis vinifera L. subsp. vinifera) is one of the most important and valuable fruit crops. Globally, 7.5 million hectares are under viticulture. The annual grape yield reached 75.8 million tons in 2016. The largest part of the harvested grapes (47.3%) sustains wine production (267 million hl). The remaining shares are sold as fresh grapes (35.8% of the annual yield), followed by raisins and the production of juice (13.5%; OIV 2017).
High-quality fruits are crucial for winemakers and the fruit processing industry. However, V. vinifera grapevine cultivars are susceptible to several diseases and pests, so viticulture depends on intense protective sprayings. The obligate biotrophic pathogens Erysiphe necator (the causal agent of powdery mildew) and Plasmopara viticola (the causal agent of downy mildew), both specific pathogens of grapevine, as well as the ubiquitous fungus Botrytis cinerea (teleomorph Botryotinia fuckeliana, the causal agent of gray mold) represent the major threats (Pertot et al. 2017). Recent grapevine breeding efforts succeeded in the introgression of resistance loci for Erysiphe necator and for Plasmopara viticola from Vitis wild species into new high-quality cultivars (Töpfer et al. 2011). Grapevine varieties with enhanced genetically determined resistance against those pathogens became available. However, this strategy is not a solution to obtain resistance to Botrytis cinerea. There is no efficient cellular defense response known against this fungus. Due to the lack of resistance donors, grapevine breeding and clonal selection for resilience to Botrytis have to rely on the utilization of physical factors, e.g., the selection of genotypes with loose cluster architecture, thick berry skin and hydrophobic berry surface (Gabler et al. 2003; Herzog et al. 2015; Shavrukov et al. 2004). Loosely structured grape clusters have enhanced resilience to B. cinerea due to improved ventilation within the grape cluster. The accelerated drying process of residual humidity after rainfall or the precipitation of dew functions as a physical barrier against infections with fungal pathogens (Hed et al. 2010; Molitor et al. 2012). Several studies underline the importance of wetness duration for the successful infection by B. cinerea (Broome et al. 1995; Nair and Allen 1993; Nelson 1956). In addition, fungicide applications can better reach the berries surface within the cluster in the case of a more open, loose cluster (Hed et al. 2010). Furthermore, spatial temperature gradients between the inner and outer berries of a cluster are less pronounced. Solar radiation can much better reach the internally situated berries. Fruit maturity thus reaches a higher rate of uniformity in a loosely structured grapevine cluster (Pieri et al. 2016; Vail and Marois 1991). The formation of micro-cracks and the subsequent loss-of-barrier effect of the berry’s epidermis against pathogens (Becker and Knoche 2012) appear reduced. According to Smart and Robinson (1991) berries may even burst due to high pressure inside of compact clusters and thereby lose any kind of barrier against pathogens. Loose cluster architecture thus contributes to healthier grapes and harmonized ripening periods for the production of supreme yield and quality.
The grade of density or openness of a grapevine cluster relates to the ratio between the volume occupied by berries and the total cluster volume. This ratio describes the free space between the berries. Cluster architecture (CA) determines the arrangement of berries in a cluster and the distribution of free space. The components of CA comprise berry traits and stalk traits. The interplay of berry traits, e.g., berry number and berry volume, and stalk traits, e.g., rachis length or pedicel length, determines the final grade of compactness (discussed in Tello and Ibáñez 2017). The International Organization of Vine and Wine (OIV) developed descriptors to score and measure morphologic grape cluster traits (OIV 2015). Based on the assessment of the available space between single berries, the descriptor “OIV204 (cluster density)” is applicable to score the cluster compactness (OIV 2015). Furthermore, cluster architecture can be assessed by measuring cluster architecture sub-traits, e.g., the length of single rachis internodes (Shavrukov et al. 2004) or berry size and number (Rist et al. 2018; Kicherer et al. 2013). These measurements of single sub-traits can be assembled into CA factors, e.g., the ratio of cluster weight by length (Tello and Ibáñez 2014).
Although environmental and management conditions affect CA traits (Li-Mallet et al. 2016; Tello and Ibáñez 2017), their expression is also under genetic control. Houel et al. (2013) studied the genetic variability of berry size in a wide range of grapevine genotypes and found an immense variation of berry volume. For berry weight, Ban et al. (2016) detected the genetic influence in the offspring of a hybrid cross. Genetic characterization of 140 F1 individuals from a table grape cross-population indicated significant genotypic effects for all of the 23 CA traits under investigation (Correa et al. 2014). Shavrukov et al. (2004) compared four grapevine genotypes and found that rachis size variation is due to rachis cells size variation. Tello et al. (2015) compared 125 genotypes in an association genetic study and described major variations concerning the lengths of the rachis and secondary branches. Fanizza et al. (2005) detected genetic variation in the offspring of a table grape cross associated with berry number per cluster. Wine grapes and table grapes belong to different gene pools and show, among other characteristics, considerable variations in berry and cluster architecture sub-traits (Migicovsky et al. 2017). The authors revealed genetic differences associated with bigger berries and less dense clusters in table grapes as compared to wine grapes. Di Genova et al. (2014) compared a genetic draft sequence of the table grape cultivar “Sultanina” with the reference genome for grapevine derived from an inbred line of “Pinot Noir,” a wine grape cultivar. In total, 2000 genes were found affected by structural variants. Among these genes, more than 50 genes are associated with the GO (gene ontology) term “anatomical structure development” (GO:0048854) providing a source of genetic diversity potentially involved in cluster architecture differences. Grimplet et al. (2017) compared clones with loose or compact CA of the same cultivar (near-isogenic lines). These authors found 470 genes differentially expressed (two loose clones vs. two compact clones). More specifically, compact clones showed a higher gene activity in genes involved in the production of cellular material and in genes of the cell cycle network. Shiri et al. (2018) performed a co-expression experiment with a compactly clustered table grape variety along the development from pre-flowering to pre-harvest. The authors identified gene expression networks with influence on cluster architecture via regulation of gibberellin abundance.
In this study, detailed phenotyping and statistics of CA sub-traits classified the investigated sub-traits according to their impact on the overall grade of compactness/openness. The linkage of phenotypic characteristics of CA with molecular markers identified quantitative trait loci (QTLs). These QTLs should be involved in the manifestation of multiple sub-traits that contribute to CA. A transfer of the genetic positions of the QTLs to the physical map by projection of the confidence interval-flanking markers onto the reference genome of PN40024 (12x) revealed clusters of overlapping confidence intervals from QTLs of strong impact on CA traits. The elucidated genomic regions, i.e., the novel knowledge about linked molecular markers, restrict the size of genomic regions for investigation in further studies. The here presented LODmax-associated markers for cluster architecture sub-traits are first steps to marker-assisted selection and could be further evaluated for their transferability in molecular breeding for cultivars with loose clusters.
Materials and methods
Plant material
The parents and 151 F1 genotypes from a controlled cross of GF.GA-47-42 × “Villard Blanc” (G × V) were used in this work. The vines were located in two neighboring vineyards at the Institute for Grapevine Breeding Geilweilerhof (N49°21.675, E8°04.433). In the first vineyard (vineyard 1), for each of the individual 151 F1 genotypes two vegetatively propagated clones were planted on their own roots with 1.8 m row spacing and 0.9 m plant spacing in the year 2000. The second vineyard (vineyard 2) with eight additional clonal replicates (made from wooden cuttings grafted on rootstock SO4) was planted in 2010. Here, the vines were grown with 2 m (row) × 1 m (plant) spacing. The vines underwent “Guyot pruning” with 10 to 12 buds remaining and were grown in a vertical shoot position trellis system. An integrated pesticide spray program according to best practice policies for viticulture (BMELV 2010) protects the plantation.
The maternal parent, the fungus-resistant breeding line GF.GA-47-42, and the paternal parent, the fungus-resistant white wine cultivar “Villard Blanc”, exhibit reduced cluster densities according to OIV204 as evaluated over 3 years at six plants each (Online Resource 1). The resulting segregating population includes transgressive phenotypes with extreme differences in CA. Two genotypes were excluded from the evaluation process since they showed no or unusually poor fruit set during consecutive growing seasons. Moreover, the population provides 45 plants with female flowers and 106 plants with hermaphrodite flowers.
Sampling
Phenotypic investigations used 3 to 12 clusters per genotype harvested from different vines per season. In the year 2013, 12 samples came from two vines, while in the years 2014 to 2017, three to six independent samples originated from different vines (Table 1). When the first vines of the population reached véraison the clusters were inspected two times per week. To avoid the loss of berries during harvest and transport of the clusters the samples were harvested when the clusters showed characteristics of maturity, but were not overripe. At this time, the berries had a sugar content of ~ 10° to 20° Brix. The clusters were strictly sampled from the basal insertions of three central shoots on the fruit cane. The analyzed clusters were cut directly at the connection with the shoot and stored at 5 °C until use.
