Abstract
In higher plants, formate dehydrogenase (FDH, EC1.2.1.2.) catalyzes the NAD-linked oxidation of formate to CO2, and FDH transcript accumulation has been reported after various abiotic stresses. By sequencing a Phaseolus vulgaris BAC clone encompassing a CC-NBS-LRR gene rich region of the B4 resistance gene cluster, we identified three FDH-encoding genes. FDH is present as a single copy gene in the Arabidopsis thaliana genome, and public database searches confirm that FDH is a low copy gene in plant genomes, since only 33 FDH homologs were identified from 27 plant species. Three independent prediction programs (Predotar, TargetP and Mitoprot) used on this large subset of 33 plant FDHs, revealed that mitochondrial localization of FDH might be the rule in higher plants. A phylogenetic analysis suggests a scenario of local FDH gene duplication in an ancestor of the Phaseoleae followed by another more recent duplication event after bean/soybean divergence. The expression levels of two common bean FDH genes under different treatments were investigated by quantitative RT-PCR analysis. FDH genes are differentially up-regulated after biotic and abiotic stresses (infection with the fungus Colletotrichum lindemuthianum, and dark treatment, respectively). The present study provides the first report of FDH transcript accumulation after biotic stress, suggesting the involvement of FDH in the pathogen resistance process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are exposed to a large variety of environmental stresses including abiotic stresses such as drought or extreme temperatures, and biotic stresses caused by pathogens. To survive, plants have evolved mechanisms to perceive these stresses in order to respond to these adverse environmental conditions (Fujita et al. 2006). Concerning biotic stresses, biotroph pathogen perception is often conditioned by major resistance (R) genes following the classic gene-for-gene model (Flor 1955). Most plant R genes cloned to date belong to the Nucleotide Binding Site (NBS)-Leucine Rich Repeat (LRR) gene family (Martin et al. 2003; Belkhadir et al. 2004; Rairdan and Moffett 2007). R genes belonging to this class have been identified in various plant species (monocotyledons as well as dicotyledons) and correspond to R genes effective against all types of pathogens and pests including fungi, bacteria, viruses, nematodes, oomycetes, and insects (Dangl and Jones 2001; Hammond-Kosack and Parker 2003; McHale et al. 2006).
R proteins have two functions: they are ‘sensors’ that directly or indirectly detect pathogens, and they activate downstream signaling leading to pathogen resistance (Chisholm et al. 2006). Upon pathogen recognition, R proteins trigger defence responses that often result in the so-called hypersensitive response (HR), a rapid induction of host cell death localized at the site of invasion of the pathogen (Hammond-Kosack and Jones 1996). This plant response is associated with massive cellular ion fluxes, activation of mitogen-activated protein (MAP) kinase (K) cascades, generation of reactive oxygen species (ROS), and strengthening of cell walls as well as the induction of the expression of many defence genes, including pathogenesis-related (PR) genes (Hammond-Kosack and Jones 1996; van Loon et al. 2006; Desveaux et al. 2006; Grant et al. 2006). Furthermore, evidence of complex interplay between defence molecules such as salicylic acid (SA), ethylene (ET), jasmonic acid (JA) and nitric oxide is emerging (Hammond-Kosack and Jones 1996; Hammond-Kosack and Parker 2003; Robert-Seilaniantz et al. 2007). During the last decade, several studies have reported evidence for points of convergence in biotic and abiotic stress signalling networks (Sessa et al. 1995; Grant et al. 2003; Chini et al. 2004; Agarwal et al. 2006; Cao et al. 2006; Jiang et al. 2008). Hormone, ROS signalling pathways and activation of MAPK cascade are common plant responses to both types of stresses, suggesting that resistance against biotic and abiotic stresses at least partially share common pathways (Fujita et al. 2006). However, as the molecular mechanisms involved in these stresses were analyzed mostly independently, convergence points between biotic and abiotic stress signaling processes remain elusive (Fujita et al. 2006). Therefore, further studies are needed to understand the key factor(s) governing the connection between the signaling pathways involved in response to biotic and abiotic stresses.
Formate dehydrogenase (FDH) is a widespread enzyme found in various organisms such as bacteria, fungi, yeast, and plants. FDH catalyzes the oxidation of formate (HCOO−) into CO2. Different types of FDHs have been described; however, only the NAD-dependent type (EC 1.2.1.2) has been reported in plants. NAD-dependent FDHs have been shown to provide NADH to the respiratory chain (Popov and Lamzin 1994). Historically, the first plant FDH activity was described in common bean (Phaseolus vulgaris) seeds (Thunberg 1921). In the Arabidopsis thaliana genome, only one FDH encoding gene has been identified, suggesting that FDH is a low copy gene in plant genomes (Li et al. 2001). In plants, FDH is nuclear-encoded as a precursor containing an NH2-terminal extension signal peptide, which contains the organelle targeting information. Direct evidence for the Solanum tuberosum FDH targeting and activity into mitochondria has been given (Colas des Francs-Small et al. 1993; Ambard-Bretteville et al. 2003a); however, FDH activity has also been detected in the chloroplasts of A. thaliana (Olson et al. 2000; Herman et al. 2002). FDH transcript accumulation has been reported under various abiotic stresses including hypoxia, chilling, drought, dark and wounding in potato, and Arabidopsis leaves (Hourton-Cabassa et al. 1998; Li et al. 2001) as well as iron deficiency and anaerobia in barley root (Suzuki et al. 1998). At the protein level, FDH activity has been reported in various organs such as pea pods, developing seeds, mature seeds, embryo, seedling and root nodule (Davison 1949), and potato tuber where it is one of the major mitochondrial proteins (Colas des Francs-Small et al. 1993). FDH protein accumulation and increased activity were observed in barley roots under iron deficiency (Suzuki et al. 1998) and in potato tuber where an increase in FDH activity was correlated with a decrease in oxygen concentration (Bykova et al. 2003). However, no evidence for the implication of FDH in biotic stress has been reported so far.
In common bean, a complex R gene cluster referred to as the B4 R gene cluster has been identified at the end of linkage group (LG) B4 (Geffroy et al. 1999). Specific R genes against the fungus Colletotrichum lindemuthianum (Geffroy et al. 1999, 2008) and R quantitative trait loci (QTL) against C. lindemuthianum and Bean yellow mosaic virus (Geffroy et al. 2000; Lopez et al. 2003) as well as expressed Coiled-coil-NBS-LRR (CNL) encoding genes (Ferrier Cana et al. 2003; Ferrier Cana et al. 2005) were mapped to the B4 R gene cluster. In preparation for sequencing of the B4 R gene cluster in common bean, we generated a physical map based on bacterial artificial chromosome (BAC) clones from the Meso-American genotype BAT93 (David et al. 2008). This analysis revealed that the B4 R gene cluster is a very large cluster spanning several megabases and that CNL sequences were heterogeneously distributed across the B4 R gene cluster, with several regions extremely rich in CNL sequences separated by large intervening regions with few or no CNL sequences.
In the present study we report the sequencing and gene annotation of one BAC clone (105 kb) of common bean, mapped at the B4 R gene cluster (David et al. 2008). We identify 17 putative genes of which four CNL and three full-length formate dehydrogenase (FDH) encoding genes (FYN24G.15, FYN24G.16 and FYN24G.17). Database searches confirm that FDH is a low copy gene in plant genomes and prediction programs suggest that mitochondrial localization of FDH might be the rule in higher plants. Expression analyses are presented for two of the three common bean FDH (FYN24G.15 and FYN24G.16) under dark stress and under pathogen infection with the fungus Colletotrichum lindemuthianum. It revealed that, although these two genes are very similar, they are differentially expressed and respond to distinct stresses, i.e., dark stress for FYN24G.15 and infection with an avirulent strain of C. lindemuthianum infection for FYN24G.16.
Materials and methods
Biological materials and plant treatments
Seeds of Phaseolus vulgaris genotype BAT93 were germinated and grown for 8 days as described previously (Geffroy et al. 1999). For the biotic stress, the two cotyledonary leaves of BAT93 seedlings were sprayed, 8 days after germination, with M126 (virulent) or C531 (avirulent) Colletotrichum lindemuthianum conidia according to Geffroy et al. (1999). Anthracnose symptoms appear 96 h after infection with the virulent strain. Control plants were sprayed with water and incubated in a control growth chamber under the same temperature (19°C) and humidity conditions (90% relative humidity) as the infected plants. For the dark treatment, 8-day-old BAT93 seedlings were placed in a dark growth chamber at 19°C, for 72 h. After 72 h in the dark, the plants were re-exposed to light for 2 days. Control plants were placed in a growth chamber under the same conditions, except that they were exposed to the fluorescent tubes (OSRAM L, 58W/830) providing light at a fluence of 166 μmol m−2 s−1. One hundred mg of cotyledonary leaves from three different plants were harvested at 10, 24, 36, 48, 72, and 96 h after C. lindemuthianum infection and at 10, 24, 36, 48, 72, 96, and 120 h after the dark treatment initiation, as well as from the control plants and immediately frozen in liquid N2 for mRNA isolation. To provide replicate RNA samples for Real-time quantitative PCR assays, each time-course was repeated twice.
BAC library screening, sequencing, and sequence annotation
A BAT93 Phaseolus vulgaris BAC library (Kami et al. 2006) was screened with PRLJ1, a NBS probe previously shown to be specific for the B4 R gene cluster (Geffroy et al. 1999; Kami et al. 2006). The positive BAC clones were organized in contigs (David et al. 2008) and BAC clone FY-N24 was selected for sequencing. Lark Genaissance (UK) sequenced 768 random clones of P. vulgaris BAT93 BAC FY-N24 from a shotgun library with inserts of 1–3.5 kb, giving approximately an eightfold sequence coverage. Since no gene prediction program specifically trained for common bean is yet available, the annotation was done manually, using a combination of gene-finding programs and sequence homology with known plant genes and proteins.
First, the FY-N24 sequence was analyzed for repetitive elements and transposon sequences using RepeatMasker (Smit and Green, http://repeatmasker.org) and The Institute for Genomic Research (TIGR) Arabidopsis repetitive element database. Second, gene models were predicted using the ab initio gene finder FGENESH (Burset and Guigo 1996) and GeneMark.hmm (Lukashin and Borodovsky 1998) using the Arabidopsis model. Additionally, BLASTX (Altschul et al. 1997) algorithm was used to search for homologous protein sequences against the Genbank non-redundant (NR) database. Third, to find correct predictions of exon/intron borders, the genomic sequence of FY-N24 was extracted to perform BLASTN analyses against all the expressed sequence tags (ESTs) available at GenBank (25 August 2008) from P. vulgaris (83,348) (Ramirez et al. 2005), P. coccineus (20,120), P. acutifolius (739), and P. angustissimus (6,672). The resulting list of ESTs overlapping with FY-N24 genomic sequence was passed onto a custom script that automatically retrieves the sequences of all matches and performs an in-depth verification of each EST using Sim4 (Florea et al. 1998). Since the three common bean FDH sequences present a high level of nucleotide identity, EST assignment to each of them was deduced from careful manual inspection of multiple alignments established with CLUSTALX version 1.83 (Thompson et al. 1994) between EST and putative FDH coding sequences. The same procedure was used for EST assignment to CNL sequences. This information was compiled with ARTEMIS (Rutherford et al. 2000) to decide on final gene structure. The resulting predicted protein sequences were used to query the PFAM database (Bateman et al. 2004) for domain prediction. The FY-N24 BAC sequence has been deposited in the NCBI database under accession number EU931620.
Predicted genes are named with the FY-N24 BAC name followed by the letter “G” for gene and X, where X is a number (from 1 to 17) corresponding to the order of the genes along the FY-N24 BAC sequence. Among the 17 annotated genes 4 correspond to CC-NBS-LRR (CNL) encoding genes, and were referred to as CNL-B8 to CNL-B11 to comply with a previous nomenclature. CNLs corresponding to pseudogene are presented with an asterisk (*) after their name. These pseudogenes contain a start methionine but present frameshift(s) or premature stop codon(s) leading to truncated predicted proteins compared with “full length” encoding CNL.
Database searches and sequence analyses
A set of nucleotide FDH sequences from other plants was identified with a TBLASTN search using the bean FDH FYN24G.16 as a query sequence against (1) the GenBank NR database (cut off evalue 1e-50, 30 March 2007) (2) the soybean whole genome shotgun (WGS) sequence (cut off evalue 1e-50, 7x draft assembly, 28 January 2008, http://phytozome.net) (3) the PUT (Putative unit transcript) of plantGDB [cut off evalue 1e-30, 30 March 2007, (Dong et al. 2004)] and the GenBank database viridiplantae ESTs division (cut off evalue 1e-30, 30 March 2007). Matching ESTs of the same species were assembled using cap3 (Huang and Madan 1999), and resulting contigs were checked manually in order to prevent wrong assignments (Supplementary Table 1). The identified FDH DNA sequences were translated with the MacMolly program (version 3.5.1, Soft gene GmbH) and only full length FDH sequences were retained for further analysis.
Predictions for protein subcellular localizations were performed using Predotar (Small et al. 2004), TargetP (Emanuelsson et al. 2007) and Mitoprot (Claros and Vincens 1996) software.
Nucleotide identities and amino acid similarities were obtained using bl2seq (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi). For phylogenetic analyses, nucleotide sequences of the 33 plant FDH sequences were aligned with CLUSTALX version 1.83 (Thompson et al. 1994) and edited in GENEDOC for manual adjustments (http://www.psc.edu/biomed/genedoc). This alignment was analyzed using Bayesian phylogenetic methods as implemented in MrBayes V. 3.1.2 (Huelsenbeck and Ronquist 2001). MrBayes analyses were run with the nucleotide model 4by4, following a gamma distribution. We used the Metropolis-coupled Markov Chains Monte Carlo method to make two runs of four chains. We ran 10 million generations, sampling every 100 generations. A consensus tree with branch lengths and posterior estimates of branch probabilities was generated with the “sumt” command of MrBayes and “burnin” parameter of 50,000. The resulting phylogenetic tree is displayed with the MEGA4.1 software (Tamura et al. 2007).
RNA extraction, cDNA preparation and amplification, cloning and sequencing of RT-PCR products
Extractions of total RNA were carried out with the Nucleospin RNA plant Kit according to manufacturer’s instructions (Macherey–Nagel, Düren, Germany). cDNA preparation was performed as described in Langlois-Meurinne et al. (2005). To confirm the FDH gene structure, three primer pairs were designed upstream of the ATG (forward oligonucleotide) and downstream of the stop codon (reverse oligonucleotide) of each of the three bean FDH nucleic sequences (Table 1). They were used to amplify fragments of 1,319; 1,370; and 1,313 bp of the first-strand cDNA product of FYN24G.17, FYN24G.16, and FYN24G.15, respectively, with the Advantage-HF 2 PCR Kit according to manufacturer’s instructions (BD Biosciences, Moutain View, CA, USA). PCR products were separated on a 1.5% agarose gel and visualized with ethidium bromide staining under ultraviolet light. Fragments of the expected size were excised from the gel, purified using Nucleospin extract 2 kit according to manufacturer’s instructions (Macherey–Nagel, Düren, Germany), and cloned with the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA, USA). Plasmid DNA of positive clones was purified using the QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA, USA) and sequenced using an automated 373A DNA sequencer and the Big Dye dideoxy chain Terminatorcycle sequencing Kit (Perkin Elmer, Applied Biosystems, Roissy, France).
Expression analysis
All primers used in the Real-time quantitative PCR (RT-qPCR) experiments are reported in Table 1. Two primer pairs specific for FYN24G.15 and FYN24G.16, respectively, were defined (Supplementary Fig. 1). However, no primer pairs following manufacturer instructions and specific to FYN24G.17 could be defined. RT-qPCR analysis of bean FDH was carried out using a Light-Cycler 480 instrument and Light CyclerR 480 SYBR Green Master mix (Roche Diagnostics, Mannheim, Germany) using primers at final concentration of 0.21 μM, 5 μl of cDNA (diluted at 1:100) as template, in a final volume of 15 μl. PCR-cycling comprises an initial polymerase activation step at 95°C for 10 min, followed by 45 cycles at 95°C for 15 s, 60°C for 20 s (except for FYN24G16 primers pairs which was 55°C), and 72°C for 15 s. After each run, a dissociation curve was acquired to confirm the absence of nonspecific amplification products, by heating the samples from 65 to 95°C. Real-time DNA amplification was monitored and analyzed using the Light Cycler® software release 1.5.0 (Roche Diagnostics, Mannheim, Germany). Two completely independent experiments were performed, i.e., based on two independent time courses. For each experiment, each sample was run in duplicates. Absence of amplification on Colletotrichum lindemuthianum genomic DNA of primer pairs designed on bean EF1-α, FDH FYN24G.15, and FYN24G.16 was controlled. Contamination by genomic DNA was checked by using primer pairs designed on FYN24G.15 intron 1 (Table 1). Quantification of gene expression was performed using the comparative Ct method (Livak and Schmittgen 2001). EF1-α, whose expression remains constant in the different conditions tested, was used as internal reference. For each condition, Ct value obtained for each gene of interest (Ct) was normalized by the internal reference Ct value (Ctr), according to the equation ΔCt = Ct − Ctr. ΔCt value obtained for each condition was then normalized by ΔCt value obtained for the calibrator (ΔCt (c)), which corresponds to untreated 8-day-old BAT93 seedlings, according to the equation ΔΔCt = ΔCt − ΔCt (c). Relative expression was then expressed as 2−ΔΔCt. Data are presented as induction fold from treated relative to control conditions.
Results
Sequence analysis of the bean FY-N24 BAC clone
The BAC clone FY-N24 was completely sequenced, providing 105,981 bp of contiguous sequence with an approximate sixfold coverage. The overall GC content of the 105,981 bp is 34.45%. The FY-N24 BAC sequence was annotated using a combination of prediction and comparative tools (see “Materials and methods”). BLAST queries were performed against ESTs and NR peptide databases. Alignments were used to optimize manually the homology regions, taking into account characteristics of splice sites and features shown by ESTs or peptides.
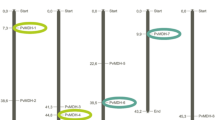
As most of the ESTs originate from different Phaseolus species (ESTs are either from P. vulgaris, P. coccineus, P. acutifolius or P. angustissimus whereas the BAC comes from P. vulgaris) or from different P. vulgaris genotypes (P. vulgaris ESTs are from 13 different genotypes, including genotype BAT93, which provides the presently sequenced BAC), we did not expect complete identity between ESTs and coding sequences of the annotated genes. The final result of the manual gene prediction is presented in Table 2; Fig. 1. In total, 17 genes named FYN24G.1 to FYN24G.17 (Fig. 1; Table 2), were predicted, providing an average gene density of one gene every 6 kb, with a large central region of 18,386 bp with no predicted gene. Four genes presenting the same transcriptional orientation (FYN24G.1, FYN24G.3, FYN24G.4, and FYN24G.6) are similar to R genes of the CNL class (Fig. 1). Two of them, FYN24G.1 and FYN24G.6, were previously identified from sequencing of phages mapped at the B4 R gene cluster, where they were referred to as CNL-B8 and CNL-B11, respectively (Geffroy et al. 2009). According to this gene nomenclature, FYN24G.3 and FYN24G.4 correspond to CNL-B9 and CNL-B10, respectively (Fig. 1; Table 2). An EST hit (FE897117) was identified that matched the N- terminal part of the CNL-B8 (Table 2). CNL-B8, CNL-B9, and CNL-B11 are intronless genes in their ORF and correspond to full length CNL encoding genes of, respectively, 3,363, 3,384, and 3,381 bp. These three CNL harbor 21 Leucine Rich Repeats (LRR) while the truncated CNL-B10* gene (2,792 bp) is composed of 13 LRRs only.
Schematic representation of the annotation of the sequence of the FY-N24 Phaseolus vulgaris BAT93 BAC clone (GenBank accession EU931620). The description of each putative gene is presented in Table 2. Arrowheads indicate the transcriptional orientation of the genes. Drawn to scale
Three regions containing similarities with known class I retroelements were found: FYN24G.2 is similar to a P. vulgaris TY-1/Copia like LTR retroelement AY533394, FYN24G.5 is similar to a Glycine max TY-3/Gypsy like retroelement polyprotein AAO23078, and FYN24G.11 is similar to a Medicago truncatula putative non-LTR retroelement RNA-directed DNA polymerase ABE85417. Apart from FYN24G.12, which is highly similar to the bean BA2 gene of unknown function (Melotto et al. 2004) and FYN24G.8, which is only a Genemark.hmm prediction, the remaining eight predicted genes are similar to plant genes with known functions. FYN24G.13 and FYN24G.14 are predicted to be basic blue protein-like encoding genes; FYN24G.9 gene is similar to the N-terminal region of an NADH dehydrogenase. The P. vulgaris EST FD791375 could be aligned with the N- terminal region of the FYN24G.9 gene. FYN24G.7 and FYN24G.10 are similar to F-box protein. Finally, a small cluster of three intact NAD-dependent formate dehydrogenase (FDH) genes (FYN24G.15, FYN24G.16 and FYN24G.17) was identified. FYN24G.15 and FYN24G.16 genes are separated by 1,287 bp and oriented in the same direction while FYN24G.17, located 4,062 bp away from FYN24G.16, is oriented in the opposite direction (head to head). The structure of the three FDH genes was established by taking into account the 17, 3, and 5 Phaseolus ESTs assigned to genes FYN24G.15, FYN24G.16, and FYN24G.17, respectively (Table 2).
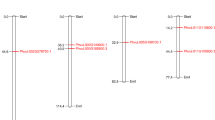
Formate dehydrogenase gene structure
To confirm common bean FDH gene structure predictions, near-full-length cDNA sequences of the BAT93 genotype were obtained by RT-PCR as described in the experimental procedure and sequenced. Three distinct transcripts corresponding to the three FDH genes (FYN24G.15, FYN24G.16 and FYN24G.17) were obtained. Nucleic acid alignments between the near-full-length cDNA and their corresponding genomic sequences confirm the predicted FDH gene structure. Six exons and five introns were identified, with introns occurring at the same positions for the three bean FDHs (Fig. 2). The most striking difference was the size of the first intron, which was 1,568, 133, and 327 bp for FYN24G.15, FYN24G.16, and FYN24G.17, respectively (Fig. 2). In all, the nucleic acid coding sequences were 1,161, 1,146, and 1,146 bp for FYN24G.15, FYN24G.16, and FYN24G.17, respectively. At the nucleotide level, the coding sequences of FYN24G.16 and FYN24G.17 are very similar, with 99% identity (only 10 bp distinguish FYN24G.16 and FYN24G.17 coding sequences). The nucleic acid coding sequence of FYN24G.15 shares 87% identity with either FYN24G.16 or FYN24G.17. The putative amino acid sequences encoded by FYN24G.16 and FYN24G.17 share 100% identity and FYN24G.15 shares 95% identity with either FYN24G.16 or FYN24G.17.
Exon-intron structure of three formate dehydrogenase (FYN24G.15, FYN24G.16 and FYN24G.17) genes from Phaseolus vulgaris. Exons (E1–E6) and introns (I1–I5) are numbered sequentially from left to right and are represented by black boxes and black lines, respectively. Gray boxes indicate putative signal peptides. Size in base pair of each exon and intron is indicated in brackets. Drawn to scale
To assess the FDH gene copy number in the P. vulgaris genome, a Southern-blot analysis was carried out (data not shown), demonstrating that only three FDH encoding genes are present in the P. vulgaris genome for both genotypes tested (BAT93: Mesoamerican; JaloEEP558: Andean). Together, these data show that three highly similar FDH sequences are present in the bean genome of BAT93 and that these three tightly linked FDH sequences are located near a cluster of CNL sequences at the end of linkage group B4.
Genome-wide exploration of FDH genes
In order to investigate the evolution of the NAD-dependent FDH gene family, we produced a comprehensive dataset of FDH sequences. FDH gene-coding sequences were searched in the GenBank (GB) non-redundant (NR) database, in Glycine max Whole Genome Shotgun (WGS) sequence and in the Plant Genome database (PlantGDB) EST Clusters. To complete this dataset with sequences from incompletely sequenced genomes, the full-length coding sequence of FDH genes was also deduced from manually established EST contigs (see “Materials and methods”) (Supplementary Table 1). Collectively, including the three present common bean FDH sequences, a total of 36 FDH sequences were identified: 33 plant FDH genes from angiosperms, gymnosperms and moss, indicating that FDHs are widespread throughout land plants and FDH sequences from bacteria (Pseudomonas syringae), yeast (Saccharomyces cerevisiae), and fungi (Neurospora crassa). The identification of NAD-dependent FDH sequences in plants, but also in diverse organisms such as bacteria, yeast and fungi indicates that NAD-dependent FDH arose early in evolution. In all the genomes of species fully sequenced or close to being completed, the number of FDH genes is low: A. thaliana (1), Medicago truncatula (1), Oryza sativa (2), Glycine max (3), Populus trichocarpa (1), Vitis vinifera (1), N. crassa (1), S. cerevisiae (2), demonstrating that FDH is a low copy gene family. In rice, the two FDH genes are separated by 24 kb on chromosome 6 and in soybean, the three FDHs identified belong to two distinct chromosomes: GmaFDH03 is located on chromosome 13 (27,112,435 to 27,116,473 bp), while GmaFDH01 (824,250 to 828,124 bp) and GmaFDH02 (829,852 to 833,359 bp) are separated by 1,728 bp on chromosome 19. At the amino acid level, alignment of the 36 FDH sequences from various taxa [bacteria (1, P. syringae), fungi (1, N. crassa), yeast (1, S. cerevisae), moss (1), monocots (7), and dicots (25)], revealed a limited level of sequence diversity except for the putative signal peptide (Supplementary Fig. 2).
Formate dehydrogenase signal peptide analysis
In order to assess the subcellular localisation of FDHs, we used three prediction programs (Predotar, TargetP and MitoProt) on the 33 plant FDH protein sequences as well as on the S. cerevisae and N. crassa protein sequences (Table 3).
For most plant sequences, all three programs gave consistent results, with high scores predicting plant FDH as mitochondrial and low scores for the chloroplast localization (Predotar). In particular, the three bean presequences get high scores for a mitochondrial localization with all three programs (0.53 for FYN24G.16 and FYN24G.17, and 0.68 for FYN24G.15 with Predotar, 0.6, 0.6 and 0.439, respectively, with TargetP and 0.974, 0.974, and 0.995, respectively with Mitoprot). It is worth noting that although Predotar gives extremely low scores for a chloroplast localization of FDHs from most species, TargetP gives quite high scores, but always lower than for a mitochondrial localization. In particular, the Predotar scores for chloroplast localization for the three bean FDHs are 0.04 for FYN24G.15 and 0.05 for FYN24G.16 and FYN24G.17, and the TargetP scores are 0.28 for FYN24G.15 and 0.43 for FYN24G.16, and FYN24G.17. This is probably due to the criteria used for the designing of the neural networks used for each program, particularly the number of serines present in the presequence. For S. cerevisae and N. crassa sequences, the scores associated with mitochondrial localization were low with the two-program tested, strongly suggesting a cytosolic localisation of FDH in these two organisms.
Phylogenetic analysis of plant FDH genes
Nucleotide coding sequences were used to conduct phylogenetic analysis because of high conservation and therefore uninformative alignments at the amino-acid level between several sequences. For example, bean FDH FYN24G.16 and FYN24G.17 presented 100% identity at the amino acid level. Bacterial, yeast, and fungal FDH sequences had to be left out of the analysis because they are too divergent to produce reliable nucleotide sequence alignments. As the basis of the phylogenetic tree constructions, we chose the entire FDH gene coding sequence except the putative signal peptide region (75 bp) and 21 additional bp (taking StuFDH01 as a reference sequence), which are too divergent (Supplementary Fig. 2). We defined clades as subtrees containing at least two sequences with greater than 70% boostrap support.
The phylogenetic tree shows five well-supported, distinct, multiplant family clades (Fig. 3). Generally, the plant FDH phylogeny reflects species phylogeny. The Poaceae clade contains FDH sequences from Triticum aesticum, Hordeum vulgare, Brachypodium distachyon, Oryza sativa, Sorghum bicolor, and Zea mays. The Taxaracum officinale and Helianthus annuus FDH sequences are part of the Asteraceae clade. The Solanaceae clade is composed of FDH sequences from Lycopersicon esculentum, Solanum tuberosum, and Nicotiana tabacum. The Brassicaceae clade is composed of three FDH sequences from Arabidopsis thaliana, Brassica oleracea, and B. napus. Finally, the last clade supported by an elevated bootstrap value (100%) is the Legume clade with the P. vulgaris, G. max, and Medicago truncatula FDH sequences. In agreement with the phylogenetic relationships between these three Legume species (Doyle and Luckow 2003), bean and soybean FDH belong to a well-supported subclade (boostrap value 100%), while the M. truncatula FDH sequence (MtrFDH01) is more distantly related. The fact that the three bean and the three G. max FDH sequences belong to the same subclade (boostrap value 100%), while a single FDH has been identified in M. truncatula, suggests that the amplification of FDH sequences in bean and soybean genome occurred after divergence of Medicago and the bean/soybean common ancestor. Two G. max FDH (GmaFDH02—chromosome 19; GmaFDH03—chromosome 13) and one bean FDH (FYN24G.15) belong to the same subclade (bootstrap value 89%), while GmaFDH01 (chromosome 19) and the two other bean FDH (FYN24G.16 and FYN24G.17) belong to another subclade with a high bootstrap value (100%). FYN24G.16 and FYN24G.17 are sisters within a strongly supported subclade (bootstrap value 100%).
Bayesian phylogenetic tree of 33 plant FDH nucleotide sequences. Numbers on the branches represent posterior probabilities after running a Markov chain Monte Carlo search for 10,000,000 generations. Sequences used in this tree are reported in Supplementary Table 1. The Glycine max chromosomes (13 and 19) on which each soybean FDH has been identified is indicated
Expression analysis of bean FDH genes
To determine if the three common bean FDH encoding genes have distinct expression patterns, quantitative RT-PCR experiments were carried out. Two primer pairs specific for FYN24.G15 and FYN24.G16, respectively, were defined for quantitative RT-PCR experiments. Given the high nucleotide identity between FYN24G.16 and FYN24G.17 (99%), no specific primers could be designed for FYN24G.17. Time-course analyses during either abiotic stresses (dark treatment) or biotic stresses (inoculation with the fungus Colletotrichum lindemuthianum) were performed. Two types of pathogen infections were performed: either with the C. lindemuthianum C531 incompatible strain (bean BAT93 genotype resistant) or with the M126 compatible strain (bean BAT93 genotype susceptible).
Dark treatment resulted in a 4.5-fold increase in FYN24G.15 mRNA levels after 72 h, while no change in FYN24G.16 mRNA levels was detected (Fig. 4a). A dark-to-light shift at 72 h after dark exposure results in a decrease in FYN24G.15 transcript accumulation, confirming that the 4.5-fold increase in FYN24G.15 mRNA levels is due to the dark treatment. After inoculation with incompatible C. lindemuthianum strain, FYