Abstract
Key message
A novel light intensity-dependent lesion mimic mutant with enhanced disease resistance was physiologically, biochemically, and genetically characterized, and the causative gene was fine mapped to a 1.28 Mbp interval containing 17 high-confidence genes.
Abstract
Lesion mimic mutants are ideal for studying disease resistance and programmed cell death photosynthesis in plants to improve crop yield. In this study, a novel light intensity-dependent lesion mimic mutant (MC21) was obtained from the wheat variety Chuannong16 (CN16) by ethyl methane sulfonate treatment. The mutant initially developed tiny lesion spots on the basal part of the leaves, which then gradually proceeded down to leaf sheaths, stems, shells, and awns at the flowering stage. The major agronomic traits were significantly altered in the mutant compared to that in the wild-type CN16. Furthermore, the mutant exhibited a lesion phenotype with degenerated chloroplast structure, decreased chlorophyll content, increased level of reactive oxygen species, and increased resistance to stripe rust and powdery mildew. Genetic analysis indicated that the lesion phenotype was controlled by a novel single semi-dominant nuclear gene. The target gene was mapped on chromosome arm 2AL located between Kompetitive Allele Specific PCR (KASP) markers, KASP-4211 and KASP-5353, and tentatively termed as lesion mimic 5 (Lm5). The fine mapping suggested that Lm5 was located in a 1.28 Mbp interval between markers KASP-5825 and KASP-9366; 17 high-confidence candidate genes were included in this genomic region. This study provides an important foundational step for further cloning of Lm5 using a map-based approach.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bread wheat (Triticum aestivum L.) is one of the most important staple foods in the world; approximately 20% of human calories are provided by wheat (Chaves et al. 2013). As, 95% of the energy comes from photosynthesis of leaves, this important organ plays a key role in increasing crop yields (Zhai et al. 2002). Lesion mimic mutants, a type of leaf-spot mutant, spontaneously produce cell death or necrotic spots on leaves, leaf sheaths, and other tissues without external pathogen infection, environmental stress, or mechanical damage (Wang et al. 2015, 2016; Lee et al. 2018). The formation of lesion spots may lead to early senescence and may even seriously influence the growth and development of plants (Kumar et al. 2005; Wang et al. 2014).
It has been reported that numerous lesion mimic mutants/genes have been mapped in various plant species (Kamlofski et al. 2007; Li and Bai 2009; Wang et al. 2020), and the formation of lesion spots could enhance plant disease resistance (Wu et al. 2008; Li et al. 2019). For example, the spotted leaf 18 mutant exhibited reddish brown spots on leaves, and SPL18 enhanced rice blast resistance through overexpression of acyltransferase (Mori et al. 2007). OsLSD1, encoding zinc finger protein, could enhance plant defense responses by negatively regulating programmed cell death (PCD) in rice (Wang et al. 2005). Similar to OsLSD1, the initiation of lesion spots indeed enhanced resistance by PCD in other crops, such as Rp1-D in maize (Collins et al. 1999), mlo in barley (Büschges et al. 1997), FZL in Arabidopsis (Landoni et al. 2013), and GmLMM1 in soybean (Wang et al. 2020).
To the best of our knowledge, only seven lesion mimic mutants/genes have been identified or mapped to date (Yao et al. 2009; Wang et al. 2016; Liu et al. 2021). For example, the recessive gene lm was located on chromosome arm 1BL, which explained 60.8% of the phenotypic variance for wheat leaf rust resistance (Li and Bai 2009). In addition, the white spots in the HLP mutant not only had little effect on agronomic traits, but also exhibited strong disease resistance to leaf rust (Kamlofski et al. 2007). The identified recessive loci of lm1 and lm2 were mapped on chromosome arms 3BS and 4BL, respectively, and the lesion mimic phenotype influenced by light was first reported in wheat (Yao et al. 2009). The semi-dominant gene Lm3 mapped on chromosome arm 3BL could enhance wheat resistance to powdery mildew, and the initiation of lesion spots was also closely related to light (Wang et al. 2016). Recent reports have shown that Lm4 mapped on chromosome arm 2DS can enhance the resistance to stripe rust (Liu et al. 2021). Lmpa1, a single dominant gene, was roughly mapped to chromosome arm 5AS (Kong et al. 2020). Although these genes have been reported, none of them has been further fine-mapped or cloned. As such, our knowledge of the molecular function of these Lm genes in wheat is highly limited (Kamlofski et al. 2007; Li and Bai 2009; Liu et al. 2021).
In this study, a genetically stable lesion mimic mutant (MC21) was identified from an ethyl methane sulfonate (EMS)-induced Chuannong16 (CN16, an elite wheat cultivar) mutant bank. The initiation of necrotic lesion spots was induced by intensive light. We investigated agronomic traits, measured physiological indices containing pigment content, photosynthesis indications, enzymatic activity, reactive oxygen species (ROS), observed cell structure, analyzed histochemical indicators of PCD, and evaluated the level of disease resistance of this mutant. Genetic analysis suggested that MC21 was controlled by a single novel semi-dominant nuclear gene (tentatively named Lm5). Lm5 was fine-mapped to a narrow region on the chromosome arm 2AL. These results may facilitate fine mapping and candidate gene screening, and ultimately contribute to the understanding of lesion spot formation in wheat.
Materials and methods
Plant materials
The wheat lesion mimic mutant (MC21) was isolated from an EMS-induced CN16 mutant bank. At the seedling stage, MC21 exhibited lesion spots on the leaves under natural conditions. Five F2 (or F2:3) populations derived from MC21/3642 (M13-1 containing 393 lines, M13-2 containing 260 lines), MC21/Shumai969 (SM969, M19-1 containing 314 lines, M19-2 containing 182 lines), and MC21/CN16 (M16 containing 260 lines) were used for genetic analysis. The F2 (and F2:3) population derived from cross M13-1 was used for gene mapping. The F2 population (3828 individual plants) derived from cross MC21/3642 was also used for fine mapping. CN16 and SM969 are commercial elite varieties and 3642 is an important advanced breeding line.
Phenotypic evaluation
M13-1 and M19-1 F2 populations were grown in Wenjiang (103° 51ʹ E, 30° 43ʹ N) in 2018, and F2:3 families were grown in Chongzhou (103° 38ʹ E, 30° 32ʹ N) in 2019 in Sichuan Province, China. M13-2 and M19-2 F2 populations were grown in Chongzhou (103° 38ʹ E, 30° 32ʹ N) in 2019. The F2 and F2:3 families of M16 were also grown in Wenjiang (103° 51ʹ E, 30° 43ʹ N) in 2019 and 2020, respectively. To maintain genealogy, the plants were individually sowed and harvested in the field. Each line was planted in a single 1.5 m row with 0.3 m between rows, and the sowing density was 15 seeds per row with 0.1 m between plants within a row (Li et al. 2020). All field trials were well irrigated and managed in accordance with the local standard practices.
In the field, a total of 100 plants from CN16 (50 plants) and MC21 (50 plants) were randomly selected to investigate agronomic traits, including effective tiller (ET) using the method reported by Liu et al. (2020), spikelet number per spike (SNS) and flowering period (FP) using the method reported by Ma et al. (2019a), plant height (PH), spike length (SL), and spike extension length (SEL) using the method reported by Li et al. (2020), flag leaf length (FLL) and flag leaf width (FLW) using the method reported by Ma et al. (2020), thousand-grain weight (TGW), grain length (GL), and grain width (GW) using the method reported by Ma et al. (2019c).
