Abstract
Responsiveness to abscisic acid (ABA) during vegetative growth plays an important role in regulating adaptive responses to various environmental conditions, including activation of a number of ABA-responsive genes. However, the relationship between gene expression and responsiveness to ABA at the seedling stage has not been well studied in wheat. In the present study, quantitative trait locus (QTL) analysis for ABA responsiveness at the seedling stage was performed using recombinant inbred lines derived from a cross between common wheat cultivars showing different ABA responsiveness. Five QTLs were found to be significant, located on chromosomes 1B, 2A, 3A, 6D and 7B. The QTL with the greatest effect was located on chromosome 6D and explained 11.12% of the variance in ABA responsiveness. The other QTLs each accounted for approximately 5–8% of the phenotypic variation. Expression analyses of three ABA-responsive Cor/Lea genes, Wdhn13, Wrab15 and Wrab17, showed that allelic differences in QTLs on chromosomes 2A, 6D and 7B influenced expression of these genes in seedlings treated with ABA. The 3A QTL appeared to be involved in the regulatory system of Wdhn13 and Wrab15, but not Wrab17. The effects of the 2A and 6D QTLs on gene expression were relatively large. The combination of alleles at the QTLs resulted in an additive or synergistic effect on Cor/Lea expression. These results indicate that the QTLs influencing ABA responsiveness are associated with ABA-regulated gene expression and suggest that the QTL on chromosome 6D with the largest effect acts as a key regulator of ABA responses including seedling growth arrest and gene expression during the vegetative stage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abscisic acid (ABA), a phytohormone, regulates many important aspects of plant growth and development including seed maturation and dormancy, stress tolerance and water use (Leung and Giraudat 1998; Finkelstein et al. 2002). A greater understanding of the mechanisms of ABA-regulated gene expression has been achieved through genetic and molecular studies in Arabidopsis thaliana (Hirayama and Shinozaki 2007). The ABA-insensitive (abi) mutants of Arabidopsis are the most extensively characterized ABA response mutants (reviewed by Finkelstein et al. 2002). These mutations confer reduced seed dormancy and decreased sensitivity to exogenous ABA for inhibition of germination. ABI1 and ABI2 encode protein phosphatase 2C, whereas ABI3, ABI4 and ABI5 encode transcription factors. ABI3 and ABI5 function as intermediates in ABA signaling to regulate seed maturation and germination, as well as expression of genes that increase desiccation tolerance during embryo dehydration at a later maturational stage (Lopez-Molina et al. 2001, 2002; Carles et al. 2002). During vegetative growth, ABA also mediates adaptive responses to various environmental conditions such as drought, salt and cold (Leung and Giraudat 1998; Finkelstein et al. 2002). Endogenous ABA concentration is increased on exposure to water deficit, and this increased ABA level is considered to be an essential mediator of the plant’s response to dehydration (Leung and Giraudat 1998). ABA-mediated signal transduction pathways, acting under abiotic stresses, are organized by various components including enzymes of ABA biosynthesis, protein kinases and phosphatases, and transcription factors regulating a number of Cor (cold-responsive)/Lea (late-embryogenesis abundant) genes (Thomashow 1999; Xiong et al. 2002; Yamaguchi-Shinozaki and Shinozaki 2006).
In wheat embryos, ABA is known as a key regulator of seed dormancy (Walker-Shimons 1987). Seed dormancy is an important agronomical trait because crop species such as wheat, barley, rice and sorghum exhibit low dormancy during grain development, leading to a susceptibility to pre-harvest sprouting. Many previous quantitative trait locus (QTL) analyses of seed dormancy and pre-harvest sprouting in common wheat have found major QTLs on homoeologous group 3 and 4 chromosomes (Imtiaz et al. 2008; Kulwal et al. 2005; Osa et al. 2003; Mori et al. 2005; Kato et al. 2001; Mares et al. 2005; Torada et al. 2008; Ogbonnaya et al. 2008). Moreover, studies using aneuploid lines of common wheat showed that chromosomes 2D and 4A carry genes for embryonic sensitivity to ABA and dormancy (Noda et al. 2002). In diploid wheat, several ABA signaling genes were isolated and mapped on chromosomes 3Am and 5Am to compare the map positions with those of QTLs for seed dormancy (Nakamura et al. 2007).
ABA responsiveness during the vegetative stage has also been studied in common wheat. A major locus regulating ABA accumulation in dehydrated leaves was mapped on the long arm of chromosome 5A using a mapping population from the cross between ‘Chinese Spring’ (CS) (low ABA accumulation) and ‘SQ1’ (high ABA accumulation) (Quarrie et al. 1994). This population was also used in QTL analysis for grain yield under various environmental conditions (Quarrie et al. 2005). ABA responsiveness of seedlings was compared between the freeze-tolerant cultivar ‘Mironovskaya 808’ (M808) and CS, which is sensitive to freezing (Kobayashi et al. 2006). M808 showed a higher ABA responsiveness and accumulated more ABA-responsive Cor/Lea transcripts after exogenous ABA treatment than CS (Kobayashi et al. 2006). Mutant analyses using two common wheat lines showed a higher induction level of ABA-responsive genes in the ABA-treated seedlings of both ABA-less-sensitive and ABA-hypersensitive mutants than in their parental lines (Kobayashi et al. 2006, 2008a). These two mutants also showed a significantly higher freezing tolerance than their parental lines, at least in seedlings without cold acclimation, the process whereby plants increase their freezing tolerance (Kobayashi et al. 2006, 2008a). These results make it possible to assume that enhanced levels of ABA-induced gene expression or unknown ABA-dependent pathways are involved in the determination of the level of freezing tolerance.
