Abstract
Key message
A new recessive powdery mildew resistance gene, Pm223899, was identified in Afghanistan wheat landrace PI 223899 and mapped to an interval of about 831 Kb in the terminal region of the short arm of chromosome 1A.
Abstract
Wheat powdery mildew, a globally important disease caused by the biotrophic fungus Blumeria graminis f.sp. tritici (Bgt), has occurred with increased frequency and severity in recent years, and some widely deployed resistance genes have lost effectiveness. PI 223899 is an Afghanistan landrace exhibiting high resistance to Bgt isolates collected from the Great Plains. An F2 population and F2:3 lines derived from a cross between PI 223899 and OK1059060-126135-3 were evaluated for response to Bgt isolate OKS(14)-B-3-1, and the bulked segregant analysis (BSA) approach was used to map the powdery mildew resistance gene. Genetic analysis indicated that a recessive gene, designated Pm223899, conferred powdery mildew resistance in PI 223899. Linkage analysis placed Pm223899 to an interval of about 831 Kb in the terminal region of chromosome 1AS, spanning 4,504,697–5,336,062 bp of the Chinese Spring reference sequence. Eight genes were predicted in this genomic region, including TraesCS1AG008300 encoding a putative disease resistance protein RGA4. Pm223899 was flanked proximally by a SSR marker STARS333 (1.4 cM) and distally by the Pm3 locus (0.3 cM). One F2 recombinant was identified between Pm3 and Pm223899 using a Pm3b-specific marker, indicating that Pm223899 is most likely a new gene, rather than an allele of the Pm3 locus. Pm223389 confers a high level of resistance to Bgt isolates collected from Pennsylvania, Oklahoma, Nebraska, and Montana. Therefore, Pm223389 can be used to enhance powdery mildew resistance in these states. Pm3b-1 and STARS333 have the potential to tag Pm223389 in wheat breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), is a globally important disease that occurs in most wheat-growing regions. Powdery mildew has occurred with an increased frequency in Europe, China, and many other countries in recent years (Morgounov et al. 2012). In the USA, the geographic range over which powdery mildew epidemics are sometimes or often severe is expanding from the traditional mid-Atlantic USA to southeastern states (Cowger et al. 2018), likely because of changing weather patterns and/or use of highly susceptible cultivars. A recent study indicated that there were substantial increases in the severity of powdery mildew overtime on a global level (Morgounov et al. 2012), and the predicted trend toward warmer winters in the eastern USA could increase the severity of Bgt epidemics by facilitating earlier epidemic onset (Cowger et al. 2018). Severe infection of powdery mildew can cause up to 40% yield loss, especially under humid rainfed and irrigated high-input conditions (Bennett 1984).
Cultivation of powdery mildew-resistant cultivars is an economical and environmentally friendly alternative to chemical control. A considerable number of powdery mildew resistance genes have been identified and used in wheat breeding (McIntosh et al. 2013, 2017). However, virulence shifts in Bgt populations lead to the rapid breakdown of powdery mildew resistance genes in mildew-prone regions. For example, Pm17 was commercially deployed in 2004 and began to lose effectiveness in the mid-Atlantic USA in 2009 (Griffey et al. 2005a, b; Cowger et al. 2009). A recent study indicated that Pm3f, Pm6, and Pm8 were entirely or largely defeated across the USA, whereas Pm2, Pm3a, Pm3b, and Pm4a were defeated in many regions. Moreover, widespread planting of cv. DG Shirley, believed to possess the previously widely effective Pm1a, has led to the emergence of Pm1a virulence in the North Carolina Bgt population in the last two years (C. Cowger, unpublished), indicating the challenge of breeding for durable resistance to powdery mildew.
The short-lived nature of powdery mildew resistance genes necessitates a continuous search for new resistance sources and pyramiding of multiple genes into a single cultivar. Although slow mildewing genes, such as Pm38 and Pm39 (Lillemo et al. 2008), are expected to offer durable powdery mildew resistance because of their race non-specific nature, they provide only partial resistance. Therefore, combination of slow mildewing genes with race-specific, highly resistant powdery mildew resistance genes is preferred for adequate and durable resistance. An alternative approach is to combine multiple race-specific genes in a single cultivar, which makes it difficult for Bgt strains to infect because mutations at multiple loci in the pathogen are required.
