Abstract
Key message
Using bulk segregant analysis (BSA) coupling with RNA-seq and DNA markers identified a potentially novel nitrogen-dependent lesion mimic gene Ndhrl1 on 2BS in wheat.
Abstract
Lesion mimic (LM) refers to hypersensitive reaction-like (HRL) traits that appear on leaf tissue in the absence of plant pathogens. In a wheat line P7001, LM showed up on the leaves under the 0 g nitrogen (N) treatment, but disappeared when sufficient N was supplied, suggesting that LM is N-responsive and N dosage dependent. Using BSA strategy together with RNA-seq and DNA markers, we identified an N-dependent LM gene (Ndhrl1) and mapped it to the short arm of chromosome 2B using an F5 recombinant inbred population developed from the cross of P7001 × P216. The putative gene was delimited into an interval of 8.1 cM flanked by the CAPS/dCAPS markers 7hrC9 and 7hr2dc14, and co-segregated with the dCAPS marker 7hrdc2. This gene is most likely a novel gene for LM in wheat based on its chromosomal location. Further analysis of RNA-seq data showed that plant–pathogen interaction, nitrogen metabolism, zeatin biosynthesis and plant hormone signal transduction pathways were significantly differentially expressed between LM and non-LM lines.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat is one of the most widely grown crops in the world with an annual global production of more than 716 million tons across over 200 million hectares. However, the wheat yield needs to increase at least 1.9 times by 2050 to meet the future food demands (http://faostat3.fao.org). To meet this challenge, development of crops with broad-spectrum and durable resistance is economically important for improving eco-adaptability and yield stability of the crops in diverse eco-regions (Johnson 1981; Wang and Bayles 2013). Plant hypersensitive reaction (HR) occurs in an incompatible host–parasite relationship in defense against pathogens. A lesion mimic (LM) trait in a leaf tissue without presence of plant pathogens, which is also called hypersensitive reaction-like (HRL) trait, may confer enhanced plant resistance to a wide range of pathogens (Bruggeman et al. 2015). Many studies have shown LM mutants that constitutively express defense response genes are essential for the revealing of defense signaling pathways (Andersson et al. 2008; Lorrain et al. 2003; Moeder and Yoshioka 2008).
LM-triggered disease resistance has been reported in several plant species such as Arabidopsis (Li et al. 2013; Yamaguchi et al. 2012), rice (Fekih et al. 2015; Yamamura et al. 2015), maize (Gray et al. 2002; Penning et al. 2004), and barley (McGrann et al. 2015). It has been shown that LM is associated with porphyrins- and phenolics-mediated chloroplast collapse (Hu et al. 1998) and with cell death that are mediated by membrane-associated proteins (Lorrain et al. 2004), ion channel proteins (Balagué et al. 2003; Borghi et al. 2011), cytochrome P450 monooxygenase (Fujiwara et al. 2010), ubiquitin/proteasome (Shirsekar et al. 2014; Wang et al. 2015), and by nitric oxide (Chen et al. 2014; Ling et al. 2012) or by reactive oxygen species (Brosché et al. 2014). These studies suggest that LM may spontaneously activate expression of defense genes. Some LM mutants spontaneously display cell death and constitutive defense responses, and have been used to decipher cell death signal pathways.
In wheat, some studies reported that LM could be induced by either a mutagen (Boyd and Minchin 2001; Boyd et al. 2002; Kamlofski et al. 2007; Sugie et al. 2007) or a transgenic approach (Anand et al. 2003, 2004). Wheat variety Ning7840 shows an LM phenotype at heading stage and adult plant resistance to leaf rust and powdery mildew diseases, and a recessive gene (lm) responsible for the LM trait was located near the proximal region of chromosome 1BL (Li and Bai 2009; Li et al. 2012). Yao et al. (2009) reported segregation of an LM trait in a wheat segregating population of Yanzhan1/Zaosui30 although both parents do not have LM phenotypes, and assumed that the LM phenotype is due to the interaction of two recessive genes lm1 (lesion mimic 1) on 3BS and lm2 (lesion mimic 2) on 4BL. Although a lot of LM mutants (Boyd and Minchin 2001; Boyd et al. 2002; Kamlofski et al. 2007; Sugie et al. 2007) and LM-related genes have been reported, only a few LM-related genes have been mapped in wheat.