Although a number of studies such as QTL analyses and isolation of ABA-responsive genes at the seed stage have been reported, information on the relationship between seedling responsiveness to ABA and expression of ABA-responsive genes during vegetative growth is still limited in wheat and related species. ABA response, including gene expression at the vegetative stage, is considered to influence the development of environmental stress tolerance. In this study, we detected QTLs associated with ABA responsiveness during seedling growth using a mapping population of recombinant inbred lines (RILs) from a cross between M808 and CS and studied expression profiles of the ABA-responsive Cor/Lea genes in RILs carrying each QTL. These analyses showed that the QTLs influenced both seedling growth inhibition and gene expression in the presence of ABA and therefore seemed to be key regulators of the ABA response in seedlings of common wheat.
Materials and methods
Plant materials
The mapping population of 210 RILs was established at the F7 generation by the single-seed descent method from an F2 family derived from the common wheat (Triticum aestivum L.) cultivars M808 and CS. M808 was bred in Mironovska Institute, Ukraine. It is reported to be one of the hardiest wheat cultivars tested (Veisz and Sutka 1990; Ohno et al. 2001; Kobayashi et al. 2004a; Kume et al. 2005), and its seedlings are more responsive to exogenous ABA than those of CS (Kobayashi et al. 2006).
Bioassay for ABA responsiveness during seedling growth
For seed germination, seeds from each line were imbibed under tap water for 5 h and kept overnight at 4°C. Imbibed seeds were placed in glass Petri dishes (90 mm in diameter and 20 mm in depth) containing filter papers (82 mm diameter) wetted with distilled water and incubated for 24 h at 20°C in the dark. Ten synchronously germinated seeds were further treated with distilled water or 20 μM ABA solution under the same conditions as for germination. After 3 days, the lengths of the shoots and primary roots were recorded. The whole experiment was repeated three times for statistical analysis.
SSR amplification and detection of polymorphisms
To amplify the PCR fragment of SSR markers, total DNA was extracted from the parents and RILs using standard procedures. A total of 102 wmc, 154 gwm, 23 gdm, 203 barc, 45 cfd, 3 cfa, 39 hbg, 16 hbe and 3 hbd (http://wheat.pw.usda.gov/GG2/index.shtml; http://nics.naro.affrc.go.jp/team/dna_marker/) primer pairs were tested for the detection of polymorphisms between CS and M808. The PCR amplification procedure was performed according to previous reports (Gupta et al. 2002; Röder et al. 1998; Pestsova et al. 2000; Song et al. 2005; Guyomarc’h et al. 2002; Torada et al. 2006). The amplification and polymorphism of the PCR products were analyzed with a high-efficiency genome scanning (HEGS) system (Nihon Eido, Tokyo, Japan) according to Hori et al. (2003).
Genetic mapping and QTL analysis
Polymorphic SSRs of the parents were genotyped and used for map construction. Genetic mapping was performed using MAPMAKER/EXP version 3.0b (Lander et al. 1987). The threshold for log-likelihood (LOD) scores was set at 3.0, and the genetic distances were calculated with the Kosambi function (Kosambi 1944). A total of 37 wmc, 83 gwm, 9 gdm, 124 barc, 21 cfd, 1 cfa, 29 hbg, 6 hbe and 3 hbd loci were used to assign chromosomes as anchor markers. Chromosomal assignment was generally conducted based on other reference maps (Gupta et al. 2002; Röder et al. 1998; Pestsova et al. 2000; Somers et al. 2004; Song et al. 2005; Guyomarc’h et al. 2002; Torada et al. 2006).
QTL analysis was carried out by composite interval mapping using Windows QTL Cartographer version 2.5 (Wang et al. 2007) with the forward and backward method. An LOD score threshold of 2.5 was determined by computing 1,000 permutations. The percentage of phenotypic variation explained by a QTL for a trait and the additive effect were also estimated using the software.
Gene expression analysis
To analyze the gene expression pattern of three Cor/Lea genes [Wdhn13 (accession number AB076807), Wrab15 (AB115913) and Wrab17 (AF255053)], 7-day-old seedlings of the 62 RILs and parental lines grown in a greenhouse under controlled temperature (23°C) and natural lighting conditions were sprayed with a solution containing 20 μM ABA and 0.1% (w/v) Tween 20. Total RNA was extracted from the seedling leaves and roots using Sepasol-RNA I (Nacalai Tesque, Kyoto, Japan). First-strand cDNA was synthesized from 1 μg of the DNaseI-treated RNA sample in a 20 μL reaction solution with oligo-dT primers using ReverTra Ace-α- (Toyobo, Osaka, Japan). The accumulation of Wdhn13, Wrab15 and Wrab17 transcripts was detected by reverse transcription (RT)-PCR amplification as previously reported (Kobayashi et al. 2006, 2008a). The transcript accumulation of each gene was also detected by real-time PCR amplification. Real-time PCR was carried out using Mx3000P (Stratagene Products Division, Agilent Technologies, Tokyo, Japan) with Brilliant II SYBR Green QPCR Master Mix (Stratagene) according to the manufacturer’s recommendations. A diluted series of pCR2.1-TOPO vectors (Invitrogen, CA, USA) containing fragments of the Cor/Lea or actin (Act) genes was used to generate the standard curve. The transcripts were amplified with the following gene-specific primer sets: 5′-AGAACCAGTGTCAGATTTCCCT-3′ and 5′-ATTCTGCAAAGTAGCGGGTC-3′ for Wdhn13, 5′-GGGATTCTTTCTTCGCGTCT-3′ and 5′-AGCCTCGGCCTTGAGTATGT-3′ for Wrab15, 5′-CGAGACGGGGCAGACCATTC-3′ and 5′-CCATCCCGAGCGTGTTCATG-3′ for Wrab17, and 5′-GCCGTGCTTTCCCTCTATG-3′ and 5′-GCTTCTCCTTGATGTCCCTTA-3′ for Act. The values for the Cor/Lea genes were normalized using Act as an internal control. The data were analyzed by Student’s t test.
