Abstract
Key message
Significant differences in defence pathway-related gene expression were observed among chickpea cultivars following A. rabiei infection. Differential gene expression is indicative of diverse resistances, a theoretical tool for selective breeding.
Abstract
A high number of Ascochyta rabiei pathotypes infecting chickpea in Australia has severely hampered efforts towards breeding for sustained quantitative resistance in chickpea. Breeding for sustained resistance will be aided by detailed knowledge of defence responses to isolates with different aggressiveness. As an initial step, the conserved and differential expressions of a suit of previously characterised genes known to be involved in fungal defence mechanisms were assessed among resistant and susceptible host genotypes following inoculation with high or low aggressive A. rabiei isolates. Using quantitative Real-Time PCR (qRT-PCR), 15 defence-related genes, normalised with two reference genes, were temporally differentially expressed (P < 0.005) as early as 2 h post inoculation of Genesis090 (resistant) or Kaniva (susceptible). The highly aggressive isolate, 09KAL09, induced vastly different expression profiles of eight key defence-related genes among resistant and susceptible genotypes. Six of these same genes were differentially expressed among ten host genotypes, inclusive of the best resistance sources within the Australian chickpea breeding program, indicating potential use for discrimination and selection of resistance “type” in future breeding pursuits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chickpea (Cicer arietinum L.) is an important food legume and break crop when grown in rotation with cereals and oilseeds, which ultimately improves yields and maintains soil fertility through atmospheric nitrogen fixation (Singh 1997; Dalal et al. 1998). Globally, 10.5 million tonnes of chickpea is produced annually (FAOSTAT 2013). However, the fungal pathogen Ascochyta rabiei (Pass.) Labr. constrains both production and quality (Nene et al. 1987; Gaur and Singh 1996). As a seed-borne pathogen, dissemination usually occurs through anthropogenic movement of seed as well as dispersal by wind and rain splash, which eventually affects all aerial plant parts (Pande et al. 2005).
Ascochyta rabiei spores germinate 12 h post inoculation (hpi) (Pandey et al. 1987). Appresoria are formed at 24 hpi and mucilaginous exudates are secreted to provide a tight contact with the host surface (Köhler et al. 1995). At this point, necrotrophic fungi are known to produce compounds, such as saponin detoxifying enzymes (Markham and Hille 2001), to suppress plant defence responses and prevent the signalling of host defence pathways (Staples and Mayer 2003). Once A. rabiei mycelia penetrate the host epidermal cells (Pandey et al. 1987), they expand and secrete cell wall degrading enzymes and toxins such as solanapyrone A, B and C (Hohl et al. 1990; Alam et al. 1989; Kaur 1995). Subsequently, pycnidia are formed in the host tissue 6–8 days later to complete the life cycle (Hohl et al. 1990).
Chickpea has both active and passive defence responses to stop initial pathogenic attacks and to prevent successful invasion and spread to neighbouring cells (Coram and Pang 2006). Passive defence mechanisms include preformed structural and chemical barriers such as glandular trichomes, which secrete antifungal isoflavones (Armstrong-Cho and Gossen 2005). Active defence systems in plants may employ R genes to recognise pathogen-specific effectors encoded by the Avr genes (McDonald and Linde 2002), leading to effector-triggered immunity (ETI) and possible programmed cell death (PCD) via a hypersensitive response (HR) (Jones and Dangl 2006).
Few sources of stable resistance to A. rabiei exist. Singh and Reddy (1993) identified five (ICC4475, ICC6328, ICC12004, ILC200 and ILC6428) from 19,343 chickpea accessions resistant to six races of A. rabiei in Syria. The accession ICC3996 was added to this list by Chen et al. (2004) and together these have become the genetic basis of A. rabiei resistance breeding programs worldwide. However, some moderate resistance has recently broken down (Tar’an et al. 2007; Kanouni et al. 2002) and in 2010, the widely adopted resistant cultivars ‘Genesis090’ and ‘PBA HatTrick’ were heavily infected indicating that, as well as optimal environmental conditions, the pathogen may have increased aggressiveness.
Although, molecular studies of the 2010 Australian A. rabiei population uncovered a relatively low genetic diversity when measured with microsatellites and compared to that observed in other countries (Phan et al. 2003; Pradhan 2006; Leo et al. 2011), the low diversity for neutral genetic markers may not accurately reflect the evolutionary adaptive potential for pathogenicity of the population. Indeed, when 24 isolates with an identical microsatellite genotype were tested for their ability to cause disease on 12 Australian chickpea genotypes, extensive pathogenic variation (aggressiveness) was observed among the Australian cultivars and caused significant damage to the most current resistant cultivars (Elliott et al. 2011). This suggests that there may be differences in the perception, signalling and defence-related pathways among resistance sources to different isolates.
The defence of chickpea to A. rabiei is multigenic and quantitative with resistance-quantitative trait loci (R-QTL) identified on linkage groups 1, 2, 3, 4, 6 and 8 (Huettel et al. 2002; Flandez-Galvez et al. 2003; Iruela et al. 2006; Tar’an et al. 2007). This indicates that several defence-related mechanisms are involved as previously postulated (Tar’an et al. 2007). However, little is known on whether chickpea selectively employs differing types and levels of defence responses when infected with isolates of different aggressiveness known to exist in the Australian population (Elliott et al. 2011). One method to investigate this is to assess and compare, among host genotypes, the responsiveness of previously characterised defence-related genes that are representative of diverse defence-related pathways and following exposure to individual isolates of differing aggressiveness.