Advances in next-generation sequencing technology provide great conveniences in genetic studies especially in development of molecular markers for the species without reference sequences. In this study, bulk segregant analysis (BSA) strategy was used together with DNA marker and RNA-seq to map a novel gene Ndhrl1 underlying nitrogen-dependent LM trait in the wheat line P7001. The results would be useful for elucidating the relationships among application of nitrogen (N), LM and disease resistance.
Materials and methods
Plant materials and evaluation of the LM phenotype
The wheat lines P7001 and P216 were planted in plastic pots filled with 3 kg of soil collected from the wheat field in Yangzhou University, China. All plants were vernalized at 4 °C in a growth chamber for four weeks and transplanted into plastic pots on a greenhouse bench at 17 ± 2 °C (night) and 22 ± 5 °C (day) with supplemental light for 12 h in Yangzhou University, Yangzhou, Jiangsu province, China. The experiment was arranged in a randomized-complete-block design with four replicates (pots), with eight plants per replicate. Plants were treated with four levels (0, 2, 4 and 6 g per pot) of N fertilizer (carbamide), where 1 g carbamide per pot roughly equal to 350 kg per hectare. The nitrogen availability in soil was determined by the distillation method (Bremner 1965). A mixture of Blumeria graminis f. sp. tritici (Bgt) isolates collected from eastern China was applied to inoculate the wheat line P7001 after heading stage when LM trait appears. Severities of powdery mildew disease on flag leaves were scored as the percentage of infected leaf area (PIA) (immune: PIA = 0; resistant: 0 < PIA ≤ 25 %; moderately resistant: 25 < PIA ≤ 50 %; moderately susceptible: 50 < PIA ≤ 75 %; susceptible: 75 < PIA ≤ 100 %).
A total of 329 recombinant inbred lines derived from the population of P7001 × P216 by single seed descent were planted in the field of Yangzhou University. No nitrogen fertilizer was applied in the field plots in the entire growing season. Phenotype was scored as presence (1) or absence (0) of LM symptoms on the flag leaf at heading stage.
Mapping of Ndhrl1
Genomic DNA was extracted following the CTAB method (Porebski et al. 1997). A set of 2133 SSR markers was screened for polymorphism between the two parents including 1109 from public database (http://wheat.pw.usda.gov/GG3), and 1024 new markers developed in our lab (unpublished results). Equal amounts of DNA from 15 homozygous F5 LM+ lines and 15 homozygous LM− lines were bulked separately to generate LM+ pool and LM− pool, respectively. Polymorphic SSR markers between the two parents were subsequently used to screen the two DNA pools to identify polymorphic markers between the pools.
BSA RNA-seq (BSR-Seq)
The same lines used for DNA bulks were used to form two RNA pools. Two biological replicates were prepared from different plants of the same lines. Total RNA was extracted from the flag leaves collected at heading stage using the TRIzol reagent (Invitrogen, USA). DNA was removed using DNase (Invitrogen) and then cleaned using the RNAeasy Mini Kit (Qiagen). The RIN (RNA integrity number) values of RNA samples were assessed using an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA). The equal amounts of RNA with RIN >7.0 from 15 F5 lines of each bulk were pooled for cDNA libraries construction and sequencing in the Beijing Genomics Institute (BGI, China).
De novo transcriptome assembly and SNP calling
After removing low-quality reads, the clean reads were then de novo assembled using the Trinity platform with the parameters set at ‘K-mer = 25 and group pairs distance = 300’ (Grabherr et al. 2011). For each library, short reads were first assembled into longer contigs based on overlapping sequences. Then different contigs from another transcript and their distance were further recognized by mapping the clean reads back to corresponding contigs based on their paired-end information, and thus the sequences of the transcripts were produced. Finally, the potential transcript sequences were clustered using the TGI clustering tool to obtain uni-transcripts (Pertea et al. 2003). Single-nucleotide polymorphism (SNP) was identified using SOAP2 (Li et al. 2009) based on the assembled transcript sequences.