Light- and temperature-dependent analyses
To investigate whether MC21 was associated with light, the leaves of wild-type (WT, CN16) and mutant (MC21) were covered with a piece of 2.0 cm-width aluminum foil to confirm whether the initiation of lesion spots was affected by the environment in the natural field conditions as described previously (Wang et al. 2016). To further study the lesion formation of mutants in different environments, wheat seeds (CN16 and MC21) were superficially sterilized with NaClO3 and then cultured in a growth chamber with different photoperiods (day/night 16/8, 12/12, and 8/16; 15,000 lux; 25 °C), light intensity (16/8; 10,000, 6700, and 4500 lux; 25 °C), and temperature (16/8; 3000 lux; 10, 15, and 20 °C), according to previous methods (Wang et al. 2016; Chen et al. 2018).
Measurement of photosynthesis and photosynthetic pigment contents
Photosynthesis plays an important role in plant development and growth. Chlorophyll a (Ch a) and chlorophyll b (Ch b) are of great significance to the normal progress of photosynthesis. Therefore, the contents of Ch a, Ch b, and carotenoid (Car) were measured at the jointing stage, as described by Wellburn (1994). Three biological replicates were used for each sample.
Furthermore, MC21 (the same female parent), CN16, Mianmai 37 (M37), SM969, 3642, and F1 hybrid plants (MC21/CN16, MC21/M37, MC21/SM969, and MC21/3642) were used to measure the relative chlorophyll using MultispeQ from 8:00 am to 10:00 am during the flowering period (Kuhlgert et al. 2016). Three biological replicates and four technical replicates were tested for each sample.
Observation of chloroplast structure
The leaves of CN16 and MC21 were cut into 2 mm × 2 mm pieces and stored at 4 °C in 2.5% glutaraldehyde fixative (the leaves could not float on the surface of the fixed solution) at the jointing stage. Afterward, these samples were sent to Baihui Organisms (Chengdu, China) to observe the cell structure of the normal phenotype (CN16) and lesion phenotype (MC21) using an electron microscope.
Histochemical characterization and physiological indices measurement
Cell death of leaves of WT (CN16) and mutant (MC21) was identified using Trypan blue staining as described by Yin et al. (2000). The samples were immersed in a mixed solution containing 25% lactic acid, 23% water-saturated phenol, 2.5 mg/ml trypan blue, and H2O at 70 °C for 10 min. Afterward, the samples were heated in boiling water for 3 min, immersed in water for 12 h, and then decolorized for 3 days using a 2.5 g/ml chloral hydrate solution.
The H2O2 and O2− contents were measured during the jointing stage. The enzymatic activities of catalase (CAT), superoxide dismutase (SOD), and peroxidase (POD) were measured using anti-oxidative enzyme detection kits (Jiancheng Biotechnology, Nanjing, China) at the same stage. The methods were performed according to the manufacturer’s instructions. Three biological replicates were tested per sample, and the means from three measurements were used for further analysis.
Evaluation of disease resistance
M37, MC21, and F1 (MC21/M37) plants planted in two rows according to the above sowing method were used to assess the stripe rust resistance at the adult stage during the crop season of 2019–2020 in the field. Inoculation and assessment of the stripe rust reaction were adopted from the method reported by Ma et al. (2019b). Briefly, all plants were inoculated with a urediniospore mixture of the predominant Puccinia striiformis Westend. f. sp. tritici Eriks. (Pst) races, including Chinese yellow rust (CYR) 32 (Hybrid46-3 or HY-3), CYR33 (Su11-14-3), CYR34 (V26/Gui22-9), V26/Gui22-14, Sull-4, and Sull-5 at the jointing stage (Ma et al. 2019b; Long et al. 2021; Wu et al. 2021). All Pst races were provided by the Gansu Institute of Plant Protection (Lanzhou, China).
Furthermore, to assess the stripe rust reactions of MC21 and CN16 at the seedling stage, inoculation was carried out under growth chamber conditions with the same Pst races using a previously described standard protocol (Ma et al. 2019b). In addition, CN16 and MC21 were inoculated with both powdery mildew and Fusarium head blight (FHB) in the growth chamber using the same races and methods as described by Chen et al. (2019) and Zhang et al. (2019), respectively. Three biological replicates were used for each sample.
RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
The relative expression of defense-related genes was further analyzed between CN16 and MC21 (Table S1). According to the manufacturer’s instructions, total RNA was extracted from the leaves using RNA simple Total RNA Kit (Tiangen Biotechnology, Beijing, China) at the jointing stage. Subsequently, 4 µl RNA was used to synthesize first-strand cDNA using PrimeScript™ RT reagent Kit (Takara Biotechnology, Dalian, China). Quantitative RT-PCR was performed in a total volume of 10 µL reaction buffer containing 2.5 µL cDNA template (300 ng), 5 µL SYBR Green Premix pro Taq (Accurate Biotechnology, Hunan, China), 0.5 µL former primer (10 µmol/µL), 0.5 µL reverse primer (10 µmol/µL) and 1.5 µL DNase/RNase-free water on a CFX96™ Real-Time System (Bio-Rad Laboratories, Inc., Hercules, USA) at 95 °C pre-denaturation for 10 min, followed by 95 °C (10 s), 60 °C (30 s), 72 °C (30 s) for 40 cycles, with a final dissociation at 95 °C (15 s), 60 °C (30 s), and 95 °C (15 s). Ta2776 (RNase L inhibitor-like protein) was selected as the reference gene (Paolacci et al. 2009) and the relative quantification formula (2−△△Ct) ± standard error of the mean (SEM) was used to further evaluate quantitative variation. Three biological replicates and four technical replicates were tested for each sample.
Genetic analysis and gene mapping
The wheat 660K single nucleotide polymorphism (SNP) array was used to genotype CN16 and MC21 to reveal their genetic background relationship (only homozygous SNPs were retained) according to a previous method (Liu et al. 2020). F1 leaves from the reciprocal cross between MC21 and SM969 were observed at the heading stage in the natural field. Five populations (M13-1, M13-2, M19-1, M19-2, and M16) were used for genetic analysis, and each F2 individual phenotype was deduced from its F3 progeny. We then performed a chi-square test (χ2) to test phenotypic data (lesion spots) for a goodness-of-fit to the ratio of 3:1 (or 1:2:1) expected for a single gene (or semi-dominant) genetic basis.
Furthermore, an F2:3 family developed from the cross between MC21 and 3642 was used to map the target gene. Leaves of 30 green and 30 lesion plants from the M13-1 F2 population during the jointing stage were collected. Before RNA extraction, the leaves were grouped into two bulks comprising green and lesion bulks. RNA extraction, library construction, and sequencing of the two bulks were performed by Novogene Company (Beijing, China). BioKanga (version 2.76.2) was used to analyze SNPs between the two bulks, as described by Ma et al. (2014). The SNPs were detected after aligning the clean reads against the high-confidence and low-confidence Chinese Spring (CS) genes (IWGSC 2018), allowing a maximum of two mismatches. Subsequently, SNPs were identified using the Biokanga subprocess with a minimum score of 85%, that is, the percentage of a given nucleotide at the SNP position in each sample was at least 85%. The SNPs distributed on each chromosome were scored further. The cetyltrimethylammonium bromide method was used to extract genomic DNA (Murray and Thompson 1980). Simple sequence repeat (SSR) markers were obtained from the Triticeae Research Institute of Sichuan Agricultural University. SNPs between 3642 and MC21 identified from BSR-Seq data were further converted into Kompetitive Allele Specific PCR (KASP) markers, as described previously (Li et al. 2020). Finally, we used JoinMap 4.0, (Kyazma BV, Wageningen, The Netherlands, https://www.kyazma.nl/index.php/JoinMap/) to construct a genetic map combining the phenotype and genotype data.
Differentially expressed genes analysis
Leaves of MC21 and CN16 during the jointing stage were collected. RNA extraction, library construction, and RNA sequencing were performed as described previously. Differentially expressed gene (DEG) analysis was adopted from Zou et al. (2020). Briefly, clean reads were aligned against high- and low-confidence Chinese Spring (CS) genes from IWGSC (2018) to estimate the gene expression level using the fragments per kilobase of transcript per million fragments mapped method. Differential expression analysis between MC21 and CN16 was performed using the DESeq fit. DEGs were determined with a false discovery rate (FDR) threshold < 0.05, and an absolute value of log2FoldChange > 1 or “inf” (the value of one comparative object was zero and the other was not). To confirm the putative biological functions and biochemical pathways of DEGs (CN16 and MC21), Blast2GO was used for Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (Conesa et al. 2005).
Isolation and analysis of candidate gene
RNA extraction and cDNA synthesis were performed as described previously. Many studies have focused on the role of lesion-mimicking genes in wheat by orthologous cloning. TraesCS2A02G426900 is orthologous to the rice RLIN1 (LOC_Os04g52130, Table S2) (Sun et al. 2011). To isolate the orthologous gene of TraesCS2A02G426900, the CDS was cloned by PCR using the corresponding specific primers on chromosome 2A. The Primer Premier software (version 5.0; Premier Biosoft International, CA, USA, http://www.premierbiosoft.com/) was used to design primers for these potential candidate genes in this study (Table S1). The PCR was performed in a total volume of 50 µL reaction buffer containing 4 µL cDNA template, 25 µL 112 Mix (Vazyme, Nanjing, China), 3 µL former primer (10 µmol/µL), 3 µL reverse primer (10 µmol/µL), and DNase/RNase-free water up to the final value. A Veriti™ 96-Well Thermal Cycler was used for gene magnification. The PCR reaction procedure was as follows: pre-denaturation at 95 °C for 5 min, followed by 95 °C for 45 s, 55–60 °C for 45 s, and 72 °C for 60 s for 40 cycles, and finally preserved at 15 °C. A gel extraction kit (Omega Bio-Tek, Inc., GA, USA), PMD™ 19-T Vector Cloning Kit (Takara Biotechnology, Dalian, China), and pEASE-T1 Cloning Kit (Transgen, Beijing, China) were used for gene cloning, and all methods were performed according to the manufacturer’s instructions. At least four colonies per cloned PCR amplicon were sent to Tsingke Biotech Co., Ltd. for Sanger sequencing.
Statistical analysis
IBM SPSS Statistics 26 (SPSS Inc., Chicago, IL, USA) was used to perform Student’s t test (P < 0.05) and correlation analysis. A Chi-square test (χ2) in Excel 2016 was used for genetic analysis.
Results
Lesion mimic phenotype and its effects on agronomic traits
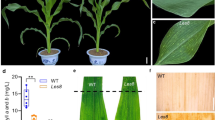
Under field conditions, lesion spots appeared first on the tip of the basal leaf (Fig. 1a). Then, both the size and number of lesion spots increased with the growth and development of plants from the tillering stage to the flowering stage in the mutant MC21 (Fig. 1a). Finally, these lesion spots also initiated on the leaf sheaths, stems, shells, and awns in mutant MC21 at the flowering stage (Fig. 1b). Compared with that in the WT CN16, PH, SL, SEL, TGW, GL, GW, ET, and FLW were significantly decreased, whereas FP and FLL were significantly increased in the mutant MC21 (Fig. 1c). No significant differences in SNS were detected between CN16 and MC21 (Fig. 1c).
Phenotypic characteristics of lesion mimic mutant MC21 and wild-type CN16. a The leaf phenotype changes of CN16 and MC21 at different development stages; b the lesion spots on awn, shell, stem, and sheath of CN16 and MC21, respectively; Scale bar = 1 cm; c The performance of agronomic traits in wild-type CN16 and mutant MC21; plant height (PH), spike length (SL), spikelet number per spike (SNS), flowering period (FP), thousand-grain weight (TGW), spike extension length (SEL), flag leaf length (FLL), flag leaf width (FLW), effective tiller (ET), grain length (GL) and grain width (GW). NS significant at no difference, *Significant at P < 0.05, **Significant at P < 0.01
Light activates lesion mimic phenotype
The newly emerged leaves from CN16 and MC21 (without lesion spots) were covered with aluminum foil to determine the effect of light on lesion spot formation under natural conditions. The results showed that the shaded leaf surface of mutant MC21 gradually became white without lesion spots similar to CN16, whereas the unshaded area had numerous lesion spots after 20-day shading treatment (Fig. 2a). Subsequently, the aluminum foil was reinstated, the light was shaded, a large number of lesion spots appeared in the unshaded area and a few necrotic spots appeared in the shaded leaf area (Fig. 2b). No necrotic lesion spots were observed before or after the shading treatment in CN16 (Fig. 2a).
Lesion spot formation, photosynthetic pigment content, and chloroplast structure observation of the mutant MC21. a The formation of lesion spots on leaves was influenced by light under the natural conditions. Null represents the leaf of CN16 or MC21 under the natural field. b The leaf was shaded after the formation of lesion spots under the natural condition. c The photosynthetic pigment contents of CN16 and MC21 under the low light condition (−) and natural condition (+), respectively, at the jointing stage. Ch a: chlorophyll a; Ch b: chlorophyll b; Car: carotenoid, Error bars means + SD. Observation of the leaf cell structure (d) and chloroplast structure (e) between wild-type CN16 and mutant MC21; Scale bar = 10 μm; e and g were; Scale bar = 1 μm; CM: cytoplasmic matrix; CP: chloroplast; CN: cell nucleus; CW: cell wall; SG: starch granule; T: thylakoids; P: plastoglobulus; ChM: chloroplast membrane; STL: stroma lamella; GRL: grana lamella; NS significant at no difference, **Significant at P < 0.01, *Significant at P < 0.05. Differences between the two groups were labeled above the significant levels
To further determine the effects of light and temperature on lesion spot formation, various photoperiod, light intensity, and temperature treatments were applied to the mutant (MC21) and the WT (CN16) in the growth chamber. The results showed that lesion spots were clearly observed under different photoperiod treatments at 25 °C with a light intensity of 15,000 lux (Fig. S1a, b, c). No lesion spots were observed on the mutant MC21 leaves under low light intensity (< 6700 lux), whereas the lesion spots gradually became severe with the increase in light intensity at 25 °C and 16/8 photoperiod (Fig. S1d, e, f). No lesions were observed on the mutant leaves with different temperature treatments under the 16/8 photoperiod and light intensity of 3000 lux (Fig. S1g, h, i). These results suggest that the initiation of lesion spots is light-intensity-dependent in mutant MC21.
Photosynthesis indications and chlorophyll contents
At the jointing stage, the levels of Ch a, Ch b, and Car were not significantly different between MC21 and CN16 under low light intensity (Fig. 2c, upper), whereas those of Ch a and Ch b were significantly different under the natural field conditions. Compared with those in CN16, Ch a and Ch b levels were significantly decreased by 56.21 and 57.14%, respectively, in MC21 under natural field conditions (Fig. 2c, lower).