A prerequisite of gene pyramiding is to identify molecular markers closely linked to the genes of interest. Simple sequence repeat (SSR) markers have been widely used in wheat linkage mapping, and many SSR markers have been used to tag powdery mildew resistance genes in wheat breeding and wheat genetic studies. Although there is increased interest in using single nucleotide polymorphism (SNP) markers, SSR markers still play a unique role in genotyping segregating populations, because the co-dominant nature of SSR markers allows for unambiguous genotyping. The recent release of the Chinese Spring reference genome sequence makes it feasible to reveal all SSR loci in any region, permitting development of adequate SSR markers for precise mapping of target genes.
Wheat landraces are important resistance sources, and at least 18 powdery mildew resistance genes have been identified in landraces, including Pm2c on chromosome 5DS (Xu et al. 2015), Pm3b on 1AS (Yahiaoui et al. 2004), Pm5d and Pm5e on 7BL (Hsam et al. 2001; Huang et al. 2003), Pm24a and Pm24b on 1DS (Huang et al. 2000a; Xue et al. 2012), Pm45 on 6DS (Ma et al. 2011), Pm47 on 7BS (Xiao et al. 2013), Pm59 on 7AL (Tan et al. 2018), and Pm61 on 4AL (Sun et al. 2018). In addition, the temporarily named genes MlHLT and PmX were mapped to chromosomes 1DS and 2AL (Wang et al. 2015; Fu et al. 2013), respectively, and another six genes, PmTm4 (Hu et al. 2008), MlXBD (Huang et al. 2000b), Mlmz (Zhai et al. 2008), pmHYM (Fu et al. 2017), PmBYYT (Xu et al. 2018a), and PmSGD (Xu et al. 2018b), were mapped to a region near the Pm5 locus on chromosome 7BL. Compared with genes originating from wild species, powdery mildew resistance genes identified in landraces can be more easily used in wheat breeding because of the lack of linkage drag.
Li et al. (2016) reported that PI 223899 (formerly Gandom), a landrace collected from Badakhshan in Afghanistan, exhibited resistance to Bgt pathotypes collected in Oklahoma and suggested that PI 223899 has potential for use in wheat improvement. The objectives of this study were to determine the genomic location of the powdery mildew resistance gene in PI 223899 and develop linked markers for wheat breeding.
Materials and methods
Plant materials
An F2 population of 221 plants developed from the cross PI 223899 × OK1059060-126135-3 was evaluated for powdery mildew response, and all plants were then transferred to a greenhouse after being vernalized for 6 weeks at 4 °C. The F3 families were evaluated in the following season. In addition, a set of control lines carrying Pm3a, Pm3b, Pm17, and Pm8 were also tested.
Evaluation of powdery mildew resistance
The F1 plants, F2 population, and F2:3 families were evaluated for powdery mildew response at the USDA-ARS Wheat, Peanut, and Other Field Crop Research Unit at Stillwater using a previously described procedure (Tan et al. 2018). In brief, the F2 population was evaluated with Bgt isolate OKS(14)-B-3-1 in 2016. Each tested plant was grown in a single cell of 135-cell growing trays containing Sunshine Redi-earth growing mix (Sun Gro Horticulture Canada Ltd.) and inoculated at the two-leaf stage as described by Li et al. (2016). ‘TAM110’ and ‘Jagalene’ were planted in each growing tray as the resistant and susceptible checks, respectively. The inoculated plants were grown under natural light at 20 ± 2 °C in a greenhouse, and powdery mildew infection types (IT) were recorded 7–10 days after inoculation using a 0–4 scale, in which 0, 0;, and 1 represented highly resistant responses, while 2, 3, and 4 indicated moderately resistant, moderately susceptible, and highly susceptible, respectively. Each plant was reexamined 2 days after the initial investigation. The criteria for each IT score were described earlier (Tan et al. 2018). A total of 221 F2:3 lines were evaluated for response to OKS(14)-B-3-1 in 2017 using a randomized complete block design with two replicates. For each F2:3 line, 16 plants were planted in two cells of a 73-cell growing tray containing Sunshine Redi-earth growing mix in each replicate, and the protocol described above was implemented. The genotype of each F2 plant was inferred from the corresponding F2:3 phenotypic data.