Results
ABA responsiveness in parental lines and RILs at the seedling stage
ABA responsiveness of CS and M808 was evaluated based on inhibition of seedling growth by 20 μM exogenous ABA. ABA greatly reduced shoot and root length in both cultivars (Fig. 1). The magnitude of inhibition estimated by the relative root growth rate (% growth in the presence of ABA relative to growth in the absence of ABA) was significantly greater in M808 (14.4%) than in CS (36.7%), while the degree of inhibition of shoot growth was also greater in M808 (21.1%) than in CS (25.2%), but the difference was not significant statistically (Fig. 1a). This result indicated that M808 is more responsive to exogenous ABA than CS at the seedling stage, in agreement with a previous result (Kobayashi et al. 2006). Since a significant difference in the relative root growth was observed between the cultivars M808 and CS (P < 0.01), a bioassay for ABA responsiveness was performed using 210 lines of the M808 × CS RIL population by measuring the relative root growth. The RILs showed a continuous distribution of trait values, from 13.9 to 56.6%, which is in agreement with the distribution expected for a polygenic and quantitatively inherited trait, and the average value of RILs was 28.0%. Among the RILs, 30 lines were less responsive to ABA than CS, and 2 lines were more responsive than M808.
Mapping and QTL analysis
A total of 558 SSR primer sets were tested for polymorphism between CS and M808, and 301 sets (51.2%) showed polymorphism for one or two alleles. The genetic map was constructed by the segregation of 313 loci using the 210 RILs. The total map length was 2,945.4 cM with an average spacing of 9.4 cM between markers.
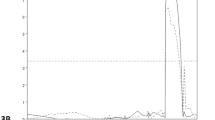
Each QTL for ABA responsiveness was detected based on the data of relative root growth and the genetic map of the RILs (Fig. S1 in ESM). Five QTLs found on chromosomes 1B, 2A, 3A, 6D and 7B showed significant LOD scores >2.5 (P < 0.05) (Figs. 2, S1). A relatively major QTL with an LOD score of 6.6 was located on chromosome 6D and contributed 11.12% of the variation in the trait (Table 1). The SSR markers Xwmc753 and Xcfd76 flanked this QTL at a 5.4 cM interval (Fig. 2). Other QTLs on chromosomes 1B, 2A, 3A and 7B explained 5.84, 5.13, 8.19 and 7.25% of the phenotypic variation, respectively (Table 1). Two QTLs on chromosomes 1B and 2A closely flanked the SSR markers Xgwm273–Xbarc240 and Xbarc208–Xgwm558, respectively, within 2-cM intervals, whereas the others on chromosomes 3A and 7B, respectively, flanked the markers Xhbg420–Xhbg227 and Xgwm333–Xbarc278 by intervals of >13cM (Fig. 2). The mean values for relative root growth of RILs carrying the M808 or CS allele at each QTL showed that RIL groups carrying the M808 allele at the 2A, 3A, 6D or 7B QTL and the group carrying the CS-type 1B QTL were more responsive to exogenous ABA than their opposite RIL groups (Table 2), indicating that M808 alleles at QTLs on chromosomes 2A, 3A, 6D and 7B, and the CS allele at the 1B QTL, contributed to higher ABA responsiveness at the seedling stage.
Expression profiles of ABA-responsive genes in M808 and CS
Exogenous ABA responsiveness for expression of Cor/Lea genes (Wdhn13, Wrab17, Wrab18 and Wrab19) and their transcription factor genes (Wdreb2, Wlip19 and Wabi5) was compared between CS and M808 in our previous studies (Kobayashi et al. 2004a, 2006, 2008b, c; Egawa et al. 2006). These results showed that exogenous ABA treatment enhanced expression of these genes, and the transcripts were more abundantly accumulated in M808 than in CS. Especially, Wdhn13 and Wrab17 showed an apparent cultivar difference in the expression levels between CS and M808 (Kobayashi et al. 2004a, 2006). In the present study, the expression profile of another Cor/Lea gene, Wrab15 (Kobayashi et al. 2004a, 2006, 2008a), was analyzed by real-time RT-PCR using ABA-treated seedlings of CS and M808. Wrab15 expression was increased by exogenous treatment with ABA, and the transcripts accumulated to reach a maximum within 2 h in M808, while the peak of transcript accumulation was observed after 5 h in CS (Fig. 3a). Although the expression level of Wrab15 at peak was almost equal in CS and M808, the transcript level was kept higher in M808 than in CS after the temporal increase (during 10–24 h) (Fig. 3a). This result showed that the expression profile of Wrab15 appeared to coincident with the higher ABA responsiveness of M808 shown by the bioassay (Fig. 1), and there was a clear cultivar difference for the expression of Wrab15 as well as those of Wdhn13 and Wrab17 (Fig. 3a).
Expression analyses of Wdhn13, Wrab15 and Wrab17 in RILs treated with exogenous ABA. a Time course of Wrab15 transcript accumulation after ABA treatment in two wheat cultivars, CS and M808, revealed by real-time RT-PCR. b Comparison of transcript accumulation levels after ABA treatment among 13 RILs grouped into five genotype classes. Numbers above the electropherograms indicate the RIL line numbers. The result was revealed by RT-PCR with the actin gene (Act) used as a control. Accumulation of Wdhn13 (c), Wrab15 (d) and Wrab17 (e) transcripts in the 13 RILs was quantified as mean values with standard deviations relative to the Act transcript by real-time PCR. Student’s t test was used to test for statistical significance (*P < 0.05) compared with the control lines, #199 and #200
Effects of QTLs on expression of ABA-responsive genes in seedlings
Comparison of the transcript levels of the ABA-responsive genes Wdreb2, Wlip19, Wabi5, Wdhn13, Wrab15, Wrab17 and Wrab18 among RILs was studied by RT-PCR using ABA-treated seedling leaves and roots. Thirteen RILs, which were classified into five genotype classes by allele type at SSR markers flanking the QTLs (Table 3), were used for the expression analysis to study the effects of each QTL on gene expression. The mean values of relative root growth showed that ABA responsiveness was higher in the genotype classes of 2A and 6D QTL than in the other classes (Table 3). No obvious differences were observed in the gene expression levels among the RILs under non-treatment condition (data not shown).