Development and verification of dCAPS markers
Primers for dCAPS markers (Supplementary Table S1) were designed using dCAPS finder 2.0 (Neff et al. 1998, Neff et al. 2002) and Primer Premier 5 (http://www.premierbiosoft.com/primerdesign) based on the assembled transcript sequences. The transcript sequences were blasted to the reference sequences of bread wheat cv. Chinese Spring (CS) (Mayer et al. 2014). The genetic positions of the transcript sequences were projected to the linkage map of CS by Poland et al. (2012) (http://wheat-urgi.versailles.inra.fr/Seq-Repository/Genes-annotations). P7001 and P216 were used for the verification of the associated SNPs from the RNA-seq data.
Construction of linkage map
A population of 329 F5 lines was used for linkage map construction using JoinMap 4 (https://www.kyazma.nl/index.php/mc.JoinMap) with an LOD value set at 3 for grouping. Only informative markers (8 SSR and 8 CAPS/dCAPS markers) were used to construct the map.
Analysis of transcripts in global pathways
To evaluate the sequencing depth, all usable reads were realigned to each uni-transcript using the SOAP aligner (http://soap.genomics.org.cn/soapaligner.html), then normalized into FPKM (fragments per kb per million reads) values (Mortazavi et al. 2008). After that, differences in uni-transcript abundance between the samples were calculated based on the ratio of the FPKM values, and the q value (Tarazona et al. 2011) was used to identify the threshold in multiple tests to determine the significance of the differences in transcript abundance between genotypes. Here, only uni-transcripts with a q value >0.8 were used for subsequent analysis. Pathway-based analysis was conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway database (Qian and Wang 2014).
Results
A single dominant gene controls the nitrogen-dependent LM phenotype
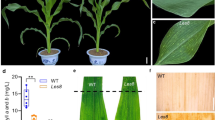
In the absence of any visible pathogen, small yellowish spots appeared randomly and dispersedly on all the leaves of P7001 at the heading stage in the treatment of 0 g carbamide per pot, whereas the spots were not observed in P7001 in the treatment of 2 g carbamide per pot. The yellowish spots did not appear on leaves of P216 irrespective of N fertilizer dosage (Supplementary Figure S1). These results suggest that the LM phenotype in P7001 is N dosage dependent and high dosage of N suppresses the expression of LM trait. P7001 with the LM trait showed a high level of resistance to powdery mildew disease with a severity of 8.4 ± 1.7 % under the treatment of 0 g N. In contrast, it had a significantly higher severity (36.8 ± 4.3 %) under sufficient N fertilization when the LM symptoms were absent.
Considering that LM trait is N dosage dependent and N deficiency favors the expression of the LM trait, P7001, P216 and their recombinant inbred lines (F4 and F5) were planted in a field without supply of any N fertilizer in the entire wheat growing season. The field originally had about 142 ± 2.3 mg/kg N content in the soil. In the F4 population, 188 families were LM type and 141 families were non-LM type. The F5 progenies from the 141 F4 non-LM families showed homozygous non-LM phenotype; whereas among the 188 F4 LM families, 158 F5 lines showed homozygous LM phenotype, nine showed homozygous non-LM phenotype, and 21 showed segregation. A total of 212 plants from the 21 segregating F5 lines had a segregation ratio of 3 (147 LM plants):1 (65 non-LM plants) (χ 2 = 3.623 < 3.814). Visual difference in LM symptoms was not observed between the LM homozygotes and heterozygotes in the segregating lines. These results suggest that LM in P7001 is controlled by a single dominant gene (designated as Ndhrl1).