PI 223899, together with Jagalene and four testers carrying Pm3a, Pm3b, Pm17, and Pm8, was evaluated for response to 18 Bgt isolates collected from different regions of the USA and maintained by the USDA-ARS Plant Science Research Unit at Raleigh, North Carolina. The detached-leaf approach described by Cowger et al. (2018) was used to assess powdery mildew responses on a 0–9 scale, which distinguished resistant (0–4), intermediate (5–6), and susceptible (7–9) reactions.
Analysis of SSR markers and genic markers
Genomic DNA was extracted from 2-week-old leaves using a previously described method (Dubcovsky et al. 1994). For each SSR assay, about 50 ng of genomic DNA was used in a volume of 10 μl containing 1.5 mM MgCl2, 0.2 mM of each dNTP, 1X PCR buffer, 0.25 unit of Taq DNA polymerase, and 0.2 mM of each primer. The reaction mixtures were denatured at 95 °C for 5 min, followed by 39 cycles of 95 °C for 30 s, 50–60 °C (depending on the annealing temperature) for 30 s, and 72 °C for 1 min, with a final extension at 72 °C for 10 min. PCR products were separated in 6–10% non-denaturing polyacrylamide gels (29:1 acrylamide/bisacrylamide ratio) and visualized under UV light after stained with ethidium bromide.
Two sequence-tagged site (STS) markers developed from the Pm3b sequence, Pm3b-1 (forward, 5′-TGCCTAGAAGATCTATGCTTATCAG; and reverse, 5′-CATGCCAGCACAGTTCAG) and Pm3b-2 (forward, 5′-TGTTCAGTTGTGGTACATCCT; and reverse, 5′-GACTGTACCAACCTATAACCTC) (Xu et al. 2006), were used to genotype the mapping population. For a 10-μl PCR, 50 ng of genomic DNA was added to a PCR mixture containing 0.2 mM dNTP, 1 × PCR buffer, 2.5 mM MgCl2, 0.25 units of Taq polymerase, 0.2 mM of each pair of STS primers. The PCR cycles consisted of an initial step of 94 °C for 5 min, followed by 36 cycles of 30 s at 94 °C, 30 s at 48 °C, and 40 s at 72 °C, with a final step of 7 min at 72 °C. PCR products were separated in 1.5% agarose gels and visualized under UV light after stained with ethidium bromide.
Bulked segregant analysis
Based on F2 genotypes inferred from F2:3 progeny’s phenotypes, DNA from each of 10 homozygous resistant and 10 homozygous susceptible F2 plants were pooled to construct resistant and susceptible bulks, respectively. The contrasting bulks and parental DNA samples were screened with more than 600 SSR markers that are evenly distributed across all wheat chromosomes to find informative markers exhibiting polymorphism between the bulks and parents.
A single informative marker was used to genotype the F2 population, leading to identification of a SSR marker associated with powdery mildew response. Additional SSR and genic markers previously mapped in the target region were also used to genotype the mapping population.
Development of SSR markers in the target region of the wheat genome
Based on Chinese Spring reference sequence IWGSC RefSeq v1.0 (https://urgi.versailles.inra.fr), all SSR loci in the genomic region around the powdery mildew resistance gene were identified, and primers were designed for a set of 36 SSR loci located in non-transposon regions using the GMATA software (Wang and Wang 2016). These new SSR markers, designated with prefix ‘STARS’ (representing Stillwater ARS) and a consecutive number (Table 1), were employed to genotype the mapping population.
Data analysis
Chi-squared tests were conducted to test the goodness of fit of observed phenotypic data to expected Mendelian ratios for a single gene. Mapmaker 3.0b (Lincoln et al. 1993) was employed to construct the genetic linkage map using the Kosambi function (Kosambi 1943), and a logarithm of the odds score of 3.0 was used as the threshold. MapDraw software (Liu and Meng 2003) was used to draw the linkage map.
Gene annotation
Genes were predicted, but were not annotated in Chinese Spring IWGSC RefSeq v1.0 (https://urgi.versailles.inra.fr). High-confidence genes in the target region were annotated using BlastX searches against the NCBI and Pfam databases for function prediction.