Investigated sub-traits
In total, data for 19 sub-traits of cluster architecture (Table 1) collected for at least two growing seasons entered this study. During the seasons of 2013 and 2014 pilot studies generated data for 12 and 8 CA traits, respectively. In the seasons of 2015 and 2016, data collection covered 16 sub-traits. Measurements assessed 3 to 12 biological replications per genotype and season. Pedicel measurements encompassed at least 60 pedicels per genotype. Cluster compactness was evaluated according to OIV204 descriptor in five classes (i.e., 1, 3, 5, 7 and 9) from grade 1 = very loose to grade 9 = very dense. A panel of four trained experts did an independent OIV204 rating to reduce the impact of subjectivity. Subsequently, the mode value of the four ratings was used. Image-based Berry Analysis Tool (BAT) generated data on berry volume and berry number according to the description in Kicherer et al. (2013). The BAT segmentation algorithm, trained with destemmed berries in BBCH79 condition as ground truth data, is able to recognize berries when presented on a standardized picture. Once the berries are individually identified, the number and the size of berries are estimated. In addition, all pictures were personally inspected and manually interpreted if the automatic assessment was not plausible. The length measurements of rachis-related sub-traits were determined using ImageJ (Schneider et al. 2012). Pictures of the rachis were taken together with a size standard to transform the pixel-based image data into SI-unit-based length values. The size standard was measured using the “straight line tool”, and the cluster architecture was measured using the “segmented line tool”. The peduncle length was measured from the cutting edge to the insertion of a wing or tendril, respectively. The wing length was measured from its insertion to the point where the pedicels separate. The rachis length was measured from the first lateral insertion to the end of the spike without the terminal pedicel. Laterals were measured from their insertion at the main rachis without the terminal pedicel. Rachis internodes were measured from the middle of the flanking nodes. Rachis diameter was measured in the middle of the second internode. Pedicels were measured from dyad or triad junctions to the contact surface where the berries have been removed. Gravimetric measurements were taken using an electronic balance, with deviance = 0.1 g (EMB 3000-1 KERN & SOHN GmbH, Balingen, Germany). °Brix measurements used an electronic refractometer (DWN2 Risun, Beijing, China).
Statistics
Statistical analyses applied R software, version 3.4.1 (R Core Team 2017), and various packages as described below. The significance level of measurement results was set at p < 0.05 as obtained by one-way ANOVA, if not stated otherwise. Data quality and model assumptions were checked by inspecting normal Q–Q plots, density distributions and scatter plots.
Measures of 16 cluster architecture sub-traits recorded in 2015 (n = 851) and 2016 (n = 896) at vineyard 2 (Table 1) were analyzed by: (i) correlation analysis between cluster architecture traits, (ii) principle component analysis (PCA) to reflect the influence of flower sex (FS) and growing season on the cluster architecture traits and (iii) random forest models and cumulative link models to assess the effect and relative importance of cluster architecture traits on visual compactness. Some genotypes did exhibit some missing data for different reasons: In 2015 for example, berry rot caused 37% missing data for “total berry volume” and “mean berry volume” and in 2016, “shoulder length” could not be recorded in 13% of the data since not all of the progeny plants produced a shoulder in each cluster. However, overall, the amount of missing values was less than 5%. Since the presence of missing data does not allow the comparison of statistical models with the “Akaike information criterion” (AIC), multiple imputations using chained equations were calculated with the R-package “mice” (van Buuren and Groothuis-Oudshoorn 2011). The averaged results of five imputations were used after visual comparison of the density distributions and the range of original and implemented data. Since metric data and ordinal data, i.e., measurements of rachis architecture sub-traits and the ordinal OIV204 descriptor scores for cluster compactness, were considered in this work, Kendall’s Taub correlation coefficient was used to perform a correlation analysis using the R-package “cormat” (Kassambara 2017) (Online Resource 2). A principle component analysis based on covariance was applied to the scaled cluster architecture traits of 2015 and 2016 using the R-packages “factoMineR” (Lê et al. 2008) and “factoextra” (Kassambara 2017). Only variables with a Kendall’s Taub correlation coefficient < 0.8 were used (Online Resource 2). To assess whether the data contain any inherent grouping structure with respect to flower sex (FS) and growing season (2015 and 2016) the clustering tendencies in the PCA scores were statistically evaluated by computing the Hopkins statistics (Ho) with the R-package “clustertend” (Han et al. 2012). Ho > 0.5 would indicate a significant cluster within a dataset (Han et al. 2012).
Random forest (RF) models and cumulative link models (CLMs) with scaled data assessed the effect and the relative importance of 15 cluster architecture traits measured in 2015 and 2016 (Table 1). Additionally, the effect of flower sex and year on OIV204 ranking was assessed. The random forests were established for an ordinal response (OIV204 descriptor) using the function “cforest” of the R-package “party” (Hothorn et al. 2006; Strobl et al. 2007, 2008). It utilizes the commonly applied random forest method introduced by Breiman (2001) (for a recent overview of the methodology, see Boulesteix et al. 2012). Prediction accuracy measurement for response levels with uniform distances was performed with ranked probability scores (RPS), appropriate for ordinal response variables, as described in Janitza et al. (2016). Variable importance measurements (VIMs) for RF were performed with RPS-based VIMs. Hence, the incorporated ordering information, contained in the ordinal responsive variable, was respected in the VIM calculation, i.e., the accelerating compactness in five classes from 1 to 9. To further study the model performance, RF calculations were repeated four times, using error rate (ER), mean standard error (MSE), mean absolute error (MAE) and RPS to compare the prediction accuracy contained in the VIM results. Cumulative link models for ordinal response were fitted with the same explanatory variables as in random forest using the R-package “ordinal” (Christensen 2018). The model selection was performed in a two-step procedure (due to processing time) and based on an information-theoretic approach (Burnham and Anderson 2002) using the R-package “glmulti” (Calcagno and de Mazancourt 2010). In a first step, various candidate models with up to eight different main terms were fitted and compared using the “Akaike information criterion” (AIC) (Burnham and Anderson 2002), where a lower AIC indicates a better fit. All variables with a model-averaged importance of > 0.75 were used in a second step to fit candidate models with main terms and two-way interactions, which were compared via AIC as above. The models within a range of delta AIC < 2 were used for interpretation of effects. The relative importance of explanatory variables was then assessed by fitting models where each explanatory variable was removed at a time and calculating the delta AIC relative to the best model. The more the delta AIC rises, the more important is the variable that was removed from the model. The overall error rate and rank-wise error rate indicated the prediction quality of a CLM. In order to assess the collinearity between the predictor variables of the best models we calculated the variance inflation factors (VIFs) with the R-package “car” using the function “vif” (Fox and Monette 1992).
Genetic evaluation
As described in Zyprian et al. (2016) a genetic map has been established based on 546 molecular markers. This map and the corresponding parental maps provided the basis in this work for the identification of QTLs related to the sub-traits of cluster architecture.
Quantitative trait locus analysis
Quantitative trait locus (QTL) analysis applied the software tool MapQTL6.0 (van Ooijen 2009). The determination of segregation of trait-linked markers and QTL detection used the interval mapping (IM) procedure with a mapping step size of 1 cM. Based on a permutation test with 1000 iterations a linkage group-specific “logarithm of the odds” (LOD) threshold was calculated (with p < 0.05). Additionally, an IM with flower sex as co-variable was computed. Regions that exceeded the LG-wide LOD threshold were recorded as QTL. This work considered QTLs that have been: (i) reproduced at least three times; or (ii) reproduced two times, but were physically co-located to other QTLs for two seasons and were found accumulated with overlapping confidence intervals on the reference genome; or (iii) identified in other crosses than in G × V according to literature references (Correa et al. 2014; Marguerit et al. 2009). For each QTL, the maximum LOD score, the percentage of explained phenotypic variation and the extension of the confidence intervals (in cM) are recorded.
The molecular markers in direct neighborhood to the LODmax − 1 positions delimited the confidence intervals. These flanking markers were used to project the QTL regions on the grapevine reference genome of (PN40024)12x V2 (Canaguier et al. 2017) as retrieved from https://urgi.versailles.inra.fr/Species/Vitis/Data-Sequences/Genome-sequences. The physical position of proximate confidence intervals assessed the accumulation of cluster architecture-linked QTLs.
Gene set enrichment analyses
The projection of confidence intervals for cluster architecture QTLs on the physical regions of the reference genome (PN40024) 12x V2 delimits gene sets that were statistically associated with cluster architecture-related traits. Genes contained in these confidence intervals were transferred to the protein classification system (PANTHER) via the gene ontology consortium online platform (Ashburner et al. 2000; The Gene Ontology Consortium 2017) available at http://geneontology.org/. The redundancy of annotated biological functions assigned to the genes within these confidence intervals was then compared to the redundancy of biological functions in the total set of genes of the reference genome. Significantly overrepresented or underrepresented (p < 0.05 Fisher’s exact with FDR multiple test correction) gene ontology (GO) terms were assessed using PANTHER, version 13.1, as described in Mi et al. (2017). The enriched GO term was used to prioritize the search for candidate genes from multiple QTLs.
Weather records
Climate data were acquired in approx. 500 m distance to the trial fields with the records of the meteorological station 88 Siebeldingen type AME 16, 192 m sea level, longitude 8.047925770315487, and latitude 49.216499765308136. Data were downloaded from http://www.am.rlp.de.