The chromosomal location of Ndhrl1
A total of 2133 SSR primer pairs were screened between P7001 and P216, and 207 primers showed polymorphism between the two parents. However, only two newly developed SSR markers (Xyzu166898 and Xyzu176836) from 2BS showed polymorphism between LM and non-LM DNA pools, and thus they were used to genotype 158 homozygous LM F5 lines and 150 (141 + 9) homozygous non-LM F5 lines (Table 1). The single marker analysis showed a significant association between LM trait and two markers on short arm of chromosome 2B (Table 1), indicating that Ndhrl1 resides on the same chromosome arm.
A dCAPS marker 7hrdc2 co-segregates with LM trait
Since only a few SSR markers were available for fine mapping of the gene of interest, the RNA-seq of the bulked F5 lines was then employed to develop closely linked markers to the gene. After trimming the adaptor sequences, ambiguous reads and low-quality reads, the four libraries generated 48 Gb of clean data from 480 million reads. The average Q20 (the percentage of bases in the reads with a Phred quality equal or bigger than 20) of the clean reads was about 97.8 %, suggesting that the sequencing results were valid (Supplementary Table S2a). A total of 143,858 transcripts were obtained using the sequence reads from the four assembled samples. The N50 (the length for which the sum of bases in the long contig of that length or longer is at least half the bases in the assembly) of transcripts was 1651 nt (Supplementary Table S2b). The size distribution of these transcripts is shown in Supplementary Table S2c.
RNA-seq analysis found 11 polymorphic SNPs between the two RNA bulks. CAPS/dCAPS (cleaved amplified polymorphic sequence/derived CAPS) markers were designed to verify these polymorphic SNPs (Table 2). Three CAPS/dCAPS markers were verified and assigned to 2BS by blasting to the draft sequences of cv. CS. These markers were then used to genotype the 329 F5 lines. The dCAPS marker 7hrdc2 developed from Unigene66718 co-segregated with the LM phenotype, and the other two markers (7hrdc5 and 7hrC9) were also associated with the LM phenotype.
Additional 30 CAPS/dCAPS markers were developed based on the trait-associated SNPs identified from the RNA-seq and were verified (Table 3). Five of them (7hr2dc1, 7hr2dc8, 7hr2dc14, 7hr2dc27 and 7hr2dc28) and six newly developed SSR markers (yzu172962, yzu175773, yzu176060, yzu182917, yzu184614 and yzu186267) were used to genotype the F5 population to construct a linkage map of the target gene (Fig. 1). To fine map Ndhrl1, 16 markers on 2BS were mapped and seven of them can separate the recombinants into seven haplotypes (Table 4). First five types had 16 lines with the LM phenotype in the field, and carried P7001-allele at the marker 7hrdc2. Haplotypes 6 and 7 showed non-LM trait and had eight lines carrying P216 alleles at markers 7hrdc5 and 7hrdc2. The results narrowed down Ndhrl1 into an interval of 8.1 cM between the markers 7hrC9 and 7hr2dc14.
The significant pathways that were enriched in LM bulk
Comparative analysis of the transcriptome data revealed that 139,392 and 140,632 transcripts were found in LM bulk and non-LM bulk, respectively. A total of 136,166 transcripts were commonly expressed in the both bulks (Supplementary Figure S2a), and many of them were quantitatively regulated. The average expression levels of the genes in LM and non-LM bulks were 5.42 and 5.29 FPKM (fragments per kb per million reads), respectively. A total of 20,910 differentially expressed genes (DEGs) were identified based on the normalized value of FPKM with 15,876 up-regulated and 5034 down-regulated in the LM bulk comparing to the non-LM bulk (Supplementary Figure S2b). Using KOBAS (KEGG Orthology-Based Annotation System), these DEGs were mapped to 127 pathways. Among them, 37 pathways showed significant differences at p < 0.05 based on the hypergeometric distribution (Supplementary Table S3). The top five enriched pathways were plant–pathogen interaction, biosynthesis of secondary metabolites, biosynthesis of stilbenoid, diarylheptanoid and gingerol, plant hormone signal transduction, and flavonoid biosynthesis.