Results
Inheritance of powdery mildew resistance in PI 223899
PI 223899 was highly resistant to Bgt isolate OKS(14)-B-3-1 with IT 0;, and OK105960-126135-3 was susceptible with IT 4. F1 plants were susceptible, and the F2 population segregated with 47 resistant and 174 susceptible plants, suggesting that resistance was conferred by a single recessive allele (χ 21:3 = 1.62, df = 1, p = 0.2).
The F3 progeny tests confirmed the single locus segregation; 47 and 58 lines were classified homogenous resistant and homogeneous susceptible, respectively, and the remaining 116 lines segregated, confirming that PI 223899 carries a recessive powdery mildew resistance gene (χ 21:2:1 = 1.64, df = 2, p = 0.44) that was designated Pm223899.
Mapping of the powdery mildew resistance gene in PI 223899
BSA using more than 600 SSR primer pairs detected a single marker, CFA2153, on chromosome 1AS that distinguished the resistant and susceptible bulks and parents. After genotyping the entire F2 population for the marker, genetic distance between CFA2153 and Pm223899 was estimated to be 8.2 cM. A set of SSR markers mapped to the terminal region of 1AS was then screened for polymorphism between the two parents, leading to identification of PSP2999. PSP2999 was 6.1 cM distal to Pm223899.
Given that PSP2999 was closely linked to the Pm3 locus (Xu et al. 2006), positioned at approximately 4.5 Mb of the Chinese Spring reference assembly, we identified all SSR loci in the genomic region possibly harboring Pm223899; this ranged from 2.99 to 7.23 Mb in the reference sequence. A total of 526 SSR loci were identified in the region, and 36 loci located in non-transposon regions were chosen for marker development. Of these, 13 markers were polymorphic between PI 223899 and OK105960-126135-3, and the remaining 23 SSR markers were monomorphic (Table 1). The polymorphic markers were subsequently used to genotype the F2 population.
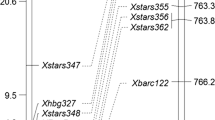
Based on F2 genotypic data, a linkage map of 25 cM was constructed. The 13 newly developed SSR markers, located from 3.12 to 7.22 Mb on the Chinese Spring physical map, covered 23.2 cM. The orders of newly developed SSRs on the linkage map were consistent with their physical positions on the Chinese Spring reference assembly (Fig. 1). Linkage analysis placed Pm223899 to an interval of 3 cM, flanked by STARS326 and STARS333 (Fig. 1).
Linkage (left) and physical map (right) of Pm223899. Marker names are shown at the right of the linkage map, and genetic distances in cM on the left. The physical positions of molecular markers are given at the far right of the physical map. The precise positions (in bp) of these markers are given in Table 1
Responses of lines containing Pm223899, Pm3a, Pm3b, Pm17, and Pm8 to Bgt isolates from different regions of the USA
Lines possessing Pm223899 and other genes located on 1AS were phenotyped with 18 Bgt isolates collected from different regions of the USA (Table 2). PI 223899 with Pm223899 showed resistance to isolates collected from Pennsylvania, Oklahoma, Nebraska, and Montana, indicating that Pm223899 can be used to enhance powdery mildew resistance in these states. However, the representative Bgt isolates from Georgia, Mississippi, North Carolina, New York, and Michigan were virulent.
Two alleles at the Pm3 locus, Pm3a and Pm3b, were also tested for responses to these differential isolates (Table 2). Both alleles were overcome by the isolates from the southeast and mid-Atlantic regions, but showed resistance to three isolates from Oklahoma and two from Montana. Pm3a exhibited resistance to a Nebraska isolate, and Pm3b conferred resistance to two Michigan isolates and one Pennsylvania isolate. In addition, Pm17, located on a wheat-rye 1AL/1RS translocation segment, exhibited either susceptible or intermediate reactions to all isolates in the panel, further confirming that Pm17 has been largely defeated in the USA. Similar results were observed with the rye-derived Pm8 gene, located on a wheat-rye 1BL/1RS translocation segment. Pm8 conferred resistance to only one isolate collected from New York, NYA-E-3-3, and showed either susceptible or intermediate reactions to the other 17 isolates.