Results
Evaluation of cluster compactness according to descriptor OIV204
The parental varieties of the G × V population were rated for their cluster density according to OIV descriptor 204 during the three seasons from 2015 to 2017. The maternal genotype GF.GA-47-42 showed a loose cluster architecture (mode for OIV204 = 3). The paternal type of the population, “Villard Blanc”, showed a very loose (mode for OIV204 = 1) cluster structure. The OIV204 scorings of the F1 individuals of the G × V population covered all classes from 1 = very loose (Fig. 1a) to 9 = very compact (Fig. 1b). The F1 progeny showed a mode value for OIV204 between 3 and 5 in the years 2013, 2015, 2016 and 2017. In 2015 the probability for a lower OIV204 score was significantly higher (p > 0.001 Pearson’s Chi-square test) as compared to 2016 (Fig. 1c). In addition, genotypes with female flowers showed significantly smaller OIV204 scores (p < 0.001; Pearson’s Chi-square test) during consecutive seasons (Fig. 1c).
Variation of cluster architecture in the cross-population GF.GA-47-42 × “Villard Blanc” during two seasons and between the flowering types female and hermaphrodite. The OIV descriptor 204 for compactness scores from a 1 = very loose, where rachis and pedicels are visible, to b 9 = very compact, where berries are non-circularly deformed (scale bar = 35 mm). c Histogram showing the relative frequency (density) of OIV204 scorings in 46 female and 103 hermaphroditic F1 genotypes from the GF.GA-47-42 × “Villard Blanc” cross measured at BBCH85 in 2015 and 2016
Cluster architecture sub-traits and their correlation
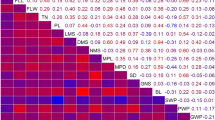
All CA sub-traits and corresponding notations are presented in Table 1. Correlation analysis (Online Resource 2) indicated the highest correlation for the CA sub-traits cluster weight and berry weight (tau-b = 1). OIV204 and berry traits were in general slightly positively correlated (tau-b = 0.1 − 0.4), while rachis traits were slightly negatively correlated to OIV204 (tau-b = − 0.1 − 0.2) in 2015 and 2016. The correlation of berry weight/rachis weight with OIV204 was positive (tau-b = 0.3 and 0.4) during the two consecutive years. The correlation among the various rachis sub-traits was found less pronounced (− 0.1 to 0.5), but stable over the 2 years. Quite in contrast, the correlation among berry traits varied between years. In 2015, the correlation between total berry volume and berry number or mean berry volume was tau-b = 0.4 and 0.7, while in 2016, it was tau-b = 0.7 and 0.3. Hence, total berry volume appeared to be determined by the components berry number and single berry volume in a contrasting way in the 2 years. The correlation between the cluster architecture sub-traits that determine OIV204 (i.e., rachis length, shoulder length, cluster weight, berry number, mean berry volume and pedicel length, see below) was generally weak and ranged between tau-b 0.0 and 0.3, with the exception of cluster weight and berry number (tau-b = 0.6) in 2015 and 2016 and RL and SL (tau-b = 0.5) in 2016 (Online Resource 2).
Identification of major components of cluster architecture and influence of flower sex
The OIV204 scores showed some influence of flower sex, indicating a shift toward higher OIV204 scores in the hermaphrodite vs. female genotypes (Fig. 1c). Therefore, a PCA was applied to the measurements of the 15 sub-traits recorded in 2015 and 2016. The PCA identified five main components that explained 69% of the variation in the data. Principal component 1 (PC1) and principal component 2 (PC2) explained 47% of the variation. PC1 was associated with berry features cluster weight, total berry volume, berry number and the rachis features rachis weight and shoulder length (Fig. 2). The contribution to PC1 was as follows: cluster weight (18.5%), total berry volume (17.3%), berry number (15.2%), rachis weigh (13.7%) and shoulder length (7.0%). PC2 was positively associated with rachis traits with a contribution of rachis length (17.7%), rachis diameter (14.2%), shoulder length (10.6%) and rachis weight (7.9%). PC2 was negatively related to the ratio of berry weight to rachis weight (20.1%) and the OIV204 score (10.9%) (Fig. 2). PCA scores displayed a pattern depending on flower sex and year. PC1 displayed higher scores for the year 2016 vs. 2015, indicating higher berry weight and volume in 2016. PC2 displayed higher scores for female genotypes, indicating elongated rachis sub-traits. However, the separation of the concentration ellipses of the PCA scores was moderate as indicated by Ho of 0.13.
Principal component analysis of cluster architecture sub-traits recorded in 2015 and 2016. The biplot shows the first principal component (PC1) where berry sub-traits are dominant contributors and the second principal component (PC2) representing mainly rachis sub-traits. The scaled cluster architecture trait values of the principal components 1 and 2 display 47% of the total variance. Concentration ellipses indicate the location of 95% of the data. a Separated by the year (growing season). b Separated by flower sex. For notation of sub-traits see Table 1
Identification of cluster architecture sub-traits that predict cluster compactness
The sub-traits (aligned according to their relevance for cluster architecture) pedicel length < rachis weight < mean berry volume < berry weight/rachis weight < shoulder length < berry number < flower sex < total berry volume < rachis length < cluster weight are important variables that predict OIV204 according to random forest (Table 2). The application of the four different prediction accuracy estimates ER, MSE, MAE and PRS for the VIM calculation showed no influence on the importance rank order (Online Resource 3).
CLMs for the prediction of OIV204 showed that the sub-traits pedicel length < shoulder length < berry number < rachis length < cluster weight had the largest impacts (in ascending order) on compactness levels (OIV204 values) of the 149 F1 genotypes of the cross-population when the season was included as predictor variable (Table 2). MBV was an important predictor variable, when the variable season was not included. The collinearity of the predictor variables in the selected models was quite low. The variance inflation factor values ranged between 1.09 for pedicel length and 3.38 for cluster weight. All sub-traits that reflect berry features were positively related to compactness, while all sub-traits measuring rachis features showed negative relationship to OIV204 scores (Online Resource 4). Genotypes with female flower organs and samples from 2015 showed a higher probability to be loosely clustered as compared to samples from 2016 and hermaphroditic flowered genotypes, respectively (Online Resource 4). The interaction between berry number and cluster weight was a predictor in CLMs regardless of whether season was in the model (Table 2). The overall error rate was 0.42 and 0.44 for the CLMs without and with season as additional predictor variable. A comparison of the error rates across OIV204 categories showed that the prediction accuracy for class three and five (loose to medium cluster architecture) was considerably higher than for the compact levels (Online Resource 5). The majority of the genotypes (over 70%) were member of these two classes (3 and 5), where the ER was 0.39 and 0.32, respectively.
According to the random forest VIM results berry weight/rachis weight and total berry volume were important sub-traits for cluster compactness, but were not included in the CLMs as predictor variable. Due to these inconsistencies, the sub-traits total berry volume and berry weight/rachis weight were not considered for further analysis. The sub-traits rachis diameter and rachis weight contributed weakly and inconsistently to CLMs when main effects only were fitted, but were not important when interactions were fitted. Interestingly, the sub-traits length of the first lateral, length of the second lateral and peduncle length were of minor importance.
QTL detection
Mean values of the cluster architecture sub-traits measurements recorded in the years 2013 to 2017 were applied for QTL analysis using interval mapping (IM) on the genetic constitutions of 149 F1 individuals and the consensus map of G × V (Zyprian et al. 2016).
IM detected 24 QTLs for CA sub-traits reproducibly (Online Resource 6). These QTLs were found on the following 10 linkage groups (LGs): LG1 (pedicel length a, pedicel length b, rachis weight, peduncle length, total berry volume), LG2 (cluster weight, rachis length, shoulder length, OIV204), LG3 (mean berry volume, shoulder length, rachis length), LG10 (cluster weight, berry number), LG11 (pedicel length), LG12 (cluster weight, mean berry volume), LG14 (peduncle length), LG15 (OIV204), LG17 (mean berry volume, cluster weight, OIV204) and LG18 (cluster weight, pedicel length).
With respect to the presence of 45 female and 106 hermaphroditic individuals in the population, flower sex was used as a co-variable in an explorative additional calculation of “IM + FS.” This approach yielded six additional QTLs on LG3 (pedicel length), LG10 (berry number, berry weight), LG14 (wing), LG17 (berry number) and LG18 (berry number) cluster architecture traits (Online Resource 6). Remarkably, three QTLs for berry number and one for berry weight were identified newly by application of flower sex as a co-factor for IM. Furthermore, a QTL for cluster complexity, i.e., the presence/absence of a shoulder at the cluster, was reproduced using flower sex as co-factor in an IM. In total, 30 QTLs for traits related to CA were reproducibly detected over two to four seasons (Online Resource 6).
The QTLs identified by IM and IM + co-variable (flower sex) showed no significant differences for the average LODmax values, the size of the average confidence interval (CI) and the explained phenotypic variance (Online Resource 7). The sub-traits rachis length, mean berry volume, berry number, cluster weight and pedicel length show high contribution to cluster density (Table 2). QTLs for these important traits were reproducible over three seasons (Table 3). For the sub-trait shoulder length, also statistically important, QTLs were reproducible over two seasons. Notably, the QTL found on LG2 for shoulder length was linked for two seasons with the same LODmax marker (VVIB23_312) than the one found for rachis length (Table 3). The major QTL for OIV204 cluster compactness was identified on LG2 in the vicinity of marker GF02-12 with an average impact explaining 20% of the variance of the OIV204 scores and LODmax of 11.07. For berry-related sub-traits the average maximum explained variance (15%) was found with a QTL on LG10 for berry weight associated with marker VRZAG7. The major QTL for rachis-related sub-traits was found on LG1 for peduncle length correlated to the SNP marker 55553gene_1_GF_WRKY. This QTL explains on average 24% of the phenotypic variance and had a LODmax value of 10.79 (Online Resource 6).