Discussion
LM in P7001 is N-dependent
Many types of LM have been reported in previous studies. These published LM phenotypes are caused by light (Arase et al. 2000; Ueno et al. 2015; Yao et al. 2009), humidity and/or temperature (Noutoshi et al. 2003; Sohn et al. 2014), day-length (Ishikawa et al. 2001) or spontaneous LM (Li and Bai 2009; Li et al. 2012). In the current study, small yellowish spots appeared at heading stage and randomly distributed on the leaves of P7001 under the 0 g N treatment, but the spots were not observed under the 2 g N treatment, implying that N deficiency results in LM phenotype in P7001 and sufficient N supply can suppress the expression of the LM phenotype. Moreover, comparative analyses of the transcriptome data between the two contrasting bulks revealed that N metabolic pathway is significantly enriched in LM bulk (Supplementary Table S3). In this pathway, ferredoxin-nitrite reductase (NiR) was significantly up-regulated in the LM bulk (Supplementary Figure S3) with 7.8-fold of that in non-LM bulk. NiR is an enzyme that converts nitrite to ammonia, a toxic compound that can cause cell death. Therefore, we may assume that high level of NiR expression may result in a high level of ammonia accumulation when abundant N is supplied, and thus LM phenotype should be expected due to more cell death caused by ammonia. However, actually this assumption is contradictory with the observation that LM phenotype was suppressed when abundant N was available. Many studies demonstrated that the feedback regulation is involved in the nitrate reductase (NR) expression and NR activity (Fan et al. 2006; Soussana et al. 2002). However, only fewer studies reported the feedback regulation of NiR. Further studies are needed to understand if the feedback regulation of NiR exists in wheat and how N is involved in the formation of LM phenotype.
BSR-seq is an effective approach to fine map Ndhrl1
To date, the genetic study of wheat has been lagging behind other plant species such as Arabidopsis, rice and maize due to its complex genome. In this study, 1109 SSR markers were used to map Ndhrl1, but we did not find significantly linked markers to the gene, which might be due to the low marker density in the map used for gene mapping. Recently, we have developed a new set of SSR markers (unpublished data) based on reference sequence of CS and used a selected set of 1024 SSRs to genotype the two DNA bulks. Two of them were linked to Ndhrl1. We used RNA-seq coupled with BSA strategy and efficiently identified closely linked, even co-segregating, SNP markers to Ndhrl1. However, only a very low proportion of SNPs from BSR-seq could be confirmed after they were converted into CAPS or dCAPS markers. Among the 41 CAPS/dCAPS primers designed from RNA-seq, only 8 were polymorphic, 9 were monomorphic, 11 failed to produce a clear band, and 13 amplified more than one band (Tables 2, 3). PCR failure was possibly due to the difference in templates between RNA and DNA. One of the most important reasons for the low conversion rate is that wheat has a high level of repeated sequences and sequence similarity among three wheat genomes can result in assembly error of short reads and thus a high proportion of false-positive SNPs between the bulks.
The genotyping technologies based on next-generation sequencing bring great conveniences to genetic studies. However, marker data generated using these technologies are strongly recommended to be validated before they can be used for marker-assisted breeding, especially for crops such as wheat with huge and complicated genomes. In the current study, towards fine mapping of Ndhrl1 was benefited from using the RNA-seq coupled with BSA for marker development. A co-segregating marker with Ndhrl1 was identified between the flanking markers at 8.1 cM apart on 2BS, which would facilitate map-based cloning of Ndhrl1. The first report of genes for LM trait on 2BS and N-dependent nature of the gene expression suggests that Ndhrl1 is more likely a novel gene for LM in wheat.
The putative pathways related to LM
Comparative analyses of the pathways between LM and non-LM bulks showed that the top one enriched pathway was plant–pathogen interaction with p = 2.79E−90. Many studies reported that the genes for LM were involved in defense signaling pathways with the constitutive expression of defense responses (Andersson et al. 2008; Lorrain et al. 2003; Moeder and Yoshioka 2008). In the current RNA-seq experiment, 5336 transcripts were involved in plant–pathogen interaction pathway. Of them, up to 1508 transcripts showed significant differences in expressions between the two bulks. P7001 without LM trait showed a moderate resistance to powdery mildew under sufficient N fertilizer, but the resistance level increased greatly under 0 g N treatment when LM showed up. These results imply that the LM phenotype mediates the resistance to powdery mildew disease. Cloning and functional characterization of Ndhrl1 may provide insights into the molecular and biochemical mechanisms underlying the interactions between N and LM genes in regulating disease resistances in wheat.