Relative chlorophyll is an important indicator of photosynthesis (Kuhlgert et al. 2016). To assess the effect of lesion spots on photosynthesis, four cross combinations (MC21/CN16, MC21/SM969, MC21/M37, and MC21/3642) were used to measure relative chlorophyll content during the flowering period (Fig. S2a). Compared with that in CN16, SM969, M37, and 3642, the relative chlorophyll content of mutant MC21 was significantly decreased by 58.04, 58.56, 52.51, and 52.84%, respectively. In addition, as expected, the heterozygotes had significantly lower (ranging from 18.67 to 40.78% with an average of 27.71%) but higher (ranging from 29.14 to 43.75% with an average of 37.89%) relative chlorophyll content than their respective wild types (CN16, SM969, M37, and 3642) and MC21 (Fig. S2a).
Photosynthesis non-photochemical exciton quenching (phiNPQ) and photosynthesis non-regulatory energy dissipation (phiNO) play key roles in the assessment of photoprotection in plants. In the mutant MC21, phiNPQ was greatly increased by 122.00 and 50.78%, whereas phiNO was significantly decreased by 37.37 and 20.60% compared with CN16 and F1 (MC21/CN16), respectively (Fig. S2b). The heterozygotes again had values that fell between CN16 and MC21, suggesting that the formation of lesion spots in the mutant had a negative effect on photosynthesis and chlorophyll content.
Ultra-structure analysis of cell and chloroplast
The decreased chlorophyll content might be related to the number of chloroplasts in the leaves. To determine the effect of chloroplast structure on lesion initiation in leaves, we observed the leaves of CN16 and mutant MC21 at the jointing stage. In the leaves of CN16, a large number of chloroplasts were regularly arranged around the cytomembrane (Fig. 2d, upper), and starch grains were clearly observed (Fig. 2e, upper). Compared with that in CN16, the number of chloroplasts was obviously decreased in mutant MC21 (Fig. 2d, lower). Furthermore, the granum-thylakoids were completely exposed to the cell matrix as the chloroplast membranes were damaged (Fig. 2e, lower); the number of plastoglobulus also increased with the destruction of chloroplasts and the increased number of vesicles in the mutant MC21 (Fig. 2e, lower).
PCD with H2O2 and O2 − accumulation
Trypan blue staining was performed to determine whether cell death occurred around the lesion spots. The result showed that numerous dark blue stains were observed around the lesion spots in MC21 but not in CN16, indicating that cell death indeed occurred with the formation of lesion spots in MC21 (Fig. 3a).
Analysis of cell death and reactive oxygen species (ROS)-associated parameters between CN16 and MC21. a Typan blue staining of MC21 and CN16; Null represents the leaf of CN16 or MC21 under the natural field; − and + represent the leaf of CN16 or MC21 was shaded before and after the lesion spot formation under the natural conditions, respectively; b The enzymatic activities of peroxidase (POD), superoxide dismutase (SOD) and catalase (CAT) in the leaves of CN16 and MC21 at jointing stage. c The content of O2− and H2O2 for CN16 and MC21 leaves at jointing stage; *Significant at P < 0.05, **Significant at P < 0.01. Differences between the two groups were labeled above the significant levels
The enzymatic activities (CAT, SOD, and POD) and the content of ROS (H2O2 and O2−) were measured to further confirm whether cell death was associated with the burst of ROS in the mutant MC21. Compared with those in WT CN16, the activities of POD (− 15.11%) and SOD (− 41.63%) were significantly decreased, whereas the activity of CAT (68.82%) was significantly increased after the formation of lesion spots in the mutant MC21 (Fig. 3b). Furthermore, the content of O2− (− 36.72%) was significantly decreased; however, the content of H2O2 (25.07%) was significantly increased after the lesion spots emerged in mutant leaves, compared with that in CN16 (Fig. 3c). The results showed that the ROS scavenging system was severely impaired, and the excessive accumulation of ROS, especially H2O2, was associated with cell death.
Formation of lesion spots enhanced disease resistance
The MC21 and CN16 reactions to stripe rust, powdery mildew, and FHB infection were evaluated in a growth chamber, and the results showed that CN16 was susceptible to powdery mildew (Fig. 4a, left), stripe rust (Fig. 4b, left), and FHB (Fig. 4c, left). Under conditions with strong light intensity, MC21 exhibited significantly enhanced resistance to powdery mildew (Fig. 4a, right) and stripe rust (Fig. 4b, right) and was susceptible to FHB after inoculation with Fusarium graminearum (Fg, 1 × 103) for 20 days (Fig. 4c, right). However, both MC21 (without lesion spots) and CN16 were susceptible to stripe rust (Fig. S3a, b), and powdery mildew (Fig. S3c) under low-light intensity conditions. Moreover, MC21, CN16, M37, and F1 of MC21/M37 were inoculated by the leaf-clipping method at the jointing stage under natural field conditions. It was clearly shown that MC21 had a good level of resistance to stripe rust (Fig. S3a, b). Therefore, CN16 was susceptible to all races (stripe rust, powdery mildew, and FHB), whereas the mutant MC21 (with lesion spots) showed significantly enhanced resistance to stripe rust and powdery mildew, and the resistance level to FHB was similar between MC21 and CN16.
The evaluation of disease resistance for CN16 and MC21 at growth chamber. The evaluation of powdery mildew resistance (a), stripe rust resistance (b) and Fusarium head blight resistance (c) for CN16 and MC21 under the growth chamber condition, respectively; − and + represent the leaf of CN16 or MC21 was uninoculated and inoculated pathogenic bacteria under the growth chamber, respectively. The evaluation of Fusarium head blight resistance for CK, 10, 15, and 20 days between CN16 and MC21, respectively
Furthermore, the expression of defense genes was also analyzed. Compared with that in CN16, all defense response genes were significantly induced in the mutant MC21 (with and without lesion spots) at the jointing stage (Fig. 5), indicating that the enhanced level of disease resistance was associated with the expression of defense response genes.
Expression analysis of defense-related genes for CN16 and MC21 at the jointing stage. + and − represent the leaves of MC21 mutant with and without lesion spots formation; The error bars represent the standard deviation (SD) between biological replicates. Mean fold-changes in the transcript abundance calculated using the 2−△△Ct method between biological replicates ± standard error of the mean (SEMs)
Genetic analysis and mapping of Lm5
The Wheat660K SNP array was used to validate the level of isogenicity between CN16 and MC21. After removing the unreliable markers, only 13,411 of 531,809 (~ 2.52%) markers were polymorphic between CN16 and MC21, suggesting that MC21 has a similar genetic background to CN16 and is indeed mutated from CN16 (Fig. 6a). To further confirm the genetic control of the lesion phenotype, MC21 was crossed with 3642, CN16, and SM969. All F1 plants derived from SM969/MC21 and MC21/SM969 showed lesion phenotypes with lesion spots, suggesting that the target locus was controlled by a nuclear gene (Fig. S4a). In addition, F2 population individuals derived from MC21/3642 and MC21/SM969 were segregated into two categories: WT-like and mutant-like. The ratio of these two types fitted to a 3:1 (χ2 M19-1 = 0.72 < χ20.05 = 3.84, χ2 M19-2 = 0.07 < χ20.05 = 3.84, χ2 M13-1 = 0.31 < χ20.05 = 3.84, χ2 M13-2 = 0.74 < χ20.05 = 3.84, Table 1). Afterwards, the F2:3 family derived from F2 individual plants was further segregated into three categories: WT-like type, intermediate type (similar to F1), and mutant-like type. The ratio of these three types fitted to a 1:2:1 (χ2 M19-1 = 7.84 < χ20.01 = 9.21, χ2 M13-1 = 6.99 < χ20.01 = 9.21, χ2 M16 = 3.16 < χ20.01 = 9.21, Table 1). Therefore, we concluded that the lesion phenotype was governed by a single semi-dominant nuclear gene.