Relevant QTLs accumulate in eight clusters
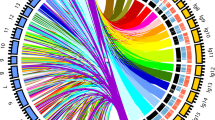
Based upon the multivariate statistical analysis of the CA data described above, the rachis features (rachis length, shoulder length and pedicel length) and specific berry sub-traits (cluster weight, berry number, mean berry volume) showed high impact on OIV204. For these traits of prominent importance, 19 QTLs were detected reproducibly. In addition, four QTLs for compactness according to OIV204 scores were identified. The major QTLs were found on LG2 (rachis length, cluster weight), LG3 (rachis length), LG11 (pedicel length), LG17 (mean berry number) and LG18 (berry number). On average, the QTLs for these traits explained approximately 14% of the total variance (ranging from 11 to 18%) (Table 3 and Online Resource 6). Beside the QTL for pedicel length on LG11, correlated to marker VMC6C3, all other high-impact QTLs were co-located in groups with two to four different QTLs for CA sub-traits. To facilitate the application of these new findings in marker-assisted grapevine breeding, these QTLs were analyzed to check whether they are spatially concentrated in a specific region of a chromosome. To this purpose the confidence intervals (positions of LODmax − 1) of the 23 QTLs were projected on the reference genome from PN40024 12x v2 (Canaguier et al. 2017) and screened for overlaps. This approach identified eight genomic regions where QTLs of cluster architecture shared the same stretch of genomic sequence as confidence interval. Twenty QTLs were co-located in reference to the PN40024 sequence (Table 4). These eight clusters cover all major QTLs for architecture sub-traits with high impact on compactness and explain 87% of the variance.
Gene set enrichment analyses
The genomic regions of the eight QTL clusters for sub-traits of cluster architecture enclose 3691 annotated genes. Using gene ontology categories related to biological processes for a GO term enrichment analysis, 3462 of the genes (93.8%) could be successfully assigned to a category. 229 genes could not be mapped to the protein database. Significant GO term enrichments were found in all confidence interval-associated gene subsets except in the cluster on LG2. Reducing the gene subset on LG2 to genes enclosed in the central 2 Mb range of the confidence interval showed that the GO term “regulation of microtubule-based process” was 50 times overrepresented in this region. VIT_202s0025g04960 was one of the GO-term-associated genes. It encodes a cell-cycle-regulated microtubule-associated protein. Moreover, this approach revealed 45 overrepresented GO terms in the gene subsets when compared to the GO annotations of all genes in the reference genome, including the category “response to auxin.” The terms “ion transport,” “anion transport” and “response to endogenous stimulus” were overrepresented in two clusters. In total, 219 genes (Online Resource 8) were assigned to at least one of the significantly overrepresented GO classes (p < 0.05 Fisher’s exact test).
Discussion
The segregating population
A population segregating for the trait of interest and a linkage map for this population are prerequisite for QTL analysis. The genetic map of the G × V population used here has been elaborated earlier and was already successfully applied to detect QTLs affecting resistance to pathogens and ripening traits of grapevines (Zyprian et al. 2016). The loose cluster architecture (CA) inherent to the parent GF.GA-47-42 (G; OIV204 = 3) and the very loose CA of the parent “Villard Blanc” (V; OIV204 = 1) suggested that the G × V population could segregate for CA. Indeed, the F1 genotypes exhibited variable and even transgressive phenotypes, showing OIV 204 density scores from very loose (1) to very dense (9). The paternal grandparent variety Seibel 6468 showed significantly lower rachis length and a higher mean berry volume in comparison with the parental varieties (data not shown). This could be used for a genetic determination of the transgressive phenotypes. The field plantation of the population was established in 2000 and in a multiplied form in 2010. The phyllotaxic phase shift inherent to grapevine development from juvenile to adult plants was completed at the time of investigation. Therefore, any phenotypic bias due to juvenile anomalism was avoided. The CA segregation pattern could be verified for consecutive seasons and thus was exploited for the detection of reproducible QTLs associated with CA.
Stability and interrelationship of cluster architecture sub-traits
The compactness of the cluster is the result of an interaction of multiple cluster architecture sub-traits (Rist et al. 2018; Tello et al. 2015; Correa et al. 2014; Shavrukov et al. 2004). This study used 16 different sub-traits for the statistical evaluation of the individual contribution to cluster compactness in two consecutive growing seasons. The correlation analysis among them showed high variation concerning the intensity and the direction of correlations between individual cluster architecture sub-traits and to the official OIV204 descriptor.
Seasonal conditions had an impact regarding the berry traits, i.e., total berry volume correlated with berry number and mean berry volume but in a divergent manner for the two seasons of 2015 and 2016 (Online Resource 2). To further assess the seasonal impact on the berry sub-traits even the traits with stronger correlation were considered in principal component analysis. Here again the berry-related sub-traits were more affected by the season compared to the rachis-related sub-traits. Climate conditions from budburst to flowering on to harvest affected berry number. However, the weather conditions recorded for this period did not provide evident differences (recorded as monthly average for air and ground temperature or for photoactive radiation) during the first weeks of growth and inflorescence development. Nevertheless, 2016 had 50% more days with rainfall compared to 2015 and therefore provided less favorable conditions for berry set during this time period. However, the berry number in 2016 was higher than in 2015. The Hopkins statistics value for clustering tendency was far below the threshold that would indicate a cluster within the dataset of measured cluster architecture sub-traits. This supports our assumption of a quantitative multiple trait genetic determinism.
The complexity of cluster architecture
Cluster density (as characterized by OIV descriptor 204) is a highly complex trait since it depends on the interaction of multiple berry and rachis sub-traits. Several previous studies concern the variability of CA sub-traits. Fanizza et al. (2005) reported berry number variation. The average berry size is highly variable from 0.5 to 11.5 cm3 according to Houel et al. (2013). Shavrukov et al. (2004) highlighted rachis internodes’ length as major contributor to CA variation. Gabler et al. (2003) and Sarooshi (1977) reported variation in CA due to elongated pedicels. Complexity of CA, i.e., the presence or absence of a “shoulder” segregated in a cross from table and wine grapes (Marguerit et al. 2009). In addition, the contribution of sub-traits to overall CA appeared to be variable among Vitis cultivars (Tello et al. 2015). In agreement with the findings of Migicovsky et al. (2017) this study here showed that there is a negative correlation of sugar content with mean berry volume evident in 2015 and in 2016 data (Online Resource 2 and Fig. 2). Hence, an important step of this work was to determine the sub-traits that substantially contribute to the CA phenotype in the given G × V cross.
Determination of the most relevant sub-traits to predict cluster architecture
Forests of regression trees and automated multi-model inference using CLMs with the CA dataset predicted the compactness level (OIV204) with CA sub-traits. Explorative, random forest VIM calculations gave an overview of the importance of single sub-traits for OIV204 prediction. The assessment of the prediction accuracy as described in Janitza et al. (2016) using four different prediction performance measures showed no impact on the VIM order (Online Resource 3). Hence, in subsequent CLMs the prediction accuracy was measured straightforward with the calculation of the error rate. This revealed that the models had a lower error rate if the compactness was lower, i.e., in season 2015, in the group of female phenotypes and the group with loose cluster OIV204 rankings (Online Resource 5). One possibility may be that the subjective visual classification of compactness might be less accurate with increasing levels of compactness which leads to a reduced predictive power of the models.
Nevertheless, within the available CA sub-traits, the best CLMs to predict the OIV204 descriptor consisted of the predictors rachis length, shoulder length, cluster weight, berry number, mean berry volume and pedicel length. Therefore, these traits were of major importance for genetic analysis. Notably, the derived measures berry weight/rachis weight and total berry volume were not included as predictor variables in the best CLMs. Instead, the models used for the ranking of the sub-traits considered original measurements as predictor variables only. The variance inflation factors for the unassembled variables in the obtained best models were quite low (between 1.09 and 3.38). The obtained values were considered to be low enough to assume no bias due to collinearity (Hair et al. 2010). However, expressed as variable importance value based on regression trees or as delta AIC value elaborated with a leave-one-out model comparison, the importance of these sub-traits in the models for compactness was diverse. In this study, rachis length and cluster weight showed the highest impact followed by the total berry volume. Tello et al. (2015) reported rachis length and berry number as highly correlated to OIV204 scorings in a wide range of cultivars over three growing seasons. Tello and Ibáñez (2014) combined up to six sub-traits to form compactness indices. In their work, the indices showing the highest correlations with the visual OIV204 classification contained the sub-traits cluster weight, rachis length, berry number and pedicel lengths (among others). Their findings are supporting our modeling results where the same traits show large effects on ordinal OIV204 values. Among cluster architecture sub-traits with elevated importance for compactness, pedicel length was least important in this study. Nevertheless, it is important enough to be recognized as determining factor for cluster compactness (Table 2). In Tello et al. (2015) the sub-trait pedicel length produced the highest correlation with cluster compactness in one of three seasons. However, the authors found a low relevance of pedicel length to the overall compactness in their study. Although our work in general corresponds to the findings of Tello et al. (2015) the study presented here revealed a higher likelihood for open cluster with longer pedicels (Online Resource 4). Supporting our notion, Gabler et al. (2003) reported that pedicel length showed impact on cluster architecture. The same was found by Sarooshi (1977) after growth regulator treatment. Additionally, on LG1, the co-localization of QTLs for compactness (OIV204) with QTLs for pedicel length supports the importance of pedicel length for compactness on genetic level (Fig. 3).