The dCAPS marker 7hrdc2 of Unigene66718 co-segregated with LM trait in the current mapping population (Table 2). Unigene66718 encodes a cytokinin trans-hydroxylase (CYP735A) belonging to cytochrome P450 monooxygenase super family that catalyzes the biosynthesis of trans-zeatin. Interestingly, the zeatin biosynthesis pathway (p = 3.86E − 09) and the downstream plant hormone signal transduction pathway (p = 9.29E−33) showed significant differences between the two bulks. Although the marker 7hrdc2 co-segregated with LM trait and the pathway of Unigene66718 was relevant to the formation of LM trait, it is still too early to claim Unigene66718 as the candidate gene for LM trait in P7001 due to the limited population size of 329 lines. Establishing the association between LM and 7hrdc2 in a much larger mapping population would reveal the candidateship of Unigene66718 for LM trait.
In conclusion, Ndhrl1 for LM trait in P7001 was mapped to the short arm of 2B chromosome. The different chromosomal location of Ndhrl1 from the published LM genes in wheat as well as the nature of the N-dependent LM trait in P7001 implied that Ndhrl1 is most likely a novel type of gene for LM in plants. Cloning and functional characterization of Ndhrl1 may provide a new insight into the interactions between N and LM trait in disease resistance in plants.
Author contribution statement
LL designed the experiments, supervised all analyses and drafted the manuscript; XS and FZ carried out the primer design and genotyping. CL carried out the phenotyping; DW carried out the analysis of RNA-seq. DG prepared the plant material, GB, JW and TL conceived the idea of the study and finalized the manuscript. All of the authors read and approved the final manuscript.
References
Anand A, Schmelz EA, Muthukrishnan S (2003) Development of a lesion-mimic phenotype in a transgenic wheat line overexpressing genes for pathogenesis-related (PR) proteins is dependent on salicylic acid concentration. Mol Plant Microbe Interact 16:916–925
Anand A, Lei Z, Sumner LW, Mysore KS, Arakane Y, Bockus WW, Muthukrishnan S (2004) Apoplastic extracts from a transgenic wheat line exhibiting lesion-mimic phenotype have multiple pathogenesis-related proteins that are antifungal. Mol Plant Microbe Interact 17:1306–1317
Andersson MX, Newman MA, Somerville SC, Thordalchristensen H (2008) A lesion-mimic syntaxin double mutant in Arabidopsis reveals novel complexity of pathogen defense signaling. Mol Plant 1:510–527
Arase S, Fujita K, Uehara T, Honda Y, Isota J (2000) Light-enhanced resistance to Magnaporthe grisea infection in the rice Sekiguchi lesion mutants. J Phytopathol 148:197–203
Balagué C, Lin B, Alcon C, Flottes G, Malmström S, Köhler C, Neuhaus G, Pelletier G, Gaymard F, Roby D (2003) HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15:365–379
Borghi M, Rus A, Salt DE (2011) Loss-of-Function of constitutive expresser of pathogenesis related genes5 affects potassium homeostasis in Arabidopsis thaliana. PLoS One 6:4131–4142
Boyd LA, Minchin PN (2001) Wheat mutants showing altered adult plant disease resistance. Euphytica 122:361–368
Boyd LA, Smith PH, Wilson AH, Minchin PN (2002) Mutations in wheat showing altered field resistance to yellow and brown rust. Genome 45:1035–1040
Bremner JM (1965) Inorganic Forms of Nitrogen1. In: Norman AG (ed) Methods of soil analysis. Part 2 chemical and microbiological properties. American Society of Agronomy, Soil Science Society of America, Madison
Brosché M, Blomster T, Salojärvi J, Cui F, Sipari N, Leppälä J, Lamminmäki A, Tomai G, Narayanasamy S, Reddy RA, Keinänen M, Overmyer K, Kangasjärvi J (2014) Transcriptomics and functional genomics of ROS-induced cell death regulation by radical-induced cell death1. PLoS Gesnet 10:e1004112