The expression profiles of several host genes, related to a range of defence mechanisms, have previously been characterised within the chickpea (ICC3996)—A. rabiei pathosystem (Coram and Pang 2005a, b, 2006). For example, pathogenesis related (PR) proteins which are induced by pathogen-derived elicitors, such as glucan and chitin within fungal cell walls, as well as fungus-secreted glycoproteins and peptides (Kombrink and Schmelzer 2001; Edereva 2005). In particular, PR-2B (β1,3-glucanase) (EC 3.2.1.39) releases glycosidic fragments that elicit host defence mechanisms, and weakens and decomposes fungal cell walls containing glucans (Kombrink and Schmelzer 2001; Edereva 2005). The speed and coordination of pathogen perception by the host is vital to achieve effective defence. Resistant hosts often respond faster and produce larger quantities of defence related compounds than susceptible ones (Yang et al. 1997). For example, PR proteins β-1,3-glucanase and chitinase (EC 3.2.1.14) are more rapidly synthesized in resistant cultivars (Vogelsang and Barz 1993; Hanselle and Barz 2001; Coram and Pang 2006; Vaghefi et al. 2013).
Meanwhile, glutathione S-transferases (GST) (EC 2.5.1.13) is a multi-gene family that protect uninfected cells from oxygen toxicity, suppress apoptosis (Coelho et al. 2010) and detoxify various compounds (Marrs 1996; Edwards et al. 2000; Dixon et al. 2002). The down-regulation of GST indicates an increase in cellular H2O2 from a possible oxidative burst (Neill et al. 2002). Another, Snakin-2 (SN2), is a broad-spectrum antimicrobial cysteine-rich peptide from potato (Solanum tubersum L.) (Segura et al. 1999), which is also known as gibberellins stimulated-like proteins (GSL2) (Meiyalaghan et al. 2014). The cysteine-rich nature of this peptide acts as both constitutive and inducible defence barriers crucial to the occurrence of disulphide bridges important in enhancing the structural stability of the plants when under stressful conditions (Berrocal-lobo et al. 2002; Pelegrini et al. 2011).
Other gene targets to asses defence to necrotrophic fungal pathogens have included the disease resistance response gene (DRRG) in pea (Pisum sativum) infected with Fusarium solani (Chiang and Hadwiger 1990) and those regulating the cellular oxidative burst in barley (Hordeum vulgare L.) infected with Botrytis cinerea. Also, members of the NAC (for NAM, ATAF1,2 and CUC2) gene family (Peng et al. 2010), and transcription factors such as those detected in Medicago truncatula following infection with Uromyces striatus (Madrid et al. 2010). One of these, TF1082, confers an ethylene response (ER) during infection (Madrid et al. 2010). Previously, the ERG (ethylene receptor gene) also known as CaETR1 (Cicer arietinum L. Ethylene receptor-like sequences) was found to be responsive to A. rabiei infection (Madrid et al. 2010). Another, TF1063, a myelobastosis (MyB) gene family member, was associated with the hypersensitive response (Madrid et al. 2010). Assessing the differential expression of these gene homologues in chickpea would aid in better understanding the complexities of defence-mechanism responses to A. rabiei.
High throughput quantitative Real-Time PCR (qRT-PCR) is an appropriate method to sensitively detect expression level changes of potentially low-abundance and previously characterised transcripts (Kakar et al. 2008). This approach was employed to compare the temporal and quantitative expression of key defence-related genes in chickpea to isolates that represent the breadth of aggressiveness within the Australian A. rabiei population. This will determine: (1) if isolates with different aggressiveness provoke differential host defence gene expressions, and the speeds in which these occur and (2) if different genes are expressed during the response, providing further evidence of different defence mechanisms among different chickpea genotypes.
Materials and methods
Plant material and fungal isolates
Ten chickpea genotypes used in this experiment (Table 1) were chosen as a representative of a differential host range based on their previously determined disease reactions to 24 Australian A. rabiei isolates (Elliott et al. 2011). These included susceptible and resistant cultivars which are commercially used, as well as parental lines used in the Australian breeding program. All genotypes were obtained from the Victorian Department of Environment and Primary Industries in Horsham, Victoria, Australia. Chickpea genotypes, ‘Genesis090’ and ‘Kaniva’ which were categorised as resistant (Pulse Australia 2009c) and susceptible (Carter 1999), respectively, were used to screen and selectively identify differentially expressed defence-related genes prior to testing the genes on other chickpea genotypes.
Ascochyta rabiei isolates; 09KAL09, 09MEL04, 09KAN19 and 09KIN11 used in this study were collected in 2009 (Leo et al. 2015). The isolates comprised two sets of two isolates with different aggressiveness based on the mean area under disease progress curve (AUDPC) in the pathotyping study by Elliott et al. (2011) which used the disease rating scale adopted from Singh et al. (1981). Briefly, the number of times each treatment (isolate on host) received a particular score was determined based on the established 1–9 scale. Scores of 1 & 3, 5 and 7 & 9 were grouped into three categories. For leaf infection, isolates which at 21 days post inoculation had a score of 7 or 9 greater than 80 % of the time were classified as high risk. Isolates with a score of 7 or 9 less than 60 % of the time were classified as low risk. For stem infection, isolates which at 21 days post inoculation had a score of 7 or 9 greater than 10 % of the time were classified as high risk. Isolates with a score of 7 or 9 less than 5 % of the time were classified as low risk. Overall severity/rank was based on the highest risk rating from either the stem or leaf data if they did not match. The highly aggressive isolates, 09KAL09 and 09KAN19 were isolated from the resistant cultivar, Genesis090 in Kalkee and Kaniva, Victoria, respectively. The low aggressive isolates, 09MEL04 and 09KIN11 were isolated from the moderately resistant cultivars, CICA0503 and Almaz in Melton and Kingsford, South Australia, respectively.
Bioassay
All isolates were passaged on sterilised chickpea leaves (autoclaved at 80 °C for 15 min) on 1 % (w/v) water agar for 1 week before being transferred onto V8 juice growth agar. Cultures were grown at 20 ± 2 °C with a 12 h photoperiod for 14 days. Spore suspensions were then prepared by adding 10 mL of sterile water and scraping the spores off the plate with a scalpel. The spore suspensions were then filtered through a muslin cloth and the concentration adjusted to 1 × 105 spores/mL using a haemocytometer.