At 2 h after ABA treatment, Wdhn13, Wrab15 and Wrab17 showed relatively clear line differences in their expression levels in leaves (Fig. 3b), while other genes did not (data not shown). Since these three genes clearly showed ABA-responsive expression in the CS and M808 (Kobayashi et al. 2006 and in Fig. 3a of this study) and differences among the genotype classes (Fig. 3b), their transcript levels were also evaluated by real-time RT-PCR analysis (Fig. 3c–e). In leaves, expression levels of these three genes were significantly higher in the RILs carrying the M808 allele at the QTLs on chromosomes 2A (#5, 8 and 198) and 6D (#53, 136 and 190) than in the others (Fig. 3c–e). On the other hand, no obvious differences in gene expression levels were observed between the control lines carrying the CS alleles at all five QTLs (RIL#199 and 200) and the RILs carrying the M808 allele at the 1B QTL (#3, 172 and 194). In RILs carrying the M808 allele at the 7B QTL (#11 and 179), Wrab15 and Wrab17 showed significantly higher expression levels as compared to the control lines (Fig. 3d, e). The Wdhn13 transcript also accumulated more abundantly in the RILs carrying the M808-type 7B QTL than in the control lines (Fig. 3c). Expression analysis using the ABA-treated roots also showed the differences in the expression levels of three genes among the RILs as well in the leaves, but the differences were not significant statistically (data not shown). These expression profiles showed correlations with the mean values of ABA responsiveness in each genotype class (Table 3). There were no data regarding the effect of the QTL on chromosome 3A, because no lines of the 3A QTL-genotype class were in the RIL population.
A total of 62 lines (the 13 original lines plus 49 new lines) were used for further expression analysis to estimate the effect of the 3A QTL in addition to those of the 2A, 6D and 7B QTLs, and moreover to evaluate whether there was an additive or synergistic effect of the QTLs on gene expression. The effects of each QTL were evaluated by the mean expression values in small RIL groups, which include 13–23 RILs sorted by allele types at marker loci defining the QTLs. The transcript level of Wrab15 was significantly higher in the group carrying the M808 allele than in the opposite group carrying the CS allele at the 3A QTL (Fig. 5b), while Wdhn13 also showed higher transcript level in the group with the M808 alleles, but that difference was not significant statistically (Fig. 4b). No obvious difference was observed in the mean value of Wrab17 expression between the groups with the M808 and CS alleles (Fig. 6b). This result suggested that allelic differences at the 3A QTL affected ABA-induced expression of Wdhn13 and Wrab15. Regarding the other QTLs, the three Cor/Lea genes showed relatively higher expression levels in the RIL groups carrying the M808 allele than in each of the opposite groups carrying the CS allele (Figs. 4, 5, 6), and especially the differences in Wdhn13 and Wrab15 expression were significant with respect to the 2A and 6D QTLs (Figs. 4a, c, 5a, c). These results corresponded to the expression profiles in each genotype class (Fig. 3c–e).
Expression of Wdhn13 in RILs after exogenous ABA treatment. The expression levels evaluated by real-time PCR were compared between the small RIL groups carrying M808 (gray bars in chart) or CS (white bars in chart) alleles at the QTLs on chromosomes 2A (a), 3A (b), 6D (c) and 7B (d), and the mean expression values of each group were compared. The alleles at the QTLs in each RIL are visualized as gray (M808) and white (CS) boxes below the chart of the expression level for each RIL
Expression of Wrab15 in RILs after exogenous ABA treatment. The expression levels evaluated by real-time PCR were compared between the small RIL groups carrying M808 (gray bars in chart) or CS (white bars in chart) alleles at the QTLs on chromosomes 2A (a), 3A (b), 6D (c) and 7B (d), and the mean expression values of each group were compared. The alleles at the QTLs in each RIL are visualized as gray (M808) and white (CS) boxes below the chart of the expression level for each RIL
Expression of Wrab17 in RILs after exogenous ABA treatment. The expression levels evaluated by real-time PCR were compared between the small RIL groups carrying M808 (gray bars in chart) or CS (white bars in chart) alleles at the QTLs on chromosomes 2A (a), 3A (b), 6D (c) and 7B (d), and the mean expression values of each group were compared. The alleles at the QTLs in each RIL are visualized as gray (M808) and white (CS) boxes below the chart of the expression level for each RIL. Triangles indicate the RILs with and without the M808 allele at the 7B QTL of the genotype classes carrying the M808-type 1B, 2A and 3A QTLs (white triangles), and the genotype classes carrying the M808-type 2A and 3A QTLs (black triangles)
The gene expression levels appeared to fluctuate with combinations of the QTL alleles and, taken altogether, M808 alleles increased the accumulation levels of three Cor/Lea transcripts after ABA treatment (Figs. 4, 5, 6). A considerable alteration of expression levels was observed in several lines with or without M808 alleles at the QTLs. For instance, transcripts of the Cor/Lea genes were more abundantly accumulated in the genotype classes carrying combinations of M808 alleles at the 1B, 2A, 3A and 6D QTLs and 1B, 2A, 3A and 7B QTLs than in the other classes; however, transcript levels obviously decreased when even one of the M808 alleles at the 2A, 3A, 6D or 7B QTLs became a CS allele (Figs. 4, 5, 6). Allele changes at the four loci exerted large influences on Wdhn13 expression (Fig. 4), while Wrab15 expression in some genotype classes was clearly affected by allele changes at the 3A and 7B QTLs (Fig. 5). The most drastic alteration was observed in Wrab17 expression, which showed a sixfold or greater difference between some RILs with the M808 versus the CS allele at the 7B QTL (Fig. 6d). The alterations were specifically observed in some lines with or without the M808-type 7B QTL of the genotype classes carrying the M808-type 1B, 2A and 3A QTLs (62.7 vs. 7.3 of the expression value; indicated with white triangles in Fig. 6d) and the genotype classes carrying the M808-type 2A and 3A QTLs (8.5 vs. 1.3; indicated with black triangles in Fig. 6d). These results suggested that allele combinations at QTLs might cause additive or synergistic effects on ABA-regulated Cor/Lea expression.