Bruggeman Q, Raynaud C, Benhamed M, Delarue M (2015) To die or not to die? Lessons from lesion mimic mutants. Front Plant Sci 6:24
Chen J, Vandelle E, Bellin D, Delledonne M (2014) Detection and function of nitric oxide during the hypersensitive response in Arabidopsis thaliana: where there’s a will there’s a way. Nitric Oxide Biol Chem 43:81–88
Fan XR, Gordon-Weeks R, Shen QR, Miller AJ (2006) Glutamine transport and feedback regulation of nitrate reductase activity in barley roots leads to changes in cytosolic nitrate pools. J Exp Bot 57:1333–1340
Fekih R, Tamiru M, Kanzaki H, Abe A, Yoshida K, Kanzaki E, Saitoh H, Takagi H, Natsume S, Undan JR, Undan J, Terauchi R (2015) The rice (Oryza sativa L.) lesion mimic resembling, which encodes an AAA-type ATPase, is implicated in defense response. Mol Genet Genom 290:611–622
Fujiwara T, Maisonneuve S, Isshiki M, Mizutani M, Chen LT, Wong HL, Kawasaki T, Shimamoto K (2010) Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J Biol Chem 285:11308–11313
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Gray J, Janick-Buckner D, Buckner B, Close PS, Johal GS (2002) Light-dependent death of maize lls1 cells is mediated by mature chloroplasts. Plant Physiol 130:1894–1907
Hu G, Yalpani N, Briggs SP, Johal GS (1998) A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10:1095–1105
Ishikawa A, Okamoto H, Iwasaki Y, Asahi T (2001) A deficiency of coproporphyrinogen III oxidase causes lesion formation in Arabidopsis. Plant J 27:89–99
Johnson R (1981) Durable resistance: definition of, genetic control, and attainment in plant breeding. Phytopathology 71:567–568
Kamlofski CA, Antonelli E, Bender C, Jaskelioff M, Danna CH, Ugalde R, Acevedo A (2007) A lesion-mimic mutant of wheat with enhanced resistance to leaf rust. Plant Pathol 56:46–54
Li T, Bai G (2009) Lesion mimic associates with adult plant resistance to leaf rust infection in wheat. Theor Appl Genet 119:13–21
Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, Wang J (2009) SNP detection for massively parallel whole-genome resequencing. Genome Res 19:1124–2132
Li T, Bai GH, Gu SL (2012) A combination of leaf rust resistance gene Lr34 and lesion mimic gene lm significantly enhances adult plant resistance to Puccinia triticina in wheat. Chin Sci Bull 57:2113–2119
Li YS, Chen LC, Mu JY, Zuo JR (2013) Lesion simulating disease1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol 163:1059–1070
Ling T, Vandelle E, Bellin D, Kleinfelder-Fontanesi K, Huang JJ, Chen J, Digby AM, Delledonne M (2012) Nitric oxide produced during the hypersensitive response modulates the plant signaling network and inhibits the pathogen’s virulence machinery. Nitric Oxide Biol Chem 27:S9
Lorrain S, Vailleau F, Balagué C, Roby D (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8:263–271
Lorrain S, Lin B, Auriac MC, Kroj T, Saindrenan P, Nicole M, Balagué C, Roby D (2004) Vascular associated death1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell 16:2217–2232
Mayer KFX, Rogers J, Doležel J, Pozniak C, Eversole K, Feuillet C, Gill B, Friebe B, Lukaszewski AJ, Sourdille P (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788
McGrann GRD, Steed A, Burt C, Nicholson P, Brown JKM (2015) Differential effects of lesion mimic mutants in barley on disease development by facultative pathogens. J Exp Bot 66:3417–3428
Moeder W, Yoshioka K (2008) Lesion mimic mutants: a classical, yet still fundamental approach to study programmed cell death. Plant Signal Behav 3:764–767
Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Neff MM, Neff JD, Chory J, Pepper AE (1998) dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J 14:387–392
Neff MM, Turk E, Kalishman M (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18:613–615