Graphical overview of co-located QTLs linked to cluster architecture sub-traits. Physical position for confidence interval regions of QTLs related to sub-traits of cluster architecture projected onto the reference genome of grapevine PN40024 12x V2. In orange the location of confidence interval clusters for QTLs calculated with interval mapping. In green the location of confidence interval clusters determined with contribution of interval mapping and interval mapping + flower sex as co-variable during QTL calculation. For trait abbreviations see Table 1. For positions and details see Table 4 and Online Resource 6
In the work of Shavrukov et al. (2004) rachis internode length was the main determinant of cluster openness of compact wine grape varieties (“Riesling” and “Chardonnay”) compared to openly structured table grape cultivars (“Exotic” and “Sultanina”). This is not in line with our findings where the length of the first and second internodes (estimated with 149 F1 genotypes of the G × V population) was not important for the prediction of compactness (OIV204 classes) with random forest and cumulative link models. Moreover, in their work they could not find significantly different pedicel lengths, discriminating compact and open cultivars, whereas in this study, elongated pedicel lengths raise the likelihood of showing loose cluster architecture (Online Resource 4). Together, this suggests that table grapes achieve their cluster openness with divergent sub-trait contributions or the highly diverse set of F1 genotypes was highlighting other genetic determinants of cluster architecture sub-traits than the wine grape versus table grape comparison.
QTLs for cluster architecture
The overall aim of this study was to reveal QTLs for cluster architecture to deduce cluster architecture-associated markers for marker-assisted selection (MAS) in grapevine breeding. Due to the complexity of the trait “cluster architecture”, several QTLs with various levels of contribution to the phenotypic variance were expected. Indeed, this investigation revealed an elevated number of 30 QTLs for cluster architecture sub-traits (Table 3 and Online Resource 6). The statistical evaluation of 16 cluster architecture sub-traits recorded in 2015 and 2016 (~ 1700 data points per trait) showed that six of the cluster architecture sub-traits had high impact on the compactness level of the cluster (OIV204).
Focusing on these statistically most relevant sub-traits for cluster architecture berry number, cluster weight, mean berry volume, pedicel length, rachis length and shoulder length reduced the number to 24 QTLs for close investigation (Table 3 and Online Resource 6). Many QTL regions accumulate in specific genomic regions. The confidence intervals of 21 QTLs were co-located on the reference genome in eight genomic regions (Table 4). This fact of clustered QTLs alleviates the task to deduce trait-linked markers for assays of applicability in grapevine breeding for loose cluster architecture. An overview of cluster architecture-related QTLs is shown in Online Resource 6.
On LG1, limited by the markers VVIN61 and VMC2B3, a cluster of the QTLs for OIV204 and pedicel length (PEDa) was detected. Pedicel length is a predictor variable in the majority of the linear models. The LODmax-associated marker for OIV204 and for pedicel length was SNP1241_207FEM. This SNP is located in the mRNA sequence of the gene VIT_201s0026g02580. The gene product, a “zinc finger DOF5.2-like” protein, is a plant-specific transcription factor of the DOF (DNA-binding One Zinc Finger) family. In the model plant A. thaliana, Fornara et al. (2015) reported that an alteration in the expression level of cycling DOF factors affected flowering and growth. However, besides VIT_201s0026g02580, there are 718 more genes encompassed in the confidence interval of the QTL; 39 of them are also found in the GO enrichment (Online Resource 8). In addition, LG1 harbors a second QTL for pedicel length (PEDb) associated with the LODmax marker GF01-24. Approximately 700 kb downstream of GF01-24 Marguerit et al. (2009) also reported a QTL for pedicel length, which was associated with the marker IRT1f in their study. Costantini et al. (2008) described a QTL for berry weight on LG1 in a table grape cross, associated with AFLP marker “mCACeATC4.” The AFLP technique of this marker prevents a precise determination of the position on the reference genome, but the closest SSR marker on their consensus map was VVIF52 at 23 Mb. In this region a QTL for peduncle length was found in the G × V cross during three seasons, but with different LODmax positions (Online Resource 6).
Incorporated on LG2, the confidence intervals of the QTLs found for rachis length, shoulder length, cluster weight and OIV204 were co-located between the markers GF02-07 and VMC5G7. The associated LODmax marker for rachis length and shoulder length was VVIB23. The QTLs for cluster weight and OIV204 share GF02-12 as common LODmax marker. In a former study Marguerit et al. (2009) found the region close to marker VVIB23 on LG2 associated with rachis sub-traits in their interspecific cross of “Cabernet Sauvignon” × V. riparia “Gloire de Montpellier,” e.g., rachis length, rachis length combined with peduncle length and the presence/absence of a wing.
The markers GF02-07 and VVIB23 are linked to cluster architecture sub-traits and also closely linked to flower sex. Using the offspring of a cross, performed with a rootstock cultivar and a wine grape breeding line, Fechter et al. (2012) pinpointed genetic determinants of flower sex within a 143 kb region between the markers VVIB23 and GF02-12. Marguerit et al. (2009) found a high association of flower sex to the marker VVIB23 in their cross. Analyzing exclusively the 103 hermaphroditic individuals of the G × V population (omitting the 46 female F1-individuals) no QTL was detectable in this region. A QTL calculation based on the paternal map (data not shown) did not show any QTL for cluster architecture in this genomic region, either. However, the QTL calculation using the maternal map showed QTLs for OIV204, rachis length and shoulder length in this region spanning the confidence interval between the markers VVIB23 and GF02-12 (data not shown). This indicates maternal heredity of these QTLs for cluster architecture sub-traits on LG2. This finding is consistent with a genetic determination for elongated rachis sub-traits and more open cluster architecture in female genotypes as visible in the PCA calculation at PC2 (Table 1 and Fig. 2).
An interval mapping using flower sex as co-variable detected a QTL for pedicel length on LG3. The marker GF03-09 was the upper limit of the LODmax − 1 confidence interval, and the marker 1044j09FEM was the lower limit and the LODmax marker at the same time (1,9 Mb). As far as we know, this is the first report for cluster architecture QTLs in this genomic region. Nevertheless, the confidence interval for this QTL harbors 170 genes; 34 of them were reported as differentially expressed between loosely and compactly clustered “Tempranillo” clones in a study of Grimplet et al. (2017). Moreover, it displays the additional power of IM using a co-variable (flower sex) for QTL calculation since the pedicel length QTL was not detectable without the application of this co-variable.
The QTL for pedicel length shares its LODmax marker with the one for mean berry volume on LG3. In grapevine, the berry size and seed number are directly related. This correlation results likely from the fact that gibberellins produced by seeds are required to promote berry growth during late berry development (Coombe 1960, 1973; Perez et al. 2000). This study here did not record seed number, but an elevated phytohormone concentration could also be the reason for longer pedicels. Gourieroux et al. (2016) discussed that phytohormones released by grape ovaries may promote the elongation of the rachis so that adequate space becomes available for the growing berries.
LG3 carries a second QTL cluster delimited by the markers VCHR03a and 2018J24 at around 16.5 Mb. This cluster covers the QTLs for rachis length and shoulder length. Both QTLs shared GF03-07 as LODmax marker. In the cross-population used by Marguerit et al. (2009), it was possible to detect QTLs for rachis length and length of the first rachis internode also on LG3, but in a different region at ~ 7.8 Mb. It remains to be explored whether these two loci correspond.
On LG10, the application of interval mapping calculation with flower sex as co-factor identified co-localized QTLs for berry number and cluster weight. Depending on the season the upper limit of the confidence interval varied considerably between 9.42 and 21.30 Mb. The lower limit and the LODmax were stable at marker VRZAG7 positioned at 23.17 Mb. The varying range of the confidence intervals over the seasons is probably a result of the influence of climate conditions on the development of berry traits, which requires two consecutive years for the full cycle [as discussed in Li-Mallet et al. (2016) or in Tello and Ibáñez (2017)]. This QTL cluster also encloses further QTLs for berry weight in 2015 and 2016, rachis weight in 2015 and 2016 and total berry volume in 2014 and 2016. In this region, with QTLs for berry-related sub-traits of cluster architecture, Tello et al. (2016) found two SNPs at around 19.17 Mb associated with the length of the first lateral. LG10 also contains QTLs for shoulder length between ~ 5 and ~ 15 Mb in the G × V cross. Associated with the marker VMC2A10 (5.98 Mb) Marguerit et al. (2009) detected QTLs for peduncle, rachis and rachis internode length on LG10 in the interspecific cross in their work. Their QTL was co-localized with AGAMOUS, a floral organ development gene. As a key finding of their work Shiri et al. (2018) have recently reported that AGAMOUS is involved in the compactness of table grape clusters.