Discussion
To identify the major loci for ABA responsiveness at the seedling stage, the QTL analysis was performed based on the magnitude of ABA-induced root growth arrest by the bioassay, in which the degree of growth inhibition by exogenous ABA was greater in root than in shoot and greater in M808 than CS (Fig. 1a). From the composite interval mapping, five QTLs for ABA responsiveness were found to be significant; these QTLs were mapped to chromosomes 1B, 2A, 3A, 6D and 7B (Figs. 2, S1; Table 1). M808 alleles at the 2A, 3A, 6D and 7B QTLs and the CS allele at the 1B QTL contributed to higher ABA responsiveness (Table 2).
Expression profiles of three Cor/Lea genes (Wdhn13, Wrab15 and Wrab17) were studied using ABA-treated seedlings of RILs. The results showed apparent line differences in the ABA responsiveness of gene expression among RILs, and transcripts of these genes were also abundantly accumulated in genotype classes carrying an M808 allele at one of the QTLs on chromosomes 2A, 6D or 7B (Fig. 3c–e). Moreover, the mean expression levels were greater in RIL groups with M808 alleles than in the opposite groups with CS alleles at these QTLs (Figs. 4, 5, 6). The allelic difference at the 3A QTL also affected expression levels of Wdhn13 and Wrab15 (Figs. 4b, 5b). Expression profiles showed that, regarding the 2A, 3A, 6D and 7B QTLs, the M808 alleles increased the expression levels of some Cor/Lea genes (Figs. 3, 4, 5, 6). These results suggest a possibility that QTLs on chromosomes 2A, 3A, 6D and 7B are involved in regulation of both root growth arrest and Cor/Lea expression in ABA-treated seedlings, but the contribution levels on these ABA responses were different among the QTLs. The 2A and 6D QTLs had a significantly larger effect on gene expression (Figs. 3c–e, 4, 5, 6), suggesting that these two QTLs are essential for ABA-regulated Cor/Lea expression. Moreover, a gene located at the 6D QTL may be a key regulator of ABA response during the seedling stage, because the 6D QTL demonstrated the largest contribution to phenotypic variation of the five QTLs (Table 1). In contrast, allelic difference at the 1B QTL did not affect the accumulation of Wdhn13, Wrab15 and Wrab17 transcripts (Fig. 3c–e). This QTL may regulate the expression of other ABA-responsive genes involved in seedling growth and associated with root growth inhibition by ABA.
Our previous transgenic and transient expression studies showed that four transcription factors, WCBF2, WDREB2, WLIP19 and WABI5, are involved in the positive regulation of expression of several Cor/Lea genes (Takumi et al. 2008; Kobayashi et al. 2008b, c, d). Wdhn13 expression was under the control of these four transcription factors, while Wrab17 was regulated by WCBF2, WDREB2 and WLIP19, but not by WABI5 (Takumi et al. 2008; Kobayashi et al. 2008b, c, d). WDREB2, WLIP19 and WABI5 are considered to act in ABA signaling pathways (Kobayashi et al. 2008b, c, d). In the present expression study using RILs, the alteration of Wdhn13, Wrab15 and Wrab17 expression levels suggests that candidate genes for the QTLs are upstream factors, including transcription factors, in ABA signaling pathways, because the three Cor/Lea genes are downstream genes in the ABA signaling pathway of common wheat. However, the transcription factor genes Wdreb2, Wlip19 and Wabi5 are located on homoeologous groups 1 and 5 chromosomes (Egawa et al. 2006; Kobayashi et al. 2008b, c) and showed no obvious differences in expression levels among RILs (data not shown). Therefore, genes located at the QTLs on chromosomes 2A, 3A, 6D and 7B seem to encode other factors involved in the regulation of Cor/Lea expression. In Arabidopsis, proteins that affect ABA responses include RNA binding proteins (RBPs) involved in RNA processing, including that of double-strand RNA, coinciding with the observations of micro or small RNAs (reviewed by Himmelbach et al. 2003; Kuhn and Schroeder 2003; Sunkar et al. 2007; Hirayama and Shinozaki 2007; Lorkovic 2009). In addition to the RBPs involved in RNA metabolism, factors for protein modification such as protein phosphatase, protein kinase and RING-finger ubiquitin E3 ligase are also involved in ABA regulation of protein processing (Himmelbach et al. 2003; Hirayama and Shinozaki 2007; Santner and Estelle 2009). Thus, it is possible that candidate genes at the QTLs encode factors controlling RNA and protein metabolism and regulating transcription factors, including WDREB2, WLIP19 and WABI5, in the ABA signaling pathways of common wheat. Since the 2A and 6D QTLs had large effects on the expression of Wdhn13, Wrab15 and Wrab17 (Figs. 3c–e, 4, 5, 6), these two loci may be implicated in the regulation of multiple transcription factors such as WDREB2, WLIP19 and WABI5 in the presence of ABA, resulting in activation of downstream genes including the Cor/Lea genes. Allelic differences at the 7B QTL also affected the expression levels of Wdhn13, Wrab15 and Wrab17 (Figs. 3c–e, 4, 5, 6). Association of the 7B QTL with the control of WDREB2, WLIP19 and WABI5 is therefore suggested; however, the association appeared to be small, because the effect of this locus was relatively minor compared with those of the 2A and 6D QTLs (Figs. 3c–e, 4, 5, 6). The 3A QTL appears to be involved in the regulation of Wdhn13 and Wrab15, but not in that of Wrab17 (Figs. 4b, 5b, 6b), the expression of which is not under control of WABI5 (Kobayashi et al. 2008c). Therefore, 3A QTL may be associated with WABI5-mediated regulation of gene expression, including that of Wdhn13 and Wrab15. The association of the QTLs with regulation of transcription factors at the posttranscriptional or posttranslational levels should be clarified in future studies.