Noutoshi Y, Ito T, Hobo T, Shinozaki K (2003) Analysis of Arabidopsis slh1 (sensitive to low humidity 1) mutant. Plant Cell Physiol 44:S81
Penning BW, Johal GS, McMullen MD (2004) A major suppressor of cell death, slm1, modifies the expression of the maize (Zea mays L.) lesion mimic mutation les23. Genome 47:961–969
Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B (2003) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19:651–652
Poland JA, Brown PJ, Sorrells ME, Jannink JL (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7:251–264
Porebski S, Bailey LG, Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15:8–15
Qian L, Wang YJ (2014) Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J Exp Bot 65:3157–3164
Shirsekar GS, Vega-Sanchez ME, Bordeos A, Baraoidan M, Swisshelm A, Fan JB, Park CH, Leung H, Wang GL (2014) Identification and characterization of suppressor mutants of spl11-mediated cell death in rice. Mol Plant Microbe Interact 27:528–536
Sohn KH, Segonzac C, Rallapalli G, Sarris PF, Woo JY, Williams SJ, Newman TE, Paek KH, Kobe B, Jones JDG (2014) The nuclear immune receptor RPS4 is required for RRS1(SLH1)-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genet 10:e1004655
Soussana JF, Minchin FR, Macduff JH, Raistrick N, Abberton MT, Michaelson-Yeates TPT (2002) A simple model of feedback regulation for nitrate uptake and N-2 fixation in contrasting phenotypes of white clover. Ann Bot 90:139–147
Sugie A, Murai K, Takumi S (2007) Alteration of respiration capacity and transcript accumulation level of alternative oxidase genes in necrosis lines of common wheat. Genes Genet Syst 82:231–239
Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A (2011) Differential expression in RNA-seq: a matter of depth. Genome Res 21:2213–2223
Ueno M, Kihara J, Arase S (2015) Tryptamine and sakuranetin accumulation in Sekiguchi lesions associated with the light-enhanced resistance of the lesion mimic mutant of rice to Magnaporthe oryzae. J Gen Plant Pathol 81:1–4
Wang J, Bayles KW (2013) Programmed cell death in plants: lessons from bacteria? Trends Plant Sci 18:133–139
Wang J, Qu B, Dou S, Li L, Yin DD, Pang ZQ, Zhou ZZ, Tian MM, Liu GZ, Xie Q, Tang DZ, Chen XW, Zhu L (2015) The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity. BMC Plant Biol 15:1–15
Yamaguchi M, Takechi K, Myouga F, Imura S, Sato H, Takio S, Shinozaki K, Takano H (2012) Loss of the plastid envelope protein AtLrgB causes spontaneous chlorotic cell death in Arabidopsis thaliana. Plant Cell Physiol 53:125–134
Yamamura C, Mizutani E, Okada K, Nakagawa H, Fukushima S, Tanaka A, Maeda S, Kamakura T, Yamane H, Takatsuji H, Mori M (2015) Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. Plant J 84:1100–1113
Yao Q, Zhou RH, Fu TH, Wu WR, Zhu ZD, Li AL, Jia JZ (2009) Characterization and mapping of complementary lesion-mimic genes lm1 and lm2 in common wheat. Theor Appl Genet 119:1005–1012
Acknowledgments
We thank the financial support provided by the National Natural Science Foundation of China (31270704), the Vital Project of Natural Science of Universities in Jiangsu Province (12KJA210002), the China Postdoctoral Science Foundation (2014M560449), and by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). Our thanks also go to Dr. Mei Han and Dr. Tao Su for improving English language of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by E. Lagudah.
Xuan Shi and Fei Zheng contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, L., Shi, X., Zheng, F. et al. A novel nitrogen-dependent gene associates with the lesion mimic trait in wheat. Theor Appl Genet 129, 2075–2084 (2016). https://doi.org/10.1007/s00122-016-2758-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2758-3