Recombination between Pm3 and Pm223899
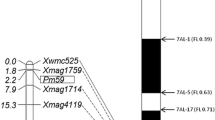
Pm223899 was mapped to a genomic region near Pm3. Therefore, it was essential to determine its relationship with Pm3. Xu et al. (2006) developed Pm3b-specific markers Pm3b-1 and Pm3b-2 from the Pm3b sequence (Yahiaoui et al. 2004). We used these markers to genotype two parents. Pm3b-1 was polymorphic and was used to genotype the F2 population. One recombination was identified between Pm3 and Pm223899 in plant 69, which was homozygous susceptible (pm223899 pm223899) and heterozygous at the Pm3b-1 locus (Pm3b-1 pm3b-1) (Fig. 2), suggesting that Pm223899 and Pm3 are different loci. The genotype of plant 69 was further confirmed by genotyping and phenotyping 16 additional F3 plants. The estimated genetic distance between Pm3 and Pm223899 was 0.3 cM (Fig. 1). Based on the physical locations of Pm3 and STARS333 on the Chinese Spring reference assembly, Pm223899 resides in an 831-Kb genomic region from 4,504,697 to 5,336,062 bp in the Chinese Spring reference.
Graphical genotypes and phenotypes of critical F2 plants and corresponding F3 phenotypes. Pm223899 was mapped to an interval flanked by Pm3b-1 and STARS333. Only one plant is shown for each genotype. R, S, HR, HS, and Seg represent resistant, susceptible, homozygous resistant, homozygous susceptible, and segregating, respectively
Predicted genes in the target region
Eight genes, TraesCS1AG008200–TraesCS1AG008900, were predicted in the genomic region spanning 4,504,697–5,336,062 bp in the Chinese Spring reference (https://urgi.versailles.inra.fr). TraesCS1AG008300 encodes an analog of putative disease resistance protein RGA4 (resistance gene analog 4), which together with RGA5 in rice directly binds with Magnaporthe oryzae avirulence proteins AVR-Pia and AVR1-CO39 to induce hypersensitive responses (Cesari et al. 2013). Another gene, TraesCS1AG008800, encodes a dirigent-like protein induced during disease response in plants. TraesCS1AG008400, TraesCS1AG008500, and TraesCS1AG008600 were annotated as 2′-deoxymugineic-acid 2′-dioxygenase, uncharacterized acetyltransferase, and alpha-humulene synthase genes, respectively, and the functions of TraesCS1AG008200, TraesCS1AG008700, and TraesCS1AG008900 are still unknown.
Discussion
Powdery mildew poses a persistent threat to wheat production worldwide. Identification and deployment of new powdery mildew resistance genes are essential for reducing large-scale yield losses caused by the breakdown of host resistance. In this study, we identified a recessive powdery mildew resistance gene, Pm223899, in an Afghanistan landrace and located it to the terminal region of chromosome 1AS.
Of the known powdery mildew resistance genes, Pm3 and Pm17 were mapped to chromosome 1AS. Pm17 is an alien resistance gene derived from rye (Mohler et al. 2001). Given that PI 223889 is a landrace, Pm223899 is unlikely Pm17. Analysis of a diagnostic marker for the Sec-1 locus of rye (Shimizu et al. 1997) indicated the absence of chromosome 1RS in PI 223899, confirming that Pm223899 is not Pm17.
Pm223899 was mapped to an interval of about 831 Kb flanked by Pm3 and STARS333. A recombinant between Pm3 and Pm223899 was identified among the F2 plants. The presence of the recombinant indicated that Pm223899 is a new gene different from the Pm3 locus. There are 18 functional alleles at the Pm3 locus (Pm3a-Pm3r) (Yahiaoui et al. 2004, 2009; Bhullar et al. 2009, 2010), and one of them, Pm3a, is widely used in the hard red winter wheat breeding programs in the Great Plains region (Li et al. 2016). A recent study indicated that 12–14% of Bgt isolates collected in this region in 2013 and 2014, as well as 90–100% of isolates collected in other regions of the USA, were virulent to Pm3a (Cowger et al. 2018), suggesting that new powdery mildew resistance genes are required in the Great Plains. PI 223899 is highly resistant to Bgt isolates collected from the Great Plains, Pennsylvania, and Montana and can be used as an alternative resistance source in these regions, but needs to be combined with other resistance genes to ensure any level of durability. Molecular markers closely linked to Pm223899, such as Pm3b-1 and STARS333, have the potential to tag Pm223389 in wheat breeding.