The genetic background analysis, BSR-Seq analysis, and rough mapping in mutant MC21. a The analysis of genetic background for CN16 and MC21 using wheat660K SNP array; the pointer represents the same rate (%) between CN16 and MC21; green color: the same SNPs; red color: the different SNPs; The credible SNPs (homozygous SNPs including AA, TT, GG, CC) from wheat660K SNP array were used to analyze. b SNPs distribution on all chromosome and 2A chromosome using BSR-Seq analysis for MC21/3642 population, respectively. c The rough genetic map and physical map of Lm5 gene on chromosome 2A
The F2:3 population derived from a cross between MC21 and 3642 was used to map the target gene using BSR-Seq analysis (Fig. S4b, c, d). The results showed that the target gene was located on chromosome 2A (Fig. 6b). SSR markers and KASP markers (Table S1) were used to map the lesion spot phenotype. Finally, the target gene was mapped between KASP markers KASP-4211 and KASP-5353, with a genetic distance of 4.3 cM and a physical distance of 23.23 Mbp (Fig. 6c). The target gene was tentatively termed lesion mimic 5 (Lm5).
Fine mapping of Lm5 candidate gene
To further fine-map and narrow down the genomic interval of the Lm5 locus, 3828 F2 individual plants derived from MC21/3642, and nine newly developed KASP markers (KASP-5825, KASP-4901, KASP-2094, KASP-7690, KASP-9366, KASP-4685, KASP-9960, KASP-7347, and KASP-3542, Table S1) were used. Finally, thirty-eight important recombinants were identified and classified into 17 categories (L1-L17). Based on their genotype and phenotype data, Lm5 was delimited to a 1.28 Mbp interval between markers KASP-5825 and KASP-9366 (Fig. 7).
Fine mapping of Lm5 gene. a Genomic location of Lm5 on chromosome arm 2AL in wheat. Black line represents the rough mapping interval of Lm5. b Physical positions of newly development nine molecular markers used for fine mapping of Lm5 gene. c Recombinant types determined by genotypes using newly development molecular markers and phenotype using the F2 plants deduced by the F2:3 families. White, black and gray segments represent regions homozygous for 3642, regions homozygous for MC21, and heterozygous regions, respectively
Furthermore, based on fine mapping result, 46 predicted genes (including 17 high-confidence genes and 29 low-confidence genes, Table S3) were searched in this physical interval of “CS” wheat reference genome (IWGSC 2018; Zhu et al. 2021).
Previous research suggested that RLIN1 (LOC_Os04g52130) is a lesion mimic gene encoding a putative coproporphyrinogen III oxidase in the tetrapyrrole biosynthesis pathway in rice (Sun et al. 2011). BLAST results showed that TraesCS2A02G426900 is an orthologous gene with RLIN1 (Table S2). The orthologous gene (TraesCS2A02G426900) was isolated from wheat, and alignment results of CDS showed that no sequence difference was detected between CN16 and MC21 (Fig. S5). Therefore, these results suggest that TraesCS2A02G426900 is not likely the candidate gene of Lm5, and the fine mapping result further confirmed that it was not located in the fine-mapped interval (Fig. 7).
In addition, DEGs obtained from RNA-Seq (MC21/CN16) analysis were used to screen for potential candidate genes. Both the downregulated and upregulated (Fig. 8) DEGs were enriched in metabolic pathways, biosynthesis of secondary metabolites, and biosynthesis of amino acids (Table S4). Of the 9096 DEGs, 509 were distributed on chromosome 2A with TraesCS2A02G421500 and TraesCS2A02G422100 encoding cysteine proteases mapped in the fine-mapping interval (Table S3, Table S6).
Discussion
In this study, we identified a novel light-dependent and semi-dominant lesion that mimicked mutant MC21. The size and number of lesion spots significantly increased under strong light intensity conditions with chloroplast degeneration and reduced chlorophyll content in leaves. The formation of lesion spots was accompanied by the accumulation of ROS, and the excessive accumulation of ROS may lead to cell death and enhance disease resistance, ultimately affecting agronomic traits. Finally, the target gene (Lm5) controlling the formation of lesion spots was fine-mapped in a 1.28 Mbp interval between KASP-5825 and KASP-9366.
Light mediated lesion phenotype
Our results suggest that light intensity plays an essential role in the formation of lesion spots, whereas photoperiod and temperature may be unnecessary for the formation of lesion spots in mutant MC21. Numerous lesion spots were induced by various environmental factors, but light remains an important factor in the induction of lesion spots. For example, the mutant spl24 (Chen et al. 2018), spl5 (Chen et al. 2012) and spl40 (Sathe et al. 2019) were light-dependent in rice, and the mutant Lm3 (Wang et al. 2016) induced by light and photoperiod in wheat.
Light is an important environmental factor that affects photosynthesis, and photorespiration is induced in leaves under strong light conditions (Wu et al. 1991; Takagi et al. 2019). Photorespiration can be defined as a process of O2 uptake and CO2 release that occurs in photosynthetic tissues, such as chloroplasts (Wu et al. 1991). Wheat, as a C3 plant, has strong photorespiration (Kozaki and Takeba 1996; Busch et al. 2013). Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is an important enzyme that plays an important role in photorespiration and CO2 assimilation (Law and Steven 1999; Portis and Parry 2007). Under strong light intensity conditions, the stomata were closed on the surface of the leaves. The content of CO2 is decreased, and the content of O2 is increased in the chloroplast, resulting in an increase in the oxidation activity of Rubisco. This may lead to the production of a large amount of ROS (H2O2) in the chloroplasts (Voss et al. 2013). Thus, photorespiration plays a major role in the readjustment of redox homeostasis (Busch 2020). Strong photorespiration may be an important reason for chloroplast structure disintegration, and a decrease in chlorophyll content and photosynthetic capacity. Altogether, despite the fact that the specific pathway responsible for this phenomenon is unclear, it is certain that light intensity acts as a trigger for the lesion spots formation of MC21 in this study.
Accumulation of ROS and enhanced disease resistance
ROS production is usually associated with cell death in plants (Gregorio and Mullineaux. 2010). The formation of lesion spots was similar to the hypersensitive response (HR) lesions caused by pathogen attack, and the most prominent feature of the necrotic lesion spots was the burst of ROS in mutants (Sun et al. 2014; Chen et al. 2018; Sathe et al. 2019). O2− is disproportionated to H2O2 under the action of superoxide dismutase (Wu et al. 1991), and the photosensitive reaction and the unnormal photorespiration may be another important reason for the production of ROS (Vellosillo et al. 2010). The excessive accumulation of ROS, especially H2O2, can lead to peroxidation of membrane lipids and destruction of the cell membrane, resulting in cell death (Hara Nishimura and Hatsugai 2011; Abdelaal et al. 2018).
It is widely accepted that the formation of lesion spots might lead to activation of defense responses and enhance resistance (Büschges et al. 1997; Kamlofski et al. 2007; Sathe et al. 2019). Lesion spot formation can prevent pathogens from infecting nearby cells, and ultimately enhance the level of disease resistance in plants (Büschges et al. 1997; Yin et al. 2000; Kamlofski et al. 2007; Wang et al. 2016; Sathe et al. 2019). The accumulation of ROS has been reported to be closely associated with disease resistance gene expression in lesion mimic mutants (Lorrain et al. 2003), such as OsPti1a (Takahashi et al. 2007) and OsSSI2 (Jiang et al. 2009) in rice, Rp1-D (Collins et al. 1999) in maize, Mlo (Büschges et al. 1997) in barley, Lm3 (Wang et al. 2016), and Lm4 (Liu et al. 2021) in wheat. Similarly, the MC21 mutant showed resistance to stripe rust and powdery mildew under natural conditions. However, the mutant was susceptible to stripe rust (Fig. S5c, d), and powdery mildew (Fig. S5 e, f) under low-light conditions. The mutant was also expectedly susceptible to FHB under growth chamber conditions, due to the absence of lesion spots in the awns and shells. We speculated that the light conditions did not satisfy the requirement of lesion spot formation, and thus the ROS did not accumulate in the awns and shells. These results further indicated that only tissues or organs of MC21 with formation of lesion spots and accumulation of ROS can enhance disease resistance to pathogens.
Furthermore, ROS also communicate with other signaling molecules and pathways, forming a network that can orchestrate downstream responses. For example, ROS (H2O2) also acts as a second messenger for the induction of defense genes, such as tomato (Orozco-Cárdenas et al. 2001), Arabidopsis (Munemasa et al. 2007), and maize (Zong et al. 2009). Therefore, ROS may upregulate the expression of defense-related genes, disrupt the bacterial membrane structure, and prevent pathogen infection in the mutant.
New type of lesion mimic gene (Lm5) affected agronomic traits in wheat
In this study, we confirmed a novel lesion mimic mutant MC21, and the target gene controlling lesion spots was finely located to a 1.28 Mbp genomic interval on the long arm of chromosome 2A. Compared with previous studies, lm (Li and Bai 2009), lm1 (Yao et al. 2009), and lm2 (Yao et al. 2009) were mapped on chromosome arms 1BL, 3BS, and 4BL, respectively, in the present study and all of them were controlled by recessive genes. A few lesion-mimicking mutants were reportedly controlled by semi-dominant genes in wheat. Lm3 mapped on chromosome arm 3BL (Wang et al. 2016) and Lm4 mapped on chromosome arm 2DS (Liu et al. 2021). To our knowledge, no semi-dominant genes related to lesion spots have been mapped to chromosome 2A. Therefore, Lm5 is a novel gene that controls the formation of lesion spots in wheat.
According to previous studies, lesion spot symptoms often affect agronomic traits in rice (Mori et al. 2007; Wu et al. 2008; Chen et al. 2018) and wheat (Wang et al. 2016). Similarly, the decrease in photosynthetic area caused by lesion spots decreased photosynthesis (Kirchhoff 2019; Woodson 2019), which resulted in a significant decrease in the majority of agronomic traits in the mutant MC21.
Candidate genes for Lm5
Based on fine mapping, we explored a narrowed interval, and 17 high-confidence genes were predicted based on this genomic region. Among the 17 high-confidence genes, five important proteins were likely associated with lesion spots, given that these encoded proteins have been reported to be involved in the formation of lesion spots in rice. These genes encode the V-type ATP synthase alpha chain (TraesCS2A01G421200), cysteine protease (TraesCS2A01G421500, TraesCS2A01G421600, TraesCS2A01G421700, TraesCS2A01G421800, TraesCS2A01G421900, and TraesCS2A01G422100), GRF zinc finger family protein (TraesCS2A01G422200), cytochrome P450 (TraesCS2A01G422300), and dehydrogenase (TraesCS2A01G422500) (Sunkar et al. 2003; Wang et al. 2005; Fujiwara et al. 2010; Cheng et al. 2011; Fekih et al. 2015; Cai et al. 2018; Zhang et al. 2020).
Transcriptome analysis further suggested that two DEGs (TraesCS2A02G421500 and TraesCS2A02G422100) encoding cysteine proteases were located in the fine mapping interval. Cysteine acts as a virulence factor which triggers cell death in the leaves of different Nicotiana species (Zhang et al. 2020). Cysteine protease and carboxylesterase enzymes may act in a synergistic manner during premature degeneration of the tapetal cell layer in tobacco (Shukla et al. 2019). Moreover, the cysteine protease CEP1 is directly involved in the clearance of cellular content during PCD and regulates secondary wall thickening during xylem development in Arabidopsis (Han et al. 2019). Altogether, these genes are important candidates for fine mapping and cloning work.
References
Abdelaal KA, Hafez YM, El-Afry MM, Tantawy DS, Alshaal T (2018) Effect of some osmoregulators on photosynthesis, lipid peroxidation, antioxidative capacity, and productivity of barley (Hordeum vulgare L.) under water deficit stress. Environ Sci Pollut Res 25:30199–30211
Busch FA (2020) Photorespiration in the context of Rubisco biochemistry, CO2 diffusion and metabolism. Plant J 101:919–939
Busch FA, Sage TL, Cousins AB, Sage RF (2013) C3 plants enhance rates of photosynthesis by reassimilating photorespired and respired CO2. Plant Cell Environ 36:200–212
Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, Töpsch S, Vos P, Salamini F, P S-L, (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88:695–705
Cai Q, Liang C, Wang S, Hou Y, Gao L, Liu L, He W, Ma W, Mo B, Chen X (2018) The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat Commun 9:1–14
Chaves MS, Martinelli JA, Wesp-Guterres C, Graichen FAS, Brammer SP, Scagliusi SM, da Silva PR, Wiethölter P, Torres GAM, Lau EY, Luciano C, Ana LSC (2013) The importance for food security of maintaining rust resistance in wheat. Food Secur 5:157–176
Chen X, Hao L, Pan J, Zheng X, Jiang G, Jin Y, Gu Z, Qian Q, Zhai W, Ma B (2012) SPL5, a cell death and defense-related gene, encodes a putative splicing factor 3b subunit 3 (SF3b3) in rice. Mol Breed 30:939–949
Chen Z, Chen T, Sathe AP, He Y, Zhang XB, Wu JL (2018) Identification of a Novel Semi-Dominant Spotted-Leaf Mutant with Enhanced Resistance to Xanthomonas oryzae pv. oryzae in Rice. Int J Mol Sci 19:3766
Chen F, Jia H, Zhang X, Qiao L, Li X, Zheng J, Guo H, Powers C, Yan L, Chang Z (2019) Positional cloning of PmCH1357 reveals the origin and allelic variation of the Pm2 gene for powdery mildew resistance in wheat. Crop J 007:771–783
Cheng Z, Sun L, Qi T, Zhang B, Peng W, Liu Y, Xie D (2011) The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol Plant 4:279–288
Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, Hulbert S, Pryor T (1999) Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11:1365–1376
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics (oxford, England) 21:3674–3676
Fekih R, Tamiru M, Kanzaki H, Abe A, Yoshida K, Kanzaki E, Saitoh H, Takagi H, Natsume S, Undan JR, Undan J, Terauchi R (2015) The rice (Oryza sativa L.) LESION MIMIC RESEMBLING, which encodes an AAA-type ATPase, is implicated in defense response. Mol Genet Genomics 290:611–622
Fujiwara T, Maisonneuve S, Isshiki M, Mizutani M, Chen L, Wong HL, Kawasaki T, Shimamoto K (2010) Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J Biol Chem 285:11308–11313
Gregorio GV, Mullineaux PM (2010) The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol Plant 138:430–439
Han J, Li H, Yin B, Zhang Y, Liu Y, Cheng Z, Liu D, Lu H (2019) The papain-like cysteine protease CEP1 is involved in programmed cell death and secondary wall thickening during xylem development in Arabidopsis. J Exp Bot 70:205–215
Hara Nishimura I, Hatsugai N (2011) The role of vacuole in plant cell death. Cell Death Differ 18:1298–1304
IWGSC (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191
Jiang C, Shimono M, Maeda S, Inoue H, Mori M, Hasegawa M, Sugano S, Takatsuji H (2009) Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol Plant Microbe Interact 22:820–829
Kamlofski C, Antonelli E, Bender C, Jaskelioff M, Danna C, Ugalde R, Acevedo A (2007) A lesion-mimic mutant of wheat with enhanced resistance to leaf rust. Plant Pathol 56:46–54
Kirchhoff H (2019) Chloroplast ultrastructure in plants. New Phytol 223:565–574
Kong W, Wang L, Cao P, Li X, Ji J, Dong P, Yan X, Wang C, Wang H, Sun J (2020) Identification and genetic analysis of EMS-mutagenized wheat mutants conferring lesion-mimic premature aging. BMC Genet 21:88
Kozaki A, Takeba G (1996) Photorespiration protects C3 plants from photooxidation. Nature 384:557–560
Kuhlgert S, Austic G, Zegarac R, Osei-Bonsu I, Hoh D, Chilvers MI, Roth MG, Bi K, TerAvest D, Weebadde P, Kramer DM (2016) MultispeQ Beta: a tool for large-scale plant phenotyping connected to the open PhotosynQ network. R Soc Open Sci 3:160592
Kumar Y, Mishra S, Tyagi M, Singh S, Sharma B (2005) Linkage between genes for leaf colour, plant pubescence, number of leaflets and plant height in lentil (Lens culinaris Medik.). Euphytica 145:41–48
Landoni M, De Francesco A, Bellatti S, Delledonne M, Ferrarini A, Venturini L, Pilu R, Bononi M, Tonelli C (2013) A mutation in the FZL gene of Arabidopsis causing alteration in chloroplast morphology results in a lesion mimic phenotype. J Exp Bot 64:4313–4328
Law RD, Steven JC-B (1999) Inhibition and acclimation of photosynthesis to heat stress is closely correlated with activation of ribulose-1, 5-bisphosphate carboxylase/oxygenase. Plant Physiol 120:173–182
Lee D, Lee G, Kim B, Jang S, Lee Y, Yu Y, Seo J, Kim S, Lee Y-H, Lee J, Lee J, Kim S, Koh H-J (2018) Identification of a spotted leaf sheath gene involved in early senescence and defense response in rice. Front Plant Sci 9:1274
Li T, Bai G (2009) Lesion mimic associates with adult plant resistance to leaf rust infection in wheat. Theor Appl Genet 119:13–21
Li Y, Cao X, Zhu Y, Yang X, Zhang K, Xiao Z, Wang H, Zhao J, Zhang L, Li G, Zheng Y, Fan J, Wang J, Chen X, Wu X, Zhao J, Dong OX, Chen X, Chern M, Wang W (2019) Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol 222:1507–1522
Li C, Tang H, Luo W, Zhang X, Mu Y, Deng M, Liu Y, Jiang Q, Chen G, Wang J, Qi P, Pu Z, Jiang Y, Wei Y, Zheng Y, Lan X, Ma J (2020) A novel, validated, and plant height-independent QTL for spike extension length is associated with yield-related traits in wheat. Theor Appl Genet 133:3381–3393
Liu J, Tang H, Qu X, Liu H, Li C, Tu Y, Li S, Habib A, Mu Y, Dai S, Deng M, Jiang Q, Liu Y, Chen G, Wang J, Chen G, Li W, Jiang Y, Wei Y, Lan X, Zheng Y, Ma J (2020) A novel, major, and validated QTL for the effective tiller number located on chromosome arm 1BL in bread wheat. Plant Mol Biol 104:173–185
Liu R, Lu J, Zheng S, Du M, Zhang C, Wang M, Li Y, Xing J, Wu Y, Zhang L (2021) Molecular mapping of a novel lesion mimic gene (lm4) associated with enhanced resistance to stripe rust in bread wheat. BMC Genomic Data 22:1
Long L, Yao F, Guan F, Cheng Y, Duan L, Zhao X, Li H, Pu Z, Li W, Jiang Q, Wei Y, Ma J, Kang H, Dai S, Qi P, Xu Q, Deng M, Zheng Y, Jiang Y, Chen G (2021) A stable QTL on chromosome 5BL combined with Yr18 conferring high-level adult-plant resistance to stripe rust in Chinese wheat Landrace Anyuehong. Phytopathology
Lorrain S, Vailleau F, Balagué C, Roby D (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8:263–271
Ma J, Stiller J, Zhao Q, Feng Q, Cavanagh C, Wang P, Gardiner D, Choulet F, Feuillet C, Zheng YL, Wei Y, Yan G, Han B, Manners JM, Liu C (2014) Transcriptome and allele specificity associated with a 3BL locus for Fusarium crown rot resistance in bread wheat. PLoS One 9:e113309
Ma J, Ding P, Liu J, Li T, Zou Y, Habib A, Mu Y, Tang H, Jiang Q, Liu Y, Chen G, Wang J, Deng M, Qi P, Li W, Pu Z, Zheng Y, Wei Y, Lan X (2019a) Identification and validation of a major and stably expressed QTL for spikelet number per spike in bread wheat. Theor Appl Genet 132:3155–3167
Ma J, Qin N, Cai B, Chen G, Ding P, Zhang H, Yang C, Huang L, Mu Y, Tang H, Liu Y, Wang J, Qi P, Jiang Q, Zheng Y, Liu C, Lan X, Wei Y (2019b) Identification and validation of a novel major QTL for all-stage stripe rust resistance on 1BL in the winter wheat line 20828. Theor Appl Genet 132:1363–1373
Ma J, Zhang H, Li S, Zou Y, Li T, Liu J, Ding P, Mu Y, Tang H, Deng M, Liu Y, Jiang Q, Chen G, Kang H, Li W, Pu Z, Wei Y, Zheng Y, Lan X (2019c) Identification of quantitative trait loci for kernel traits in a wheat cultivar Chuannong16. BMC Genet 20:77
Ma J, Tu Y, Zhu J, Luo W, Liu H, Li C, Li S, Liu J, Ding P, Habib A, Mu Y, Tang H, Liu Y, Jiang Q, Chen G, Wang J, Li W, Pu Z, Zheng Y, Wei Y, Kang H, Chen G, Lan X (2020) Flag leaf size and posture of bread wheat: genetic dissection, QTL validation and their relationships with yield-related traits. Theor Appl Genet 133:297–315
Mori M, Tomita C, Sugimoto K, Hasegawa M, Hayashi N, Dubouzet JG, Ochiai H, Sekimoto H, Hirochika H, Kikuchi S (2007) Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol Biol 63:847–860
Munemasa S, Oda K, Watanabe-Sugimoto M, Nakamura Y, Shimoishi Y, Murata Y (2007) The coronatine-insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in Arabidopsis guard cells. Specific impairment of ion channel activation and second messenger production. Plant Physiol 143:1398–1407
Murray M, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Orozco-Cárdenas ML, Narváez-Vásquez J, Ryan CA (2001) Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13:179–191
Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10:11
Portis AR, Parry MAJ (2007) Discoveries in Rubisco (Ribulose 1,5-bisphosphate carboxylase/oxygenase): a historical perspective. Photosynth Res 94:121–143
Sathe AP, Su X, Chen Z, Chen T, Wei X, Tang S, Zhang X, Wu J (2019) Identification and characterization of a spotted-leaf mutant spl40 with enhanced bacterial blight resistance in rice. Rice 12:68
Shukla P, Gautam R, Singh NK, Ahmed I, Kirti PB (2019) A proteomic study of cysteine protease induced cell death in anthers of male sterile tobacco transgenic plants. Physiol Molecul Biol Plants 25:1073–1082
Sun C, Liu L, Tang J, Lin A, Zhang F, Fang J, Zhang G, Chu C (2011) RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J Genet Genomics 38:29–37
Sun Y, Wenyi LU, Zhang Y, Wang G, Sun J, Xue D, Zhang X (2014) Genetic and Physiological Analyses of Barley Lesion Mimic Mutant bspl1. J Hangzhou Normal Univ (in Chinese)