Combinations of the QTL alleles influenced accumulation levels of Wdhn13, Wrab15 and Wrab17 transcripts (Figs. 4, 5, 6). The alterations as a whole represented an increase of expression levels caused by additive effects of M808 alleles. A considerable increase of Wrab17 expression was induced by the addition of the M808 allele at the 7B QTL (Fig. 6d). Although 7B QTL had a relatively minor effect on Wrab17 expression (Fig. 3e), the allele combination of 7B QTL with other loci such as 2A and 3A QTLs in some lines might cause a synergistic effect in the activation of Wrab17 (Fig. 6d). These observations suggest that the QTLs act cooperatively in the activation of ABA-regulated Cor/Lea expression, and that this cooperation may bring about synergistic effects in the activation of gene expression. The data from expression analyses suggest interactions among the QTLs; however, no significant epistatic interactions among the loci were detected from multiple interval mapping analyses (data not shown). These results suggest that gene expression and seedling growth inhibition under ABA-treated condition are controlled by additive effects and by cooperation among the QTLs, leading to a synergistic effect.
Previous QTL analyses for seed dormancy and pre-harvest sprouting in wheat detected a large number of QTLs on all 21 chromosomes of hexaploid wheat. Comparison between the QTL maps showed that each locus for ABA responsiveness in the present study appeared to overlap with at least one locus for seed dormancy or pre-harvest sprouting (Figs. 7, S2, S3, S4, S5). The largest QTL on chromosome 6D was in the vicinity of the pre-harvest sprouting locus QPhs.cnl-6D.1 (Fig. 7; Munkvold et al. 2009). The closest marker flanking the peak of QPhs.cnl-6D.1 was the SSR marker Xcfd37 that was involved in the 6D QTL region in the present study (Fig. 7). QPhs.cnl-6D.1 was detected as a significant locus for pre-harvest sprouting; however, no QTLs for seed dormancy were detected around QPhs.cnl-6D.1 (Munkvold et al. 2009). These results suggest that this chromosomal region may be associated with seedling-specific ABA responsiveness, but there is no direct evidence for a relationship between QPhs.cnl-6D.1 and ABA sensitivity in seeds. The 3A QTL was related to QTLs for seed dormancy, including the minor locus QPhs.ocs-3A.2 and two QTLs on chromosome 3Am in diploid wheat (Fig. S4; Osa et al. 2003; Nakamura et al. 2007). The two minor QTLs on chromosome 3Am were co-localized with the ABA signaling genes TmABI8 and TmABF (Nakamura et al. 2007). Therefore, these genomic regions on chromosome 3A in diploid and hexaploid wheat may influence ABA responsiveness during both the seed and seedling stages. The other QTLs on chromosomes 1B, 2A and 7B were also located around the QTL regions for pre-harvest sprouting (Figs. S2, S3, S5) (Zanetti et al. 2000). However, these overlapping loci were relatively minor QTLs for seed dormancy. These findings suggest that the five QTLs function predominantly in the vegetative stage, and that the ABA responsiveness in seedlings is controlled by a genetic mechanism distinct from that at the seed stage.
Comparative maps of QTL regions for ABA responsiveness, pre-harvest sprouting and cold tolerance on wheat chromosome 6D. The QTL region for ABA responsiveness detected in the present study is indicated as a striped bar. The black triangle shows the peak of the QTL. The positions of the QTLs for pre-harvest sprouting (Munkvold et al. 2009) and cold tolerance (Båga et al. 2007) are indicated by gray and black boxes, respectively
ABA contributes to tolerance of abiotic stresses such as drought, salt and cold (Leung and Giraudat 1998; Finkelstein et al. 2002; Xiong et al. 2002). Mutant analyses of common wheat showed that ABA responsiveness is not associated with cold acclimation, though ABA is involved in determination of the basal level of freezing tolerance (Kobayashi et al. 2006, 2008a). Two major loci for winter hardiness, Fr-1 (frost resistance 1) and Fr-2, were identified on homoeologous group 5 chromosomes in wheat and barley (Cattivelli et al. 2002; Galiba et al. 2009). However, the Fr genes are presumed to be involved in ABA-independent regulatory systems (Kobayashi et al. 2004b; Kobayashi and Takumi 2007). QTL analyses to identify genomic regions for low temperature tolerance other than the Fr loci found a major locus on the short arm of chromosome 6A (Börner et al. 2002) and minor loci on chromosomes 1D, 2A, 2B, 6D and 7B (Båga et al. 2007). Among these loci, a QTL peak on chromosome 6D was detected at the locus of the SSR marker Xcfd76 by single marker analysis (Båga et al. 2007). This was the closest marker flanking the QTL for ABA responsiveness (Figs. 2, 7), while other QTLs on chromosomes 2A and 7B did not overlap with the loci for ABA responsiveness (Figs. S3, S5). The overlap of the genomic region around the 6D QTL suggests a relationship between ABA responsiveness and freezing tolerance.