Seeds were surface sterilized in 5 % (w/v) sodium hypochlorite for 15 min and washed three times with sterile distilled water prior to sowing in 15 cm diameter pots (containing three seeds each) in sterile soil. A total of four biological replicates per chickpea line per isolate including mock controls was used. All plants were grown at 20 ± 4 °C for 14 days (until six to eight leaf stage) before inoculation. Plants were then sprayed with A. rabiei spore suspensions until run-off (approximately 5 mL/plant). Mock-inoculated controls were sprayed with sterile distilled water until run off. Following inoculation, each pot was covered with a disposable plastic cup for maximum darkness and sealed in a plastic storage box in a 20 ± 4 °C growth room to maintain humidity.
RNA extraction, cDNA preparation and development of qRT-PCR-based markers
Main stem and young leaf tissue weighed 100 mg were collected from mock and spore inoculated plants at 2, 6, 12, 24, 48 and 72 h post inoculation (hpi) for total RNA extraction using the RNeasy® Plant Mini Kit (Qiagen, CA, USA). RNase-Free DNase (Qiagen, CA, USA) was added to eliminate gDNA contamination. RNA concentration and integrity (RQI) values were determined on an Experion with RNA StdSens Chips (Bio-Rad Laboratories, CA, USA). RQI values higher than eight were used for downstream applications (Fleige and Pfaffl 2006). Total RNA (1 µg) was reverse-transcribed with a combination of Oligo(dT)20 and random primer using the iScript™ select cDNA synthesis kit (Bio-Rad Laboratories, NSW, Australia). The quality of cDNA and absence of gDNA were assessed on agarose gel.
Seventeen genes including transcription factors which were highly and differentially expressed in various legume defence mechanisms (to mostly biotic but in some cases abiotic stress factors) were selected from the literature (Table 2). Sequences were derived from GenBank and three sets of qRT-PCR primers were designed from each using Primer3 v.0.4.0. (Rozen and Skaletsky 2000). The primers were designed with the following criteria: T m of 60 ± 1 °C and PCR amplicon size of 55–250 bp, primer sequences length of 18–27 nucleotides and GC contents of 45–65 %. To normalise the relative quantities (NRQs) of these genes, three reference genes (PUBQ, RIB, PP2A) (Table 2) previously proven to give stable expressions after biotic stresses to Fusarium oxysporum f. sp. ciceris and A. rabiei in chickpea were assessed (Castro et al. 2012). All primers were synthesised at Sigma-Genosys Ltd (NSW, Australia). All primers were tested with both randomly pooled cDNA and gDNA samples, and cycle sequenced three times at the Australian Genome Research Facility (AGRF, Melbourne, Australia) to determine the correct expected amplicon size and BLASTn to ensure the amplicons were of the target sequences.
All PCR were carried out with the iQ5 Real-Time PCR detection System (Bio-Rad Laboratories, NSW, Australia). A standard curve was produced for each of the target and reference genes. The 25 µL reaction comprised 4 µL of DNA template, 13.5 µL of 1× iQSYBR Green Supermix (Bio-Rad Laboratories, NSW, Australia) and the specified primer concentration (Table 2). Thermal cycling conditions were: Initial denaturation at 95 °C for 1 min; 40 cycles of 95 °C for 10 s, 60 °C for 15 s, 72 °C for 30 s, 83 °C for 10 s (fluorescence reading), followed by melt curve analysis at 60–95 °C every 0.5 °C for 10 s. All reactions were performed in triplicate and the sample maximisation layout strategy was employed (Hellemans et al. 2007). The cDNA samples for each gene were preferably run within a single plate to reduce technical, run-to-run variation. However, inter-run calibrators (IRC) were used whenever all samples could not be analysed in the same run. Minus reverse transcription control (-RTC) and no template control (NTC) were carried out for every gene to detect the presence of contaminating DNA and/or primer dimers.
Data analysis
Data and PCR efficiency of each gene were analysed using Bio-Rad iQ5 v2.0 software (Bio-Rad, CA, USA). Reactions with more than one melt curve peak and not within the PCR efficiency range of 95–110 % were discarded.
Reference genes were analysed and selected based on stable expression using geNormPLUS (Hellemans et al. 2007). Normalization of expression values from targeted genes were calculated using qbase PLUS software, and were reported as normalized relative quantities (NRQs) (Hellemans et al. 2007).
General linear model was performed using SAS and Minitab 16 to determine differentially expressed genes at P < 0.05. A mean fold change of 2.0 was used as the cut-off point. Differentially expressed genes between genotypes, treatments, or genotype × treatment interactions, were clustered using an hierarchical cluster analysis. A data matrix for each genotype with the expression ratio was used to calculate an Euclidean distance matrix. The UPMG method was used to generate a dendogram using K-means clustering with Cluster v3.0 (Eisen et al. 1998) and viewed with Treeview v1.60 (Page 1996) as a heat map.
To assess for differences in host gene expression levels when infected with different isolates, the mean expressions derived from each interaction were compared and an analysis of variance (ANOVA) was then performed with qbase PLUS software (P = 0.05). The same analyses determined significant gene expression differences among ten chickpea genotypes (Table 1) infected with the aggressive isolate, 09KAL09. A total of eight defence genes which were up-regulated in ‘Genesis090’ when infected with 09KAL09 (CARNAC, GST, PR2B, SN2, ERG, PAMP, RGA4, TF1082) were selected to identify and determine potential responses induced among four other resistant, two moderately resistant and two moderately susceptible chickpea genotypes relative to the susceptible genotypes (Table 1) and compared using ANOVA.
Results
Single fragments of 80–250 bp were amplified with efficiencies of 90–110 % from 15 of the 17 target genes and were used to assess expression levels with qRT-PCR (Table 2). Of the sequences tested for suitability for expression normalisation, PUBQ and RIB were the most stable with M (gene stability) values of 1.102 and coefficients of variation (CV) values of 0.414 and 0.412, respectively (Hellemans et al. 2007). The M value for PP2A was 1.411 and thus excluded.
Timing and expression levels of defence-related host genes based on interactions with different levels of isolate aggressiveness
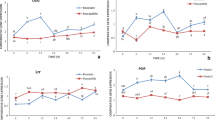
All 15 defence-related genes were differentially expressed in at least one time point following inoculation of ‘Genesis090’ and ‘Kaniva’ when compared to the un-inoculated controls. Of the four isolates assessed, the highly aggressive 09KAL09, produced a grossly different expression profile across all 15 genes. The most down-regulation, was consistent in timing and levels across both the resistant (Genesis090) and susceptible (Kaniva) genotypes (Fig. 1). Conversely, the expression profile produced by the other highly aggressive isolate (09KAN19) was not largely different in either timing or magnitude to the two less aggressive isolates (09MEL04 and 09KIN11). Again, this was consistent among host genotypes (Figs. 1, 2). In general, the majority of differentially expressed genes were up-regulated as early as 2 hpi and started to be down-regulated at 72 hpi (Supplementary Material 1).
Kinetic trends of differential gene expression for Genesis090 (resistant) and Kaniva (susceptible) genotypes over the time course of infection: a 2 hpi; b 6 hpi; c 12 hpi; d 24 hpi; e 48 hpi; f 72 hpi, with 4 A. rabiei isolates, 09KAL09, 09MEL04, 09KAN19 and 09KIN11. The vertical axis indicates the number of up-regulated (red) and down-regulated genes (green) at each time point
Heatmap of 15 genes expression profiles for Genesis090 (resistant) and Kaniva (susceptible) over the time course after infection with 4 A. rabiei isolates, 09KAL09, 09MEL04, 09KAN19 and 09KIN11. Up-regulation is indicated in red, down-regulation is indicated in green, normalised expression values close to the mean are in black. No detectable expression is in grey. The Log2 values of the expression profile for each treatment and genotype were normalised with two reference genes and non-inoculates samples
Similarities in host gene expression trends among isolate interactions
Cluster analysis shown similarities in individual gene expression profiles following exposure to each of the four isolates (Fig. 2). Mega Cluster I contained PR2B, up-regulated as early as 12 hpi in ‘Kaniva’ and ‘Genesis090’ regardless of isolate applied. Cluster II was divided into six subgroups. Cluster II.1:2 comprised ERG and LZP genes, which were not up-regulated when either host genotype was inoculated with either 09KAL09 or 09MEL04, but were up-regulated when inoculated with 09KAN19 or 09KIN11 (as early as 2 hpi). Cluster II.2:3 comprised CARNAC, GST and SDCCP. CARNAC was up-regulated in all interactions except when Kaniva was inoculated with 09KAL09, suggesting that lack of expression may lead to susceptibility when inoculated with the highly aggressive isolates. GST and SDCCP were up-regulated in at least one time point in both genotypes when exposed to any of the isolates. Cluster II.3:5 comprised PAMP, RGA4, RGA7, TF1063 and SPK. Cluster II.4:1 contained TF1082, Cluster II.5:2 and Cluster II.6:1 comprised RGA10&5, and SN2, respectively. Genes in Cluster II 3, 4, 5 and 6 were up-regulated in both genotypes infected with 09KAN19 and 09KIN11 as early as 6 hpi. When infected with 09MEL04, these genes were up-regulated at later infection stages (>12 hpi), but remained at either a basal expression rate or were down-regulated when the genotypes were infected with 09KAL09.
Differentially expressed defence-related genes and their relations to different levels of host susceptibility
Following inoculation with isolate 09KAL09, major differences in the gene differential expression profiles were observed among the ten host genotypes assessed, which ranged in classification from resistant to susceptible (Supplementary material 2; Fig. 3). The mean expression profiles of each genotype at a 95 % confidence interval identified six genes, CARNAC, ERG, GST, RGA4, SN2 and TF1082, differentially expressed across all ten host genotypes (Supplementary material 2). The expression of PR2B was not differential but consistently highly up-regulated among all hosts. PAMP was up-regulated in all hosts except for the highly susceptible Kaniva, perhaps indicating a lack of recognition. To further identify which genes were up-regulated in the resistant and moderately resistant genotypes in comparison to the susceptible genotypes, the mean expression profiles of each genotype were categorised into their susceptibility levels and compared (Supplementary materials 3, 4). Between moderately resistant and resistant genotypes, only SN2 was differentially expressed, more highly in moderately resistant genotypes. Interestingly, no genes were differentially expressed between moderately resistant and moderately susceptible classified genotypes at any of the time points assessed, however, four genes, SN2, GST, ERG and RGA4, were differentially expressed between resistant/moderately resistant and susceptible genotypes. Three, SN2, GST, and ERG, were expressed at higher levels in resistant/moderately resistant than susceptible genotypes and one, RGA4, at a higher level in susceptible genotypes. Following validation across a broader germplasm and in response to a larger number of isolates, the differential expression of these four genes may be useful as tools for future molecular selection of resistance within breeding programs.
Heatmap of eight genes expression profiles for 10 chickpea genotypes (Table 1) over the time course after infection with the most pathogenic A. rabiei isolate, 09KAL09. Up-regulation is indicated in red, down-regulation is indicated in green, normalised expression values close to the mean are in black, no detectable expression is in grey. The Log2 values of the expression profile for each treatment and genotype were normalised with two reference genes and non-inoculates samples
Discussion
For the first time, they study has demonstrated that A. rabiei isolates of a similar high aggressiveness level are able to cause different host responses within the same chickpea genotype. One might postulate that 09KAL09 is able to evade detection and recognition and then goes on to suppress host defence responses whilst it establishes itself and begins to evade and colonise the tissues. Meanwhile 09KAN19, also highly aggressive, is detected and recognised almost immediately (and certainly by 2 hpi), causing the up-regulation of the spectrum of defence responses related to the genes under study. The question remains regarding what differentiates the ability for the 09KAL09 isolate to be highly aggressive compared to the two less aggressive isolates. Perhaps other isolate-related fitness characteristics are important in establishing and maintaining infection ahead of host defences? Certainly, the timing of gene expressions was largely indifferent following exposure to the highly aggressive 09KAN19 or either of the less aggressive isolates, indicating that molecular evidence of pathogenicity differences among these three isolates was not captured in this study and on these cultivars, hence a wider range of defence-related genes and cultivars would need to be assessed. This would be more feasible with whole genome transcriptomics in response to A. rabiei inoculation.
Another plausible reason to the down regulation of most differentially expressed genes in both susceptible and resistant genotypes is the production, deletion or selection of fungal effectors in 09KAL09 that impact on pathogen recognition. This may trigger different host defence mechanisms. Positive selection occurring within the effector proteins has been observed quite extensively for Phytophtora sojae of soybean (Jiang et al. 2008). This was postulated as a mechanism employed to enable escape from host resistance protein detection and potentially adapt to different host virulence targets (Ellis et al. 2009). Indeed, mutation of motifs in the C-terminus of an Avr1b protein reduced the ability of the pathogen to suppress programmed cell death (PCD) and also abolished the avirulence interaction of Avr1b with the Rps1b resistance gene in soybean (Dou et al. 2008). A similar mechanism may be occurring within A. rabiei isolate 09KAL09, enabling it to evade detection by chickpea.
During industry establishment, selection of a narrow gene pool and subsequent inbreeding has led to a lack of genome diversity across cultivated chickpea, which has also likely constricted the potential diversity of defence mechanisms retained within Australian chickpea cultivars. This low diversity of defence mechanisms was shown in the considerably fewer number of disease resistance gene homologues in chickpea in comparison to other legume species (Varshney et al. 2013). However, significant differences in expression levels and timings of the 15 defence-related genes assessed in the current study were detected among the 10 host genotypes assessed. At very early time points (2–6 hpi) these are likely related to differences in the timing of pathogen recognition and subsequent speed to signal down-stream defence mechanisms.
Indeed, the faster expression of GST in ‘Kaniva’ (6 hpi) compared to ‘Genesis090’ (24 hpi) is likely associated with the earlier accumulation of H2O2 in the susceptible cultivar to trigger a rapid hypersensitive response. However, across genotypes, the susceptible genotypes produced significantly less GST than the resistant ones at earlier time points and greater expression in the resistant genotypes later on (24–48 hpi). This may indicate that although the hypersensitive response is employed by susceptible genotypes this is not effectual for containing the pathogen and that resistant genotypes only instigate this defence response after other first-line defence responses have been triggered. Indeed, other reactive oxygen species involved in the precursors to the hypersensitive response have been detected in resistant genotypes at earlier time points of the interaction (Hohl et al. 1990; Coram and Pang 2006).
The pattern of expression of SN2 was similar to GST (another antioxidant) with greater quantities detected in resistant/moderately resistant genotypes than susceptible genotypes. This is in accordance to the up-regulation of SN2 previously detected in ICC3996 (Coram and Pang 2005b, 2006). Sequence similarities of SN2 peptides to GIP2 (GASA-like protein) from Petunia hybrida suggests involvement in redox regulations which regulate the production of reactive oxygen species in pathogenesis and wounding (Berrocal-lobo et al. 2002; Wigoda et al. 2006; Balaji and Smart 2012).
Meanwhile, CaETR1 (Cicer arietinum L. ethylene receptor-like sequences) was the first ethylene receptor discovered in chickpea associated with A. rabiei resistance (Madrid et al. 2010). The ERG locus is closely linked to a major QTL, QTLAR1 proposed to condition resistance to pathotype II (Iruela et al. 2006; Madrid et al. 2012). Recently, the CaETR1 and CaETR-1a/CaETR-1b alleles from resistant and susceptible chickpea genotypes (Madrid et al. 2012) were used to negatively select and eliminate susceptible individuals from a breeding program (Madrid et al. 2013). The differential expression of the allele (unknown) observed in the current study between resistant/moderately resistant and susceptible genotypes may further indicate its suitability for resistance selection across broad range of germplasm.
A spectrum of differences in levels and timings of the CARNAC transcription factor was observed among the 10 genotypes. Down- or unaltered expression in the majority of genotypes may be related to involvement in developmental processes such as apical meristem development, flowering and secondary wall formation (Peng et al. 2010). This may be anticipated in response to a pathogen attack, as reserves are rerouted to defence-related activities (Coram and Pang 2006). However, up-regulation witnessed in Almaz, Genesis114, Genesis090 and Kaniva may have been directly related to defence responses through participation in signalling pathways and regulatory networks (Nuruzzaman et al. 2013). Indeed, NAC proteins activate PR genes, induce the hypersensitive response and cause cell death at the infection site (Kaneda et al. 2009; Seo et al. 2010). NAC proteins also have the ability to form alliances with certain host regulatory complexes, enabling them to act as negative regulators of the defence response by suppressing defence-related genes (Wang et al. 2009).
Meanwhile, PR proteins are pathogen-induced proteins classified into 17 families from PR-1 to PR-17, based on biochemical properties (Van Loon et al. 2006). As observed for PR2B in the current study, they may be expressed prior to infection due to involvement in plant development (Edereva 2005), accumulated and synthesized for a long lag period. They may then be translocated from the site of induction to other plant parts during pathogenic attack (Matsuoka and Ohashi 1986). Ultimately, PR2B (β1,3-glucanase) produces glycosidic fragments which weakens and decomposes fungal cell walls containing glucans, chitin and proteins (Kombrink and Schmelzer 2001; Edereva 2005). As previously reported, the PR2B gene was significantly up-regulated at 48–72 hpi compared to other time points, particularly in ‘Genesis114’, ‘PBA HatTrick’, ‘Almaz’, ‘Genesis090’ and ‘Kaniva’ (Hanselle and Barz 2001; Coram and Pang 2006; Cho and Muehlbauer 2004).
The gene deemed to regulate polymorphic antigen proteins (PAMP) was up-regulated at 12 and 48 hpi in all genotypes except for the susceptible ‘Kaniva’. This gene is likely to be a homologue of Enolase phosphatise E1 protein, a bifunctional enzyme of methionine salvage that regenerates methionine from 5′-methylthioadenosine (MTA) (Wang et al. 2005). Its function in the resistance response is likely via its metabolism that utilises polyamines (PAs), nicotianamines (NAs) and interacts with ethylene biosysnthesis (Waduwara-Jayabahu et al. 2012). PAs are associated with cell division as a response to abiotic and biotic stress (Takahashi and Kakehi 2010; Vera-Sirera et al. 2010). NAs act as chelators for long distance ion transport and defence signalling processes (Curie et al. 2009), and ethylene is a phytohormone capable of signalling within defence pathways.
The transcription factor TF1082 was increasingly upregulated in several resistant and moderately resistant genotypes from 48 hpi onwards after exposure to the highly aggressive isolate 09KAL09. Previously, in Medicago truncatula infected with Uromyces striatus, this gene was up-regulated in resistant genotypes and down-regulated in susceptible genotypes, thought to bind to the GCC box of PR gene promoters and confer ethylene responsiveness (Madrid et al. 2010).
In conclusion, this study showed that chickpea has a number of defence-related mechanisms which are activated simultaneously to mount defence to A. rabiei, confirming that it is a race-nonspecific resistance controlled by genes with minor to intermediate and additive effects. Although a small subset of genes was assessed, several were differentially expressed among cultivars, further indicating the potential of different defence mechanisms in chickpea under controlled conditions where all plants are subjected to the same environment conditions. Further studies such as RNA sequencing and identifying sequence polymorphisms of within or upstream or downstream signalling regions of the differentially expressed genes in susceptible and resistant cultivars may identify potential allelic differences that, once functionally validated, could be converted into stable markers for future selective breeding purposes. Breeding chickpea genotypes containing several defence strategies will improve durability against the pathogenic diversity of the pathogen population.
Author contribution statement
AEL participated in the design of the experiment, carried out the experimental work, performed the statistical analyses, and drafted the manuscript. CCL assisted in revising the manuscript. RF participated in the design of the experiment and drafted the manuscript. All authors read and approved the final manuscript.
References
Alam SS, Bilton JM, Slavin AMZ, Williams DJ, Sheppard RN, Strange RN (1989) Chickpea blight: production of the phytotoxins solanapyrones A and C by Ascochyta rabiei. Phytochemist 28:2627–2630
Armstrong-Cho C, Gossen BD (2005) Impact of glandular hair exudates on infection of chickpea by Ascochyta rabiei. Can J Bot 83:22–27
Balaji V, Smart CD (2012) Over-expression of snakin-2 and extensin-like protein genes restricts pathogen invasiveness and enhances tolerance to Clavibacter michiganensis subsp. michiganensis in transgenic tomato (Solanum lycopersicum). Transgenic Res 21:23–37
Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-1 FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29:23–32
Carter JM (1999) Chickpea growers guide: a guide for the production of chickpeas. Department of Natural Resources and Environment, Agriculture, Victoria, Australia, p 20
Castro P, Román B, Rubio J, Die JV (2012) Selection of reference genes for expression studies in Cicer arietinum L.: analysis of cyp81E3 gene expression against Ascochyta rabiei. Mol Breed 29:261–274
Chen W, Coyne CJ, Peever TL, Muehlbauer FJ (2004) Characterization of chickpea differentials for pathogenicity assay of ascochyta blight and identification of chickpea accessions resistant to Didymella rabiei. Plant Pathol 53:759–769
Chiang CC, Hadwiger LA (1990) Cloning and characterization of a disease resistance response gene in pea inducible by Fusarium solani. Mol Plant Microbe Interact 3:78–85
Cho S, Muehlbauer FJ (2004) Genetic effect of differentially regulated fungal response genes on resistance to necrotrophic fungal pathogens in chickpea (Cicer arietinum L.). Physiol Mol Plant Pathol 64:57–66
Coelho LP, Peng T, Robert FM (2010) Quantifying the distribution of probes between subcellular locations using unsupervised pattern unmixing. Bioinformatics 26:i7–i12
Coram TE, Pang EC (2005a) Isolation and analysis of candidate ascochyta blight defence genes in chickpea. Part I. Generation and analysis of an expressed sequence tag (EST) library. Physiol Mol Plant Pathol 66:192–200
Coram TE, Pang EC (2005b) Isolation and analysis of candidate ascochyta blight defence genes in chickpea. Part II. Microarray expression analysis of putative defence-related ESTs. Physiol Mol Plant Pathol 66:201–210
Coram TE, Pang EC (2006) Expression profiling of chickpea genes differentially regulated during a resistance response to Ascochyta rabiei. Plant Biotech J 4:647–666
Curie C, Cassin G, Couch D, Divol F, Higuchi K, Le Jean M, Misson J, Schikora A, Czernic P, Mari S (2009) Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot 103:1–11
Dalal RC, Strong VM, Weston EJ, Cooper JE, Wildermuth GB, Lehane KJ, King AJ, Holmes CJ (1998) Sustaining productivity of a Vertisol at Warra, Queensland, with fertilisers, no-tillage, or legumes. 5. Wheat yields, nitrogen benefits and water-use efficiency of chickpea-wheat rotation. Anim Prod Sci 38:489–501
Dixon DP, Lapthorn A, Edwards R (2002) Plant glutathione transferases. Genome Biol 3:3004
Dou D, Kale SD, Wang X, Jiang RH, Bruce NA, Arredondo FD, Zhang X, Tyler BM (2008) RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell Online 20:1930–1947
Edereva A (2005) Pathogenesis-related proteins: research progress in the last 15 years. Gen Appl Plant Physiol 31:105–124
Edwards R, Dixon DP, Walbot V (2000) Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci 5:193–198
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95:14863–14868
Elliott VL, Taylor PW, Ford R (2011) Pathogenic variation within the 2009 Australian Ascochyta rabiei population and implications for future disease management strategy. Australas Plant Pathol 40:568–574
Ellis JG, Rafiqi M, Gan P, Chakrabarti A, Dodds PN (2009) Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Curr Op Plant Biol 12:399–405
FAOSTAT (2013) Food and Agriculture Organization of the United Nations. http://faostat.fao.org/. Accessed 13 Feb 2014
Flandez-Galvez H, Ades PK, Ford R, Pang ECK, Taylor PWJ (2003) QTL analysis for ascochyta blight resistance in an intraspecific population of chickpea (Cicer arietinum L.). Theor Appl Genet 107:1257–1265
Fleige S, Pfaffl MW (2006) RNA integrity and the effect on the realtime qRT-PCR performance. Mol Aspects Med 27:126–139
Gaur RB, Singh RD (1996) Effects of Ascochyta blight on grain yield and protein in chickpea. Indian J Mycol Plant Pathol 26:259–262
Hanselle T, Barz W (2001) Purification and characterisation of the extracellular PR-2b β-1, 3-glucanase accumulating in different Ascochyta rabiei infected chickpea (Cicer arietinum L.) cultivars. Plant Sci 161:773–781
Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19
Hohl B, Pfautsch M, Barz W (1990) Histology of disease development in resistant and susceptible cultivars of chickpea (Cicer arietinum L.) inoculated with spores of Ascochyta rabiei. J Phytopathol 129:31–45
Huettel B, Santra D, Muehlbauer FJ, Kahl G (2002) Resistance gene analogues of chickpea (Cicer arietinum L.): isolation, genetic mapping and association with a Fusarium resistance gene cluster. Theor Appl Genet 105:479–490
Iruela M, Rubio J, Barro F, Cubero JI, Millán T, Gil J (2006) Detection of two quantitative trait loci for resistance to ascochyta blight in an intra-specific cross of chickpea (Cicer arietinum L.): development of SCAR markers associated with resistance. Theor Appl Genet 112:278–287
Jiang RH, Tripathy S, Govers F, Tyler BM (2008) RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci 105:4874–4879
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329
Kakar K, Wandrey M, Czechowski T, Gaertner T, Scheible WR, Stitt M, Torres-Jerez I, Xiao Y, Redman JC, Wu HC, Cheung F, Town CD, Udvardi MK (2008) A community resource for highthroughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Meth 4:18
Kaneda T, Taga Y, Takai R, Iwano M, Matsui H, Takayama S, Isogai A, Che FS (2009) The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J 28:926–936
Kanouni H, Arbat HK, Moghaddam M, Neyshabouri MR (2002) Selection of chickpea (Cicer arietinum L) entries for drought resistance. Agri Sci (Tabriz) 12:109–121
Kaur S (1995) Phytotoxicity of solanapyrones produced by the fungus Ascochyta rabiei and their possible role in blight of chickpea (Cicer arietinum). Plant Sci 109:21–29
Köhler G, Linkert C, Barz W (1995) Infection studies of Cicer arietinum (L.) with GUS-(E. coli β-glucuronidase) transformed Ascochyta rabiei strains. J Phytopathol 143:589–595
Kombrink E, Schmelzer E (2001) The hypersensitive response and its role in local and systemic disease resistance. Euro J Plant Pathol 107:69–78
Leo AE, Ford R, Linde CC, Shah RM, Oliver R, Taylor PWJ, Lichtenzveig J (2011) Characterization of fifteen newly developed microsatellite loci for the chickpea fungal pathogen Ascochyta rabiei. Mol Ecol Res 11:418–421
Leo AE, Ford R, Linde CC (2015) Genetic homogeneity of a recently introduced pathogen of chickpea, Ascochyta rabiei, to Australia. Biol Inv 17:609–623
Madrid E, Gil J, Rubiales D, Krajinski F, Schlereth A, Millán T (2010) Transcription factor profiling leading to the identification of putative transcription factors involved in the Medicago truncatula–Uromyces striatus interaction. Theo Appl Gen 121:1311–1321
Madrid E, Rajesh PN, Rubio J, Gil J, Millán T, Chen W (2012) Characterization and genetic analysis of an EIN4-like sequence (CaETR-1) located in QTLAR1 implicated in ascochyta blight resistance in chickpea. Plant Cell Rep 31:1033–1042
Madrid E, Chen W, Rajesh PN, Castro P, Millán T, Gil J (2013) Allele-specific amplification for the detection of ascochyta blight resistance in chickpea. Euphytica 189:183–190
Markham JE, Hille J (2001) Host-selective toxins as agents of cell death in plant–fungus interactions. Mol Plant Pathol 2:229–239
Marrs KA (1996) The functions and regulation of glutathione S-transferases in plants. Ann Rev Plant Biol 47:127–158
Matsuoka M, Ohashi Y (1986) Induction of pathogenesis-related proteins in tobacco leaves. Plant Physiol 80:505–510
McDonald BA, Linde C (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Ann Rev Phytopathol 40:349–379
Meiyalaghan S, Thomson SJ, Fiers MW, Barrell PJ, Latimer JM, Mohan S, Eirian Jones E, Conner AJ, Jacobs JM (2014) Structure and expression of GSL1 and GSL2 genes encoding gibberellin stimulated-like proteins in diploid and highly heterozygous tetraploid potato reveals their highly conserved and essential status. BMC Genom 15:2
Nasir M, Bretag TW, Kaiser WJ, Meredith KS, Brouwer JB (2000) Screening chickpea germplasm for ascochyta blight resistance. Australas Plant Pathol 29:102–107
Neill SJ, Desikan R, Clarke A, Hurst RD, Hancock JT (2002) Hydrogen peroxide and nitric oxide as signalling molecules in plants. J Exp Botany 53:1237–1247
Nene YL, Reddy MV, Saxena MC, Singh KB (1987) Chickpea diseases and their control. Chickpea 233–270
Nuruzzaman M, Sharoni AM, Kikuchi S (2013) Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol 4:248
Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Pande S, Siddique KHM, Kishore GK, Bayaa B, Gaur PM, Gowda CLL, Gowda TW, Bretag TW, Crouch JH (2005) Ascochyta blight of chickpea (Cicer arietinum L.): a review of biology, pathogenicity, and disease management. Crop Pasture Sci 56:317–332
Pandey BK, Singh US, Chaube HS (1987) Mode of infection of Ascochyta blight of chickpea caused by Ascochyta rabiei. J Phytopathol 119:88–93
Palomino C, Fernández-Romero MD, Rubio J, Torres A, Moreno MT, Millán T (2009) Integration of new CAPS and dCAPS-RGA markers into a composite chickpea genetic map and their association with disease resistance. Theor Appl Genet 118(4):671–82
Pelegrini PB, Perseghini del Sarto RP, Silva ON, Franco OL, Grossi-de-Sa MF (2011) Antibacterial peptides from plants: what they are and how they probably work. Biochem Res Int. doi:10.1155/2011/250349
Peng H, Yu XW, Cheng HY, Shi QH, Zhang H, Li JG, Ma H (2010) Cloning and characterization of a novel NAC family gene CarNAC1 from Chickpea (Cicer arietinum L.). Mol Biotechnol 44:30–40
Phan HTT, Ford R, Taylor PWJ (2003) Population structure of Ascochyta rabiei in Australia based on STMS fingerprints. Fungal Div 13:111–129
Pradhan P (2006) Studies of Ascochyta rabiei in Australia. M Agr Sci thesis, University of Melbourne
Pulse Australia (2009a) Almaz. Pulse variety management package. http://www.pulseaus.com.au/storage/app/media/crops/2009_VMP-Kchickpea-Almaz.pdf. Accessed 24 July 2015
Pulse Australia (2009b) Flipper. Pulse variety management package. http://www.pulseaus.com.au/storage/app/media/crops/2009_VMP-Dchickpea-Flipper.pdf. Accessed 24 July 2015
Pulse Australia (2009c) Genesis090. Pulse variety management package. http://www.pulseaus.com.au/storage/app/media/crops/2011_VMP-Kchickpea-Genesis090.pdf. Accessed 24 July 2015
Pulse Australia (2009d) Genesis114. Pulse variety management package. http://www.pulseaus.com.au/storage/app/media/crops/2010_VMP-Kchickpea-Genesis114.pdf. Accessed 24 July 2015
Pulse Australia (2009e) Genesis509. Pulse variety management package. http://www.pulseaus.com.au/storage/app/media/crops/2009_VMP-Dchickpea-Genesis509.pdf. Accessed 24 July 2015
Pulse Breeding Australia (2009) PBA HatTrick. http://www.pulseaus.com.au/storage/app/media/crops/2011_VMP-Dchickpea-PBAHatTrick.pdf. Accessed 24 July 2015
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Segura A, Moreno M, Madueno F, Molina A, Garcia-Olmedo F (1999) Snakin-1, a peptide from potato that is active against plant pathogens. Mol Plant Microbe Interact 12:16–23
Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, Kim J, Park CM (2010) Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J 61:661–671
Singh KB (1997) Chickpea (Cicer arietinum L.). Field Crops Res 53:161–170
Singh KB, Reddy MV (1993) Sources of resistance to ascochyta blight in wild Cicer species. Neth J Plant Pathol 99:163–167
Singh KB, Hawtin GC, Nene YL, Reddy MV (1981) Resistance in chickpea to ascochyta blight. Plant Dis 65:586–587
Staples RC, Mayer AM (2003) Suppression of host resistance by fungal plant pathogens REVIEW. Israel J Plant Sci 51:173–184
Takahashi T, Kakehi JI (2010) Polyamines: ubiquitous polycations with unique roles in growth and stress responses. Ann Bot 105:1–6
Tar’an B, Warkentin T, Tullu A, Vandenberg A (2007) Genetic relationships among Chickpea (Cicer arietinum L.) genotypes based on the SSRs at the quantitative trait loci for resistance to Ascochyta blight. Eur J Plant Pathol 119:39–51
Vaghefi N, Mustafa BM, Dulal N, Selby-Pham J, Taylor PWJ, Ford R (2013) A novel pathogenesis-related protein (LcPR4a) from lentil, and its involvement in defence against Ascochyta lentis. Phytopathol Mediterranea 52:192–201
Van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162
Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Sharpe AG, Cannon S et al (2013) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nature Biotech 31:240–246
Vera-Sirera F, Minguet EG, Singh SK, Ljung K, Tuominen H, Blázquez MA, Carbonell J (2010) Role of polyamines in plant vascular development. Plant Physiol Biochem 48:534–539
Vogelsang R, Barz W (1993) Purification, characterization and differential hormonal regulation of a β-1, 3-glucanase and two chitinases from chickpea (Cicer arietinum L.). Planta 189:60–69
Waduwara-Jayabahu I, Oppermann Y, Wirtz M, Hull ZT, Schoor S, Plotnikov AN, Sauter M, Moffatt BA (2012) Recycling of methylthioadenosine is essential for normal vascular development and reproduction in Arabidopsis. Plant Physiol 158:1728–1744
Wang H, Pang H, Bartlam M, Rao Z (2005) Crystal structure of human E1 enzyme and its complex with a substrate analog reveals the mechanism of its phosphatase/enolase activity. J Mol Biol 348:917–926
Wang XE, Basnayake BVS, Zhang H, Li G, Li W, Virk N, Mengiste T, Song F (2009) The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol Plant-Microbe Interact 22:1227–1238
Wigoda N, Ben-Nissan G, Granot D, Schwartz A, Weiss D (2006) The gibberellin-induced, cysteine-rich protein GIP2 from Petunia hybrida exhibits in planta antioxidant activity. Plant J 48:796–805
Yang Y, Shah J, Klessig DF (1997) Signal perception and transduction in plant defense responses. Genes Dev 11:1621–1639
Acknowledgments
The study was supported by an Australian Research Council-Linkage grant (LP0990385).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by D. A Lightfoot.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Leo, A.E., Linde, C.C. & Ford, R. Defence gene expression profiling to Ascochyta rabiei aggressiveness in chickpea. Theor Appl Genet 129, 1333–1345 (2016). https://doi.org/10.1007/s00122-016-2706-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2706-2