On LG12, the QTLs for mean berry volume and cluster weight co-localized between 17.92 and 23.76 Mb. Within this 5.84-Mb-wide region, an additional QTL for OIV204 was detected, but only in the season of 2017. During 2 years (2015 and 2016) the LODmax for the QTL for OIV204 was located also on LG12, but at different positions of VV_12_3836836FEM (3.88 Mb) and VV_12_6764538FEM (6.05 Mb), respectively. Trying to explain the positional shift of the OIV204 QTL in the year 2017, the climatic conditions around the time of flowering were compared between the three seasons (14 days pre-bloom until 14 days post-bloom counted from the median of the flowering time range of a given season). The most prominent climatic event between the seasons was a heavy rain storm on June 3, 2017 (31 l/m2 in 6 h), at the beginning of the flowering time of the cross-population with the potential to affect the pollination rate. Such an event could have influenced the expression of the trait. Interestingly, Costantini et al. (2008) reported a QTL for berry weight in the region of the confidence interval for OIV204 at 5.44 Mb. Berry weight is significantly correlated with OIV204 in the population investigated here over 2 years. Assuming that the QTL for OIV204 reported here is influenced by berry weight Costantini et al. (2008) may thus have indirectly confirmed the position of the QTL for OIV204 in the range of 3.88-6.05 Mb by their finding. In the work of Tello et al. (2016) a SNP associated with cluster compactness was located in this region also, directly supporting the QTL position for OIV204 in the upper third of the chromosome.
On LG17, QTLs for berry number, cluster weight, mean berry volume and OIV204 were found between the LODmax markers SCU06 (3.29 Mb) and UDV092 (9.61 Mb) in this work. Several studies using populations with diverse genetic background reported QTLs for cluster architecture traits in this chromosomal region. Fanizza et al. (2005) found a QTL for berry number associated with an AFLP marker (17mCTG eATC8) at the very top of LG17. Correa et al. (2014) reported a QTL for rachis traits linked to the marker VMC2H3 at 3.68 Mb. Linked to the marker VVIN73 (5.63 Mb), Doligez et al. (2013) reported a QTL for berry weight. Marguerit et al. (2009) reported VVIN73 as LODmax marker for rachis internode length. Hence, the region on LG17 seems to be strongly engaged in the genetic determination of cluster architecture. The fact that the same marker was linked to rachis as well as to berry traits, in two different studies, could probably be explained by the dependency of rachis traits on the manifestation of flower and berry traits as explained in Gourieroux et al. (2016). With the resolution of QTL analysis it is not feasible to dissect underlying candidate genes for single sub-traits. It remains elusive to suggest a pleiotropic effect of a locus on several phenotypic features. Indeed, the proximity of QTLs for berry- and rachis-related sub-traits in this region provides the opportunity for marker-assisted selection. It may be possible to take advantage of this situation by applying a small range of molecular markers from this QTL region to select less berry volume with large rachis features tagging several traits that might be co-inherited.
On LG18, the confidence interval of the QTL for cluster weight flanks the confidence interval for the QTL for pedicel length. Both confidence intervals were co-located additionally with the confidence interval for berry number, when flower sex was used as a co-factor in IM calculation. This QTL-saturated region is flanked by markers VMC2A3 (0.95 Mb) and VV18_8582805FEM (9.58 Mb). In addition, the sub-trait QTLs for berry weight and rachis weight were co-located in this cluster.
Several recent reports for cluster architecture sub-traits identified QTLs on LG18. In the studies of Correa et al. (2014), Doligez et al. (2013), Costantini et al. (2008) and Cabezas et al. (2006) the marker VMC7F2 at 30.31 Mb was linked to berry volume, berry weight and seed traits. In the close vicinity of this marker Tello et al. (2016) reported a SNP in the 5′UTR of a MADS-box SEEDSTICK encoding gene correlated with ramification length. Correa et al. (2014) could show the linkage of rachis node number to the markers VMC2A7 and VMCNG2F12 at 13.39 and 22.85 Mb. Downstream of this region, in proximity of the marker UDV108, they reported the QTL position for berry number and berry number after gibberellic acid treatment.
On LG18, all so far reported QTLs for berry-related cluster architecture sub-traits from table grape crosses were located at the lower arm of the group. Quite in contrast, the QTLs detected in this work were exclusively located on the upper arm of LG18. Doligez et al. (2013) used three cross-populations to investigate the coupling of berry size and seed content. Two of these were table grape crosses and one was a wine grape cross. Only in the cross of wine grape cultivars they found a QTL for berry sub-traits, also on the upper arm linked to marker VVIN83 at 10.67 Mb.
Survey of GO classes enriched in the QTL cluster regions
Looking at the highly enriched GO classes and the corresponding annotated genes reveals six groups of GO-term-related genes enriched between 30- and 90-fold in the QTL clusters for cluster architecture-associated traits (Online Resource 8). The first group comprises a set of genes encoding a component of menaquinone biosynthesis, a 2-oxoglutarate decarboxylase hydro-lyase magnesium ion binding protein and a gene encoding naphthoate synthase, enriched 45-fold in the QTL cluster on chromosome 1. These genes are involved in the formation of co-factors for the electron transfer machinery of photosystem I (PSI) (Gross et al. 2006). At a similar level of enrichment (36-fold) there are copper transporter systems encoded in cluster 3.2. Copper is a crucial element in electron transport, but may also be implicated in other processes like free radical elimination, signaling and hormone perception (Sancenón et al. 2003). It remains to be elucidated whether electron transfer systems of PSI are particularly involved in cluster architecture determination. The role of copper transporters may be ambiguous with the possibility to contribute to PSI or to participate in signaling during cellular development. In cluster 2 there is a strong enrichment (50-fold) for genes encoding a cell-cycle-regulated microtubule-associated protein and armadillo repeat-containing kinesin-like protein 2. The products of these genes are involved in cell division and intracellular transport along microtubuli using motor proteins like kinesins. This function is in line with the strong enrichment (90-fold) of as yet uncharacterized proteins assigned to the GO classes for bidirectional movement of large protein complexes along microtubules (GO:0035721 and 42073) found in cluster 10. These functions are intrinsic to cell development and may be an important part of the formation of the cluster architecture sub-traits. The genes strongly enriched (37-fold) in cluster 18 encode flavonol synthase (FLS1), an iron-binding light-responsive oxidoreductase that contributes to flavonoid biosynthesis. It acts on dihydroflavonols to yield quercetin, kaempferol and myricetin in grapevine. These substances serve as UV protectants. Five FLS genes have been shown to be expressed in flower buds and flowers of grapevine. Two FLS genes keep on being expressed from véraison (the transition point of berry growth from hard, green berries to berry softening and sugar accumulation) to harvest stage (Fujita et al. 2006). Heijde and Ulm (2012) reported enhanced FLS expression after UV-B photon perception by the UV-B photoreceptor (UVR8) pathway in A. thaliana. Also Hayes et al. (2014) reported for A. thaliana that the perception of UV-B radiation was maintained with the UVR8-mediated UV-B responses. They could link the UVR8 pathway to growth patterns, i.e., shade avoidance responses in Arabidopsis thaliana by antagonizing the phytohormones auxin and gibberellin. Nevertheless, how a higher level of UV protectants may be beneficial for a more loosely structured inflorescence remains to be revealed. The cluster 3.1 contains a prominent group of SAUR family proteins and auxin-induced genes in 33.5-fold enrichment. SAURs (“Small Auxin Up” RNAs) are early auxin-responsive genes that play a role in the regulation of plant cell growth (cell expansion and cell division). The plant-specific SAUR genes are generally present in tandem arrays with high redundancy and arranged in large genomic blocks due to segmental duplications of very closely related genes. These genes are induced by auxins, but may also be regulated by brassinosteroids, gibberellins, abscisic acid and jasmonate. They are involved in cell differentiation and patterning. The SAURs also respond to environmental conditions (light, drought) and may modulate auxin transport (Ren and Gray 2015). From all the genes enriched in the QTL clusters, this block in cluster 3.1, together with the finding of highly enriched intracellular microtubule-guided transporter functions involved in cell development in the cluster on chromosome 2, provides the best candidates to explain different growth patterns that result in the phenotypes of loose or compact cluster architecture traits. However, their functional relevance awaits further investigation.
Conclusions
The combination of statistical methods, i.e., correlation analysis, PCA, RF and CLM modeling, enabled the determination of the most relevant sub-traits that determine cluster architecture in the evaluated G × V cross. For those highly effective sub-traits of cluster architecture, it was possible to identify 19 reproducible QTLs. As compared to literature references, some QTLs already reported could be verified and new QTLs in yet unreported regions became accessible. Co-localized QTLs determined 87% of the total phenotypic variation of traits with high impact on cluster architecture detected in this study. Projection of confidence intervals of co-localized QTLs onto the reference genome for grapevine (PN40024) revealed eight QTL clusters, and the QTL clustering facilitates marker deduction for MAS. GO term enrichment analysis suggested accumulation of genes related to biological functions as first ideas on the molecular basis underlying the phenotype of cluster architecture.
Author contribution statement
EZ designed the study, acquired funding and supervised the work. RR performed the experiments, measurements and calculations. FR contributed phenotypic data. DG provided statistical expertise and tools. RT provided all plant materials, infrastructure and special advice. RR and EZ wrote the paper. All authors read the manuscript.