In this study, we used root tissue for the QTL analysis and shoot tissue for the gene expression study. However, we believe that most of the detected QTLs are involved in the ABA responsiveness of both shoot and root, because expression of the three Cor/Lea genes in the seedling leaves was affected by the allelic differences at the QTLs on chromosomes 2A, 3A, 6D and 7B (Figs. 3, 4, 5, 6). The difference in the responsiveness to ABA between shoot and root shown in Fig. 1a may result from the effects of some minor factors that function in a tissue-specific manner. To improve the correlation between the growth inhibition and transcript assays, we aim to carry out QTL and gene expression studies using the same tissue in our future work, to reveal the detailed mechanism of ABA responsiveness in wheat growth. Moreover, expression data from the RILs was not clear because the genetic background was not uniform except for the QTL region in each RIL. Therefore, near-isogenic lines for each QTL should be produced and used in further analyses, including expression studies, to reveal the roles of each QTL.
References
Båga M, Chodaparambil SV, Limin AE, Pecar M, Fowler DB, Chibbar RN (2007) Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Funct Integr Genomics 7:53–68
Börner A, Schumann E, Fürste A, Cöster H, Leithold B, Röder MS, Weber WE (2002) Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor Appl Genet 105:921–936
Carles C, Bies-Etheve N, Aspart L, Leon-Kloosterziel KM, Koonneef M, Echeverria M, Delseny M (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30:373–383
Cattivelli L, Baldi P, Crosatti C, Di Fonzo N, Faccioli P, Grossi M, Mastrangelo AM, Pecchioni N, Stanca AM (2002) Chromosome regions and stress-related sequences involved in resistance to abiotic stress in Triticeae. Plant Mol Biol 48:649–665
Egawa C, Kobayashi F, Ishibashi M, Nakamura T, Nakamura C, Takumi S (2006) Transcript accumulation and alternative splicing of a DREB2 homolog are differentially regulated by cold and drought stresses in common wheat. Genes Genet Syst 81:77–91
Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl):S15–S45
Galiba G, Vágújfalvi A, Li C, Soltész A, Dubcovsky J (2009) Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci 176:12–19
Gupta PK, Balyan HS, Edwards KJ, Isaac P, Korzun V, Röder M, Gautier MF, Joudrier P, Schlatter AR, Dubcovsky J, DelaPena RC, Khairallah M, Penner G, Hayden MJ, Sharp P, Keller B, Wang RCC, Hardouin JP, Jack P, Leroy P (2002) Genetic mapping of 66 new microsatellite (SSR) loci in bread wheat. Theor Appl Genet 105:413–422
Guyomarc’h H, Sourdille P, Charmet G, Edwards KJ, Bernard M (2002) Characterization of polymorphic microsatellite markers from Aegilops tauschii and transferability to the D-genome of bread wheat. Theor Appl Genet 104:1164–1172
Himmelbach A, Yang Y, Grill E (2003) Relay and control of abscisic acid signaling. Curr Opin Plant Biol 6:470–479
Hirayama T, Shinozaki K (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12:344–351
Hori K, Kobayashi T, Shimizu A, Sato K, Takeda K, Kawasaki S (2003) Efficient construction of high-density linkage map and its application to QTL analysis in barley. Theor Appl Genet 107:806–813
Imtiaz M, Ogbonnaya FC, Oman J, Van Ginkel M (2008) Characterization of quantitative trait loci controlling genetic variation for preharvest sprouting in synthetic backcross-derived wheat lines. Genetics 178:1725–1736
Kato K, Nakamura W, Tabiki T, Miura H (2001) Detection of loci controlling seed dormancy on group 4 chromosomes of wheat and comparative mapping with rice and barley genomes. Theor Appl Genet 10:980–985
Kobayashi F, Takumi S (2007) Contribution of ABA signal pathways on development of freezing tolerance in wheat. Curr Top Plant Biol 8:33–43
Kobayashi F, Takumi S, Nakata M, Ohno R, Nakamura T, Nakamura C (2004a) Comparative study of the expression profiles of the Cor/Lea gene family in two wheat cultivars with contrasting levels of freezing tolerance. Physiol Plant 120:585–594
Kobayashi F, Takumi S, Nakamura C (2004b) Regulation of cold-responsive Cor/Lea genes and their transcription factors by the major freezing tolerance locus Fr-1 in wheat. Recent Res Dev Plant Sci 2:249–266
Kobayashi F, Takumi S, Egawa C, Ishibashi M, Nakamura C (2006) Expression patterns of low temperature responsive genes in a dominant ABA-less-sensitive mutant line of common wheat. Physiol Plant 127:612–623
Kobayashi F, Takumi S, Nakamura C (2008a) Increased freezing tolerance in an ABA-hypersensitive mutant of common wheat. J Plant Physiol 165:224–232
Kobayashi F, Maeta E, Terashima A, Kawaura K, Ogihara Y, Takumi S (2008b) Development of abiotic stress tolerance via a bZIP-type transcription factor LIP19 in common wheat. J Exp Bot 59:891–905
Kobayashi F, Maeta E, Terashima A, Takumi S (2008c) Positive role of an HvABI5 homolog in abiotic stress response of wheat seedlings. Physiol Plant 134:74–86
Kobayashi F, Ishibashi M, Takumi S (2008d) Transcriptional activation of Cor/Lea genes and increase in abiotic stress tolerance through expression of a wheat DREB2 homolog in transgenic tobacco. Transgenic Res 17:755–767
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Kuhn JM, Schroeder JI (2003) Impacts of altered RNA metabolism on abscisic acid signaling. Curr Opin Plant Biol 6:463–469
Kulwal PL, Kumar N, Gaur A, Khurana P, Khurana JP, Tyagi AK et al (2005) Mapping of a major QTL for pre-harvest sprouting tolerance on chromosome 3A in bread wheat. Theor Appl Genet 111:1052–1059