A set of eight genes were predicted in the interval in which Pm223899 was located, including TraesCS1AG008300, an R gene encoding an RGA4 protein. To date, five dominant powdery mildew seedling resistance genes, Pm2, Pm3b, Pm8, Pm21, and Pm60 (Yahiaoui et al. 2004; Hurni et al. 2013; Sánchez-Martín et al. 2016; Xing et al. 2017; Zou et al. 2018), have been cloned. Of these, Pm2 and Pm3b were identified in bread wheat, while Pm8, Pm21, and Pm 60 originated from rye, Haynaldia villosa, and Triticum urartu, respectively. All of them are R genes encoding coiled-coil nucleotide binding site leucine-rich repeat (CC-NBS-LRR) domain proteins (Yahiaoui et al. 2004; Hurni et al. 2013; Xing et al. 2017; Zou et al. 2018). Pm223899 is a recessive gene, and the underlying mechanism may be different from these dominant genes. A previous study indicated that the rice resistance protein pair RGA4/RGA5 directly binds with Magnaporthe oryzae avirulence proteins AVR-Pia and AVR1-CO39 to induce hypersensitive responses (Cesari et al. 2013). Thus, TraesCS1AG008300 is likely a candidate gene for Pm223899. In addition, another gene in the target region, TraesCS1AG008800, is also involved in plant defense, and the functions of three other genes are still unknown. Further cloning of Pm223899 is essential for understanding the mechanism of powdery mildew resistance in wheat.
Author contribution statement
XX, GL, BFC, and GB designed the research; GL performed the research; CC evaluated responses of differential lines to Bgt isolates; XX wrote the paper. All authors read, revised, and approved the manuscript.
Change history
01 November 2018
Unfortunately, the caption of Fig. 2 was incorrectly published in the original publication. The complete correct caption should read as follows.
References
Bennett FGA (1984) Resistance to powdery mildew in wheat: a review of its use in agriculture and breeding programs. Plant Pathol 33:279–300
Bhullar NK, Street K, Mackay M, Yahiaoui N, Keller B (2009) Unlocking wheat genetic resources for the molecular iden-tification of previously undescribed functional alleles at the Pm3 resistance locus. Proc Natl Acad Sci USA 106:9519–9524
Bhullar NK, Mackay M, Keller B (2010) Genetic diversity of the Pm3 powdery mildew resistance alleles in wheat gene bank accessions as assessed by molecular markers. Diversity 2:768–786
Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, Morel JB (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25:1463–1481
Cowger C, Parks R, Marshall D (2009) Appearance of powdery mildew of wheat caused by Blumeria graminis f. sp. tritici on Pm17-bearing cultivars in North Carolina. Plant Dis 93:1219
Cowger C, Mehra L, Arellano C, Meyers E, Murphy JP (2018) Virulence differences in Blumeria graminis f. sp. tritici from the central and eastern United States. Phytopathology 108:402–411
Dubcovsky J, Galvez AF, Dvořák J (1994) Comparison of the genetic organization of the early salt-stress-response gene system in salt tolerant Lophopyrum elongatum and salt-sensitive wheat. Theor Appl Genet 87:957–964
Fu B, Chen Y, Li N, Ma H, Kong Z, Zhang L, Jia H, Ma Z (2013) pmX: a recessive powdery mildew resistance gene at the Pm4 locus identified in wheat landrace Xiaohongpi. Theor Appl Genet 126:913–921
Fu BS, Zhang ZL, Zhang QF, Wu XY, Wu JZ, Cai SB (2017) Identification and mapping of a new powdery mildew resistance allele in the Chinese wheat landrace Hongyoumai. Mol Breed 37:133
Griffey CA, Rohrer WL, Pridgen TH, Brooks WS, Chen J, Wilson JA, Nabati D, Brann DE, Rucker EG, Behl HD, Vaughn ME (2005a) Registration of’ McCormick wheat. Crop Sci 45:417–420