Sunkar R, Bartels D, Kirch HH (2003) Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J 35:452–464
Takagi D, Ihara H, Takumi S, Miyake C (2019) Growth light environment changes the sensitivity of photosystem I photoinhibition depending on common wheat cultivars. Front Plant Sci 10:686
Takahashi A, Agrawal GK, Yamazaki M, Onosato K, Miyao A, Kawasaki T, Shimamoto K, Hirochika H (2007) Rice Pti1a negatively regulates RAR1-dependent defense responses. Plant Cell 19:2940–2951
Vellosillo T, Vicente J, Kulasekaran S, Hamberg M, Castresana C (2010) Emerging complexity in reactive oxygen species production and signaling during the response of plants to pathogens. Plant Physiol 154:444–448
Voss I, Sunil B, Scheibe R, Raghavendra A (2013) Emerging concept for the role of photorespiration as an important part of abiotic stress response. Plant Biol 15:713–722
Wang L, Pei Z, Tian Y, He C (2005) OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol Plant Microbe Interact 18:375–384
Wang Z, Wang Y, Hong X, Hu D, Liu C, Yang J, Li Y, Huang Y, Feng Y, Gong H, Li Y, Fang G, Tang H, Li Y (2014) Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J Exp Bot 66:973–987
Wang J, Ye B, Yin J, Yuan C, Zhou X, Li W, He M, Wang J, Chen W, Qin P, Ma B, Wang Y, Li S, Chen X (2015) Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice. Plant Physiol Biochem 97:44–51
Wang F, Wu W, Wang D, Yang W, Sun J, Liu D, Zhang A (2016) Characterization and genetic analysis of a novel light-dependent lesion mimic mutant, lm3, showing adult-plant resistance to powdery mildew in common wheat. PloS one 11:e0155358
Wang D, Liang X, Bao Y, Yang S, Zhang X, Yu H, Zhang Q, Xu G, Feng X, Dou D (2020) A malectin-like receptor kinase regulates cell death and pattern-triggered immunity in soybean. EMBO Rep e50442
Wellburn R (1994) The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 144:307–313
Woodson JD (2019) Chloroplast stress signals: regulation of cellular degradation and chloroplast turnover. Curr Opin Plant Biol 52:30–37
Wu J, Neimanis S, Heber U (1991) Photorespiration is more effective than the Mehler reaction in protecting the photosynthetic apparatus against photoinhibition. Bot Acta 104:283–291
Wu C, Bordeos A, Madamba MRS, Baraoidan M, Ramos M, Wang G, Leach JE, Leung H (2008) Rice lesion mimic mutants with enhanced resistance to diseases. Mol Genet Genomics 279:605–619
Wu Y, Wang Y, Yao F, Long L, Li J, Li H, Pu Z, Li W, Jiang Q, Wang J (2021) Molecular mapping of a novel quantitative trait locus conferring adult plant resistance to stripe rust in Chinese wheat landrace guangtoumai. Plant Dis. PDIS-07-20-1544-RE
Yao Q, Zhou R, Fu T, Wu W, Zhu Z, Li A, Jia J (2009) Characterization and mapping of complementary lesion-mimic genes lm1 and lm2 in common wheat. Theor Appl Genet 119:1005–1012
Yin Z, Chen J, Zeng L, Goh M, Leung H, Khush GS, Wang G (2000) Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol Plant Microbe Interact 13:869–876
Zhai H, Cao S, Wan J, Zhang R, Lu W, Li L, Kuang T, Min S, Zhu D, Cheng S (2002) Relationship between leaf photosynthetic function at grain filling stage and yield in super high-yielding hybrid rice (Oryza sativa. L). Sci China Ser C 45:637
Zhang Y, Chen Q, Liu C, Lei L, Li Y, Zhao K, Wei M, Guo Z, Wang Y, Xu B, Jiang Y, Kong L, Liu Y, Lan X, Jiang Q, Ma J, Wang J, Chen G, Wei Y, Zheng Y, Qi P (2019) Fusarium graminearum FgCWM1 encodes a cell wall mannoprotein conferring sensitivity to salicylic acid and virulence to wheat. Toxins 11:628
Zhang Q, Li W, Yang J, Xu J, Meng Y, Shan W (2020) Two Phytophthora parasitica cysteine protease genes, PpCys44 and PpCys45, trigger cell death in various Nicotiana spp. and act as virulence factors. Mol Plant Pathol 21:541–554
Zhu T, Wang L, Rimbert H, Rodriguez JC, Deal KR, De Oliveira R, Choulet F, Keeble-Gagnère G, Tibbits J, Rogers J, Eversole K, Appels R, Gu YQ, Mascher M, Dvorak J, Luo M (2021) Optical maps refine the bread wheat Triticum aestivum cv Chinese Spring genome assembly. Plant J. 101111/tpj15289
Zong X-j, Li D-p, Gu L-k, Li D-q, Liu L-x, Hu X-l (2009) Abscisic acid and hydrogen peroxide induce a novel maize group C MAP kinase gene, ZmMPK7, which is responsible for the removal of reactive oxygen species. Planta 229:485–495
Zou Y, Tang H, Li T, Sun M, Qu X, Zhou J, Yang C, Mu Y, Jiang Q, Liu Y, Chen G, Chen G, Zheng Y, Wei Y, Lan X, Ma J (2020) Identification and characterization of mRNAs and lncRNAs of a barley shrunken endosperm mutant using RNA-seq. Genetica 1–14
Acknowledgements
This work is supported by the National Natural Science Foundation of China (31970243 and 31971937), the Basic Research Project of Science and Technology Plan of Guizhou Province (ZK [2021] general 131), National Undergraduate Innovation Training Program (202010626001), the International Science and Technology Cooperation and Exchanges Program of Science and Technology Department of Sichuan Province (2021YFH0083), and the Applied Basic Research Programs of Science and Technology Department of Sichuan Province (2020YJ0140 and 2021YJ0503), We thank the anonymous referees for reading and revising this manuscript.
Author information
Authors and Affiliations
Contributions
CL and HL performed the entire study and drafted this manuscript. JW, QP, KYW, YW, and PYJ did phenotype measurement and data analysis. YM and HPT did field work and data analysis. QX, QTJ, YXL, PFQ, XJZ, LH collected and analyzed data. GYC, JRW, and YMW, YLZ did QTL analysis and manuscript revision. LLG and QFY revised the manuscript. XJL guided the study and revised the manuscript. JM designed the experiments, guided the entire study, participated in data analysis, wrote and extensively revised this manuscript. All authors participated in the research and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that there is no conflict of interest.
Ethical approval
All experiments and data analyses were conducted in Sichuan. All authors contributed to the study and approved the final version for submission. The manuscript has not been submitted to any other journal.
Additional information
Communicated by Reem Aboukhaddour.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, C., Liu, H., Wang, J. et al. Characterization and fine mapping of a lesion mimic mutant (Lm5) with enhanced stripe rust and powdery mildew resistance in bread wheat (Triticum aestivum L.). Theor Appl Genet 135, 421–438 (2022). https://doi.org/10.1007/s00122-021-03973-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03973-1