Kume S, Kobayashi F, Ishibashi M, Ohno R, Nakamura C, Takumi S (2005) Differential and coordinated expression of Cbf and Cor/Lea genes during long-term cold acclimation in two wheat cultivars showing distinct levels of freezing tolerance. Genes Genet Syst 80:185–197
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49:199–222
Lopez-Molina L, Mongrand S, Chua NH (2001) A post-germination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci 98:4782–4787
Lopez-Molina L, Mongrand S, McLachlin D, Chait B, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32:1–12
Lorkovic ZJ (2009) Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci 14:229–236
Mares D, Mrva K, Cheong J, Williams K, Watson B, Storlie E et al (2005) A QTL located on chromosome 4A associated with dormancy in white- and red-grained wheats of diverse origin. Theor Appl Genet 111:1357–1364
Mori M, Uchino N, Chono M, Kato K, Miura H (2005) Mapping QTLs for grain dormancy on wheat chromosome 3A and the group 4 chromosomes, and their combined effect. Theor Appl Genet 110:1257–1266
Munkvold JD, Tanaka J, Benscher D, Sorrells ME (2009) Mapping quantitative trait loci for preharvest sprouting resistance in white wheat. Theor Appl Genet 119:1223–1235
Nakamura S, Komatsuda T, Miura H (2007) Mapping diploid wheat homologues of Arabidopsis seed ABA signaling genes and QTLs for seed dormancy. Theor Appl Genet 114:1129–1139
Noda K, Matsuura T, Maekawa M, Taketa S (2002) Chromosomes responsible for sensitivity of embryo to abscisic acid and dormancy in wheat. Euphytica 123:203–209
Ogbonnaya FC, Imtiaz M, Ye G, Hearnden PR, Hernandez E, Eastwood RF et al (2008) Genetic and QTL analyses of seed dormancy and preharvest sprouting resistance in the wheat germplasm CN10955. Theor Appl Genet 116:891–902
Ohno R, Takumi S, Nakamura C (2001) Expression of a cold-responsive Lt-Cor gene and development of freezing tolerance during cold acclimation in wheat (Triticum aestivum L.). J Exp Bot 52:2367–2374
Osa M, Kato K, Mori M, Shindo C, Torada A, Miura H (2003) Mapping QTLs for seed dormancy and the Vp1 homologue on chromosome 3A in wheat. Theor Appl Genet 106:1491–1496
Pestsova E, Ganal MW, Röder M (2000) Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome 43:689–697
Quarrie SA, Gulli M, Calestani C, Steed A, Marmiroli N (1994) Location of a gene regulating drought-induced abscisic acid production on the long arm of chromosome 5A of wheat. Theor Appl Genet 89:794–800
Quarrie SA, Steed A, Calestani C, Semikhodskii ALebreton C, Chinoy C et al (2005) A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring × SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor Appl Genet 110:865–880
Röder M, Korzun V, Wendehake K, Plaschke J, Tixer MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Santner A, Estelle M (2009) Recent advances and emerging trends in plant hormone signaling. Nature 459:1071–1078
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Song QJ, Shi JR, Singh S, Fickus EW, Costa JM, Lewis J, Gill BS, Ward R, Cregan PB (2005) Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet 110:550–560
Sunkar R, Chinnusamy V, Zhu J, Zhu JK (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12:301–309
Takumi S, Shimamura C, Kobayashi F (2008) Increased freezing tolerance through up-regulation of downstream genes via the wheat CBF gene in transgenic tobacco. Plant Physiol Biochem 46:205–211
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599
Torada A, koike M, Mochida K, Ogihara Y (2006) SSR-based linkage map with new markers using an intraspecific population of common wheat. Theor Appl Genet 112:1042–1051
Torada A, Koike M, Ikeguchi S, Tsutsui I (2008) Mapping of a major locus controlling seed dormancy using backcrossed progenies in wheat (Triticum aestivum L.). Genome 51:426–432
Veisz O, Sutka J (1990) Frost resistance studies with wheat in natural and artificial conditions. In: Panayotov I, Pavlova S (eds) Proceedings of the international symposium on cereal adaptation to low temperature stress, Albena, Bulgaria, pp 12–17
Walker-Shimons MK (1987) ABA levels and sensitivity in developing wheat embryo of sprouting resistant and susceptible cultivars. Plant Physiol 84:61–66
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14(Suppl):S165–S183
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Zanetti S, Winzeler M, Keller M, Keller B, Messmer M (2000) Genetic analysis of pre-harvest sprouting resistance in a wheat × spelt cross. Crop Sci 40:1406–1417
Acknowledgments
We are grateful to Dr. K. Hori for the kind help with genetic mapping and QTL analysis. We also thank Ms. E. Okano for her skillful technical assistance. We wish to acknowledge Dr. N. Iriki for providing the wheat SSR marker information. We are also grateful to Dr. C. Nakamura for providing seeds for the mapping population. This work was supported by a Grant-in-Aid for Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists to F. K.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Snape.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kobayashi, F., Takumi, S. & Handa, H. Identification of quantitative trait loci for ABA responsiveness at the seedling stage associated with ABA-regulated gene expression in common wheat. Theor Appl Genet 121, 629–641 (2010). https://doi.org/10.1007/s00122-010-1335-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1335-4