Griffey CA, Rohrer WL, Pridgen TH, Brooks WS, Chen J, Wilson JA, Nabati D, Brann DE, Rucker EG, Behl HD, Vaughn ME (2005b) Registration of Tribute wheat. Crop Sci 45:419–421
Hsam SLK, Huang XQ, Zeller FJ (2001) Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.) 6. Alleles at the Pm5 locus. Theor Appl Genet 102:127–133
Hu TZ, Li HJ, Xie CJ, You MS, Yang ZM, Sun QX, Liu ZY (2008) Molecular mapping and chromosomal location of powdery mildew resistance gene in wheat cultivar Tangmai 4. Acta Agron Sin 34:1193–1198
Huang XQ, Hsam SLK, Zeller FJ et al (2000a) Molecular mapping of the wheat powdery mildew resistance gene Pm24 and marker validation for molecular breeding. Theor Appl Genet 101:407–414
Huang XQ, Hsam SLK, Zeller FJ (2000b) Chromosomal location of powdery mildew resistance genes in Chinese wheat (Triticum aestivum L. em. Thell.) landraces Xiaobaidong and Fuzhuang 30. J Genet Breed 54:311–317
Huang X, Wang L, Xu M, Röder M (2003) Microsatellite mapping of the powdery mildew resistance gene Pm5e in common wheat (Triticum aestivum L.). Theor Appl Genet 106:858–865
Hurni S, Brunner S, Buchmann G, Herren G, Jordan T, Krukowski P, Wicker T, Yahiaoui N, Mago R, Keller B (2013) Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J 76:957–969
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Li G, Xu X, Bai G, Carver BF, Hunger R, Bonman JM (2016) Identification of novel powdery mildew resistance sources in wheat. Crop Sci 56:1817–1830
Lillemo M, Asalf B, Singh RP, Huerta-Espino J, Chen XM, He ZH, Bjørnstad Å (2008) The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar. TheorAppl Genet 116:1155–1166
Lincoln SE, Daly MJ, Lander ES (1993) Constructing genetic linkage maps with MAPMAKER/EXP Version 3.0: a tutorial and reference manual. A Whitehead Institute for Biomedical Research Technical Report, 3
Liu RH, Meng JL (2003) MapDraw: a Microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 25:317–321
Ma HQ, Kong ZX, Fu BS, Li N, Zhang LX, Jia HY, Ma ZQ (2011) Identification and mapping of a new powdery mildew resistance gene on chromosome 6D of common wheat. Theor Appl Genet 123:1099–1106
McIntosh RA, Yamazaki Y, Dubcovsky J et al (2013) Catalogue of gene symbols for wheat. In: Ogihara Y (ed) Proccedings of the 12th international wheat genet symposium, Yokohama, Japan 8–13 Sept 2013, pp 8–13
McIntosh RA, Dubcovsky J, Rogers WJ et al (2017) Catalogue of gene symbols for wheat. Supplement. Annu Wheat Newsl 53:1–20
Mohler V, Hsam SL, Zeller FJ, Wenzel G (2001) An STS marker distinguishing the rye-derived powdery mildew resistance alleles at the Pm8/Pm17 locus of common wheat. Plant Breed 120:448–450
Morgounov A, Tufan HA, Sharma R et al (2012) Global incidence of wheat rusts and powdery mildew during 1969–2010 and durability of resistance of winter wheat variety Bezostaya 1. Eur J Plant Pathol Dordr 132:323–340
Sánchez-Martín J, Steuernagel B, Ghosh S et al (2016) Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol 17(1):221
Shimizu Y, Nasuda S, Endo TR (1997) Detection of the Sec-1 locus of rye by a PCR-based method. Genes Genet Syst 72:197–203
Sun H, Hu J, Song W (2018) Pm61: a recessive gene for resistance to powdery mildew in wheat landrace Xuxusanyuehuang identified by comparative genomics analysis. Theor Appl Genet. https://doi.org/10.1007/s00122-018-3135-1
Tan C, Li G, Cowger C, Carver BF, Xu X (2018) Characterization of Pm59, a novel powdery mildew resistance gene in Afghanistan wheat landrace PI 181356. Theor Appl Genet 131:1145–1152
Wang X, Wang L (2016) GMATA: an integrated software package for genome-scale SSR mining, marker development and viewing. Front Plant Sci 7:1350
Wang Z, Li H, Zhang D, Guo L, Chen J, Chen Y, Wu Q, Xie J, Zhang Y, Sun Q, Dvorak J (2015) Genetic and physical mapping of powdery mildew resistance gene MlHLT in Chinese wheat landrace Hulutou. Theor Appl Genet 128:365–373
Xiao M, Song F, Jiao J et al (2013) Identification of the gene Pm47 on chromosome 7BS conferring resistance to powdery mildew in the Chinese wheat landrace Hongyanglazi. Theor Appl Genet 126:1397–1403
Xing L, Hu P, Liu J, Cui C, Wang H, Di Z, Zhou S, Xu J, Gao L, Huang Z, Cao A (2017) NLR1-V, a CC-NBS-LRR encoding gene, is a potential candidate gene of the wheat powdery mildew resistance gene Pm21. bioRxiv 114058
Xu XY, Bai GH, Carver BF, Shaner GE, Hunger RM (2006) Molecular characterization of a powdery mildew resistance gene in wheat cultivar Suwon 92. Phytopathology 96:496–500
Xu H, Yi Y, Ma P, Qie Y, Fu X, Xu Y, Zhang X, An D (2015) Molecular tagging of a new broad-spectrum powdery mildew resistance allele Pm2c in Chinese wheat landrace Niaomai. Theor Appl Genet 128:2077–2084
Xu XD, Feng J, Fan JR, Liu ZY, Li Q, Zhou YL, Ma ZH (2018a) Identification of the resistance gene to powdery mildew in Chinese wheat landrace Baiyouyantiao. J Integr Agric 17:37–45
Xu XD, Li Q, Ma ZH, Fan JR, Zhou YL (2018b) Molecular mapping of powdery mildew resistance gene PmSGD in Chinese wheat landrace Shangeda using RNA-seq with bulk segregant analysis. Mol Breed 38:23
Xue F, Wang C, Li C et al (2012) Molecular mapping of a powdery mildew resistance gene in common wheat landrace Baihulu and its allelism with Pm24. Theor Appl Genet 125:1425–1432
Yahiaoui N, Srichumpa P, Dudler R, Keller B (2004) Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 37:528–538
Yahiaoui N, Kaur N, Keller B (2009) Independent evolution of functional Pm3 resistance genes in wild tetraploid wheat and domesticated bread wheat. Plant J 57:846–856
Zhai WW, Duan XY, Zhou YL, Ma HQ (2008) Inheritance of resistance to powdery mildew in four Chinese landraces. Plant Prot 34:37–40
Zou S, Wang H, Li Y, Kong Z, Tang D (2018) The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol 218:298–309
Acknowledgements
We thank M. Hargrove and R. Whetten for excellent technical assistance and Dr. Robert McIntosh of Sydney University for reviewing this paper. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. The USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by Xianchun Xia.
Unfortunately, the caption of Figure 2 was incorrectly published in the original publication. The complete correct caption should read as follows.
Fig. 2 Graphical genotypes and phenotypes of critical F2 plants and corresponding F3 phenotypes. Pm223899 was mapped to an interval flanked by Pm3b-1 and STARS333. Only one plant is shown for each genotype. R, S, HR, HS, and Seg represent resistant, susceptible, homozygous resistant, homozygous susceptible, and segregating, respectively.
Also, under the “Discussion section”, 3rd paragraph, the following sentence was incorrectly published and the complete correct sentence is given below.
There are 18 functional alleles at the Pm3 locus (Pm3a-Pm3r) (Yahiaoui et al. 2004, 2009; Bhullar et al. 2009, 2010), and one of them, Pm3a, is widely used in the hard red winter wheat breeding programs in the Great Plains region (Li et al. 2016).
Rights and permissions
About this article
Cite this article
Li, G., Carver, B.F., Cowger, C. et al. Pm223899, a new recessive powdery mildew resistance gene identified in Afghanistan landrace PI 223899. Theor Appl Genet 131, 2775–2783 (2018). https://doi.org/10.1007/s00122-018-3199-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-018-3199-y