References
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT et al (2000) Gene ontology: tool for the unification of biology. Nature Genet 25:25
Ban Y, Mitani N, Sato A, Kono A, Hayashi T (2016) Genetic dissection of quantitative trait loci for berry traits in interspecific hybrid grape (Vitis labruscana × Vitis vinifera). Euphytica 211(3):295–310
Becker T, Knoche M (2012) Water induces microcracks in the grape berry cuticle. Vitis 51(3):141–142
BMELV (2010) Gute fachliche Praxis im Pflanzenschutz: Bundesministerium für Ernährung Landwirtschaft und Verbraucherschutz (BMELV)
Boulesteix AL, Janitza S, Kruppa J, König I (2012) Overview of random forest methodology and practical guidance with emphasis on computational biology and bioinformatics. Wiley Interdiscip Rev Data Min Knowl Discov 2(6):493–507
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Broome JC, English JT, Marois JJ, Latorre BA, Aviles JC (1995) Development of an infection model for Botrytis bunch rot of grapes based on wetness duration and temperature. Phytopath 85(1):97–102. https://doi.org/10.1094/Phyto-85-97
Burnham KP, Anderson DR (2002) Model selection and multimodel inference: a practical information-theoretic approach, 2nd edn. Springer, New York
Cabezas JA, Cervera MT, Ruiz-Garcia L, Carreno J, Martinez-Zapater JM (2006) A genetic analysis of seed and berry weight in grapevine. Genome 49(12):1572–1585. https://doi.org/10.1139/G06-122
Calcagno V, de Mazancourt C (2010) glmulti: an R package for easy automated model selection with (generalized) linear models. J Stat Softw 34(12):1–29
Canaguier A, Grimplet J, Di Gaspero G, Scalabrin S, Duchêne E, Choisne N, Mohellibi N, Guichard C, Rombauts S, Le Clainche I, Bérard A, Chauveau A, Bounon R, Rustenholz C, Morgante M, Le Paslier MC, Brunel D, Adam-Blondon AF (2017) A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genom Data 14:56–62. https://doi.org/10.1016/j.gdata.2017.09.002
Christensen RHB (2018) Ordinal-regression models for ordinal data. R package version 2018, pp 4–19
Coombe BG (1960) Relationship of growth and development to changes in sugars, auxins and gibberellins in fruit of seeded and seedless varieties of Vitis vinifera. Plant Physiol 35(2):241–250
Coombe BG (1973) The regulation of set and development of the grape berry. International Society for Horticultural Science (ISHS), Leuven, Belgium, p 261
Correa J, Mamani M, Munoz-Espinoza C, Laborie D, Munoz C, Pinto M, Hinrichsen P (2014) Heritability and identification of QTLs and underlying candidate genes associated with the architecture of the grapevine cluster (Vitis vinifera L.). Theor Appl Genet 127:1143–1162
Costantini L, Battilana J, Lamaj F, Fanizza G, Grando MS (2008) Berry and phenology-related traits in grapevine (Vitis vinifera L.): from quantitative trait loci to underlying genes. BMC Plant Biol 8(1):38. https://doi.org/10.1186/1471-2229-8-38
Di Genova A, Almeida AM, Munoz-Espinoza C, Vizoso P, Travisany D, Moraga C, Pinto M, Hinrichsen P, Orellana A, Maass A (2014) Whole genome comparison between table and wine grapes reveals a comprehensive catalog of structural variants. BMC Plant Biol 14:7. https://doi.org/10.1186/1471-2229-14-7
Doligez A, Bertrand Y, Farnos M, Grolier M, Romieu C, Esnault F, Dias S, Berger G, Francois P, Pons T, Ortigosa P, Roux C, Houel C, Laucou V, Bacilieri R, Peros JP, This P (2013) New stable QTLs for berry weight do not colocalize with QTLs for seed traits in cultivated grapevine (Vitis vinifera L.). BMC Plant Biol 13:217. https://doi.org/10.1186/1471-2229-13-217
Fanizza G, Lamaj F, Costantini L, Chaabane R, Grando (2005) QTL analysis for fruit yield components in table grapes (Vitis vinifera). Theor Appl Genet 111(4):658–664. https://doi.org/10.1007/s00122-005-2016-6
Fechter I, Hausmann L, Daum M, Sorensen TR, Viehöver P, Weisshaar B, Töpfer R (2012) Candidate genes within a 143 kb region of the flower sex locus in Vitis. Mol Genet Genomics 287(3):247–259. https://doi.org/10.1007/s00438-012-0674-z
Fornara F, de Montaigu A, Sánchez-Villarreal A, Takahashi Y, Loren Ver, van Themaat E, Huettel B, Davis SJ, Coupland G (2015) The GI–CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J 81(5):695–706
Fox J, Monette G (1992) Generalized collinearity diagnostics. J Am Stat Assoc 87(417):178–183. https://doi.org/10.1080/01621459.1992.10475190
Fujita A, Goto-Yamamoto N, Aramaki I, Hashizume K (2006) Organ-specific transcription of putative flavonol synthase genes of grapevine and effects of plant hormones and shading on flavonol biosynthesis in grape berry skins. Biosci Biotechnol Biochem 70(3):632–638
Gabler FM, Smilanick JL, Mansour M, Ramming DW, Mackey BE (2003) Correlations of morphological, anatomical, and chemical features of grape berries with resistance to Botrytis cinerea. Phytopathology 93(10):1263–1273. https://doi.org/10.1094/Phyto.2003.93.10.1263
Gourieroux AM, McCully ME, Holzapfel BP, Scollary GR, Rogiers SY (2016) Flowers regulate the growth and vascular development of the inflorescence rachis in Vitis vinifera L. Plant Physiol Biochem 108:519–529
Grimplet J, Tello J, Laguna N, Ibanez J (2017) Differences in flower transcriptome between grapevine clones are related to their cluster compactness, fruitfulness, and berry size. Front Plant Sci 8:17
Gross J, Cho WK, Lezhneva L, Falk J, Krupinska K, Shinozaki K, Seki M, Herrmann RG, Meurer J (2006) A plant locus essential for phylloquinone (Vitamin K1) biosynthesis originated from a fusion of four eubacterial genes. J Biol Chem 281:17189–17196. https://doi.org/10.1074/jbc.M601754200
Hair J, Anderson R, Tatham R, Black W (2010) Multivariate data analysis, 7th edn. Pearson, New York
Han J, Kamber M, Pei J (2012) Data Mining: concepts and techniques. In: Data mining. 3rd edn. Morgan Kaufmann, Boston, pp 443–541. https://doi.org/10.1016/B978-0-12-381479-1.00016-2
Hayes S, Velanis CN, Jenkins GI, Franklin KA (2014) UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc Natl Acad Sci 111:11894
Hed B, Ngugi HK, Travis JW (2010) Use of gibberellic acid for management of bunch rot on Chardonnay and Vignoles grape. Plant Dis 95(3):269–278. https://doi.org/10.1094/PDIS-05-10-0382
Heijde M, Ulm R (2012) UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci 17:230–237
Herzog K, Wind R, Töpfer R (2015) Impedance of the grape berry cuticle as a novel phenotypic trait to estimate resistance to Botrytis cinerea. Sensors 15(6):12498–12512. https://doi.org/10.3390/s150612498
Hothorn T, Buehlmann P, Dudoit S, Molinaro A, Van Der Laan M (2006) Survival ensembles. Biostat 7(3):355–373
Houel C, Martin-Magniette ML, Nicolas SD, Lacombe T, Le Cunff L, Franck D, Torregrosa L, Conejero G, Lalet S, This P, Adam-Blondon AF (2013) Genetic variability of berry size in the grapevine (Vitis vinifera L.). Aust J Grape Wine Res 19(2):208–220. https://doi.org/10.1111/ajgw.12021
Janitza S, Tutz G, Boulesteix A-L (2016) Random forest for ordinal responses: prediction and variable selection. Comput Stat Data Anal 96:57–73
Kassambara A (2017) Practical guide to cluster analysis in R: unsupervised machine learning. vol 1. STHDA
Kicherer A, Roscher R, Herzog K, Šimon S, Förstner W, Töpfer R (2013) BAT (Berry Analysis Tool): a high-throughput image interpretation tool to acquire the number, diameter, and volume of grapevine berries. Vitis 52(3):129–135
Lê S, Josse J, Husson F (2008) FactoMineR: an R package for multivariate analysis. J Stat Softw 25(1):18. https://doi.org/10.18637/jss.v025.i01
Li-Mallet A, Rabot A, Geny L (2016) Factors controlling inflorescence primordia formation of grapevine: their role in latent bud fruitfulness? A review. Botany 94(3):147–163. https://doi.org/10.1139/cjb-2015-0108
Marguerit E, Boury C, Manicki A, Donnart M, Butterlin G, Nemorin A, Wiedemann-Merdinoglu S, Merdinoglu D, Ollat N, Decroocq S (2009) Genetic dissection of sex determinism, inflorescence morphology and downy mildew resistance in grapevine. Theor Appl Genet 118(7):1261–1278. https://doi.org/10.1007/s00122-009-0979-4
Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD (2017) PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45(Database issue):D183–D189. https://doi.org/10.1093/nar/gkw1138
Migicovsky Z, Sawler J, Gardner KM, Aradhya MK, Prins BH, Schwaninger HR, Bustamante CD, Buckler ES, Zhong G-Y, Brown PJ, Myles S (2017) Patterns of genomic and phenomic diversity in wine and table grapes. Hortic Res 4:17035
Molitor D, Behr M, Hoffmann L, Evers D (2012) Impact of grape cluster division on cluster morphology and bunch rot epidemic. Am J Enol Vitic 63(4):508
Nair NG, Allen RN (1993) Infection of grape flowers and berries by Botrytis cinerea as a function of time and temperature. Mycol Res 97(8):1012–1014. https://doi.org/10.1016/S0953-7562(09)80871-X
Nelson KE (1956) The effect of Botrytis infection on the tissue of Tokay grapes. Phytopath 46(4):223–229
OIV (2017) 2017 World Vitiviniculture Situation. OIV Statistical Report on World Vitiviniculture. International Organisation of Vine and Wine, Paris. http://www.oiv.int/. Accessed Dec 2018
OIV (2015) 2nd edition of the OIV descriptor list for grape varieties and Vitis species. http://www.oiv.int/. Accessed Dec 2018
Perez FJ, Viani C, Retamales J (2000) Bioactive gibberellins in seeded and seedless grapes: identification and changes in content during berry development. American J Enoland Vitic 51(4):315–318
Pertot I, Caffi T, Rossi V, Mugnai L, Hoffmann C, Grando MS, Gary C, Lafond D, Duso C, Thiery D, Mazzoni V, Anfora G (2017) A critical review of plant protection tools for reducing pesticide use on grapevine and new perspectives for the implementation of IPM in viticulture. Crop Prot 97:70–84. https://doi.org/10.1016/j.cropro.2016.11.025
Pieri P, Zott K, Gomes E, Hibert G (2016) Nested effects of berry half, berry and bunch microclimate on biochemical composition in grape. Oeno One 50(3):145–159
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.r-project.org. Accessed Sept 2018
Ren H, Gray WM (2015) SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol Plant 8(8):1153–1164. https://doi.org/10.1016/j.molp.2015.05.003
Rist F, Herzog K, Mack J, Richter R, Steinhage V, Töpfer R (2018) High-precision phenotyping of grape bunch architecture using fast 3D sensor and automation. Sensors 18(3):763
Sancenón V, Puig S, Mira H, Thiele DJ, Peñarrubia L (2003) Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol Biol 51:577–587
Sarooshi R (1977) Some effects of girdling, gibberellic acid sprays, bunch thinning and trimming on the sultana. Austr J Exp Agric 17(87):700–704. https://doi.org/10.1071/EA9770700
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7):671–675. https://doi.org/10.1038/nmeth.2089
Shavrukov YN, Dry IB, Thomas MR (2004) Inflorescence and bunch architecture development in Vitis vinifera L. Aust J Grape Wine Res 10(2):116–124. https://doi.org/10.1111/j.1755-0238.2004.tb00014.x
Shiri Y, Solouki M, Ebrahimie E, Emamjomeh A, Zahiri J (2018) Unraveling the transcriptional complexity of compactness in sistan grape cluster. Plant Sci 270:198–208
Smart R, Robinson M (1991) Sunlight into wine. A handbook for winegrape canopy management. Winetitles, Adelaide
Strobl C, Boulesteix A-L, Zeileis A, Hothorn T (2007). Bias in random forest variable importance measures: illustrations, sources and a solution. BMC Bioinfo 8(25). http://www.biomedcentral.com/1471-2105/8/25
Strobl C, Boulesteix A-L, Kneib T, Augustin T, Zeileis A (2008) Conditional variable importance for random forests. BMC Bioinforma 9(307). http://www.biomedcentral.com/1471-2105/9/307
Tello J, Ibáñez J (2014) Evaluation of indexes for the quantitative and objective estimation of grapevine bunch compactness. Vitis 53(1):9–16
Tello J, Ibáñez J (2017) What do we know about grapevine bunch compactness? A state-of-the-art review. Austr J Grape Wine Res. https://doi.org/10.1111/ajgw.12310
Tello J, Aguirrezabal R, Hernaiz S, Larreina B, Montemayor MI, Vaquero E, Ibáñez J (2015) Multicultivar and multivariate study of the natural variation for grapevine bunch compactness. Austr J Grape Wine Res 21(2):277–289. https://doi.org/10.1111/ajgw
Tello J, Torres-Perez R, Grimplet J, Ibanez J (2016) Association analysis of grapevine bunch traits using a comprehensive approach. Theor Appl Genet 129:227–242
The Gene Ontology Consortium (2017) Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res 45(Database issue):D331–D338. https://doi.org/10.1093/nar/gkw1108
Töpfer R, Hausmann L, Harst M, Maul E, Zyprian E, Eibach R (2011) New horizons for grapevine breeding. In: Flachowsky H, Hanke M-V (eds) Methods in temperate fruit breeding. Global Science Books Ltd, Ikenobe, pp 79–100
Vail ME, Marois JJ (1991) Grape cluster architecture and the susceptibility of berries to Botrytis cinerea. Phytopath 81(2):188–191. https://doi.org/10.1094/phyto-81-188
van Buuren S, Groothuis-Oudshoorn K (2011) mice: Multivariate imputation by chained equations in R. J Stat Softw 45(3):1–67
van Ooijen J (2009) MapQTL 6, software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma b. V, The Netherlands
Zyprian E, Ochssner I, Schwander F, Simon S, Hausmann L, Bonow-Rex M, Moreno-Sanz P, Grando MS, Wiedemann-Merdinoglu S, Merdinoglu D, Eibach R, Töpfer R (2016) Quantitative trait loci affecting pathogen resistance and ripening of grapevines. Mol Gen Genom 291(4):1573–1594. https://doi.org/10.1007/s00438-016-1200-5
Acknowledgements
We wish to thank Sarina Elser for expert technical assistance. This work was funded by the “Federal Program for Ecologic Landuse and other forms of Sustainable Agriculture” (Bundesprogramm Ökologischer Landbau und andere Formen nachhaltiger Landwirtschaft, BÖLN) of BLE (Bundesanstalt für Landwirtschaft und Ernährung) Federal Office for Agriculture and Food under the title “MATA- Molekulare Analyse der Traubenarchitektur” (Molecular analysis of cluster architecture) FKZ 2811NA056 (www.ble.de).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Matthias Frisch.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Pictures of the parental types of the cross GF.GA-47-42 × ‘Villard Blanc’. Both varieties showed reduced cluster compactness. A) maternal parent GF.GA-47-42 B) paternal parent ‘Villard Blanc’. (PDF 358 kb)
Online Resource 2
Comparison of the correlation among cluster architecture sub-traits in a correlation matrix. Cluster architecture sub-traits measured in the growing season 2015 (A), 2016 (B) and combined 2015 + 2016 (C) as Kendall’s tau-b correlation coefficient. Nonsignificant correlations are depicted as 0. (see Table 1 for full sub-trait names) (PDF 270 kb)
Online Resource 3
Overview of the importance of cluster architecture variables for the prediction of the OIV204 compactness descriptor using the “cforest” function for random forest calculation with the R-package “party.” The quality of the importance prediction was assessed with error estimates, i.e., error rate, ranked probability scores (RPS), mean absolute error (MAE), mean standard error (MSE). The combined 2015 and 2016 dataset was used with season as predictor variable (A) and without season as predictor variable (B). (PDF 246 kb)
Online Resource 4
x-axis: Manifestation of cluster architecture sub-trait. y-axis: predicted probability of OIV class membership. The 2015 and 2016 data of 149 F1 genotypes of the cross GF.GA-47-42 × ‘Villard Blanc’ was grouped according to growing season (15, 16) and flower sex (hermaphrodite (H) and female (F) in the left column and analyzed without regard to growing season (right column). (PDF 742 kb)
Online Resource 5
Error rate (ER) assessment for the prediction accuracy in cumulative link models (CLMs) over OIV204 classes and flower sex. For CLMs with the lowest AIC values (see Table 2) the ER was used to assess the prediction accuracy. OIV204 classes and flower sex members exhibited different error rates. All model variants were assessed with a mixed dataset neglecting the season information (-season) or using season as additional factor variable for modeling (+season) (PDF 180 kb)
Online Resource 6
QTLs related to cluster architecture in 149 F1 individuals of the segregating population of the cross GF.GA-47-42 × ‘Villard Blanc’ calculated with interval mapping (IM) and interval mapping with flower sex as co-factor (FS) during four growing seasons. Cluster architecture traits in bold do statistically contribute to a high extent to cluster architecture (PDF 304 kb)
Online Resource 7
QTL comparison. Attributes of QTLs reproducibly calculated in the cross-population with interval mapping (IM) and with interval mapping applying flower sex as co-variable for interval mapping (IM + FS). (PDF 177 kb)
Online Resource 8
Table of Gene ontology (GO) enrichment analysis with genes positioned in confidence intervals of cluster architecture-related QTLs, compared to all the genes in the reference genome PN40024 12x V2 (PDF 283 kb)
Rights and permissions
About this article
Cite this article
Richter, R., Gabriel, D., Rist, F. et al. Identification of co-located QTLs and genomic regions affecting grapevine cluster architecture. Theor Appl Genet 132, 1159–1177 (2019). https://doi.org/10.1007/s00122-018-3269-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-018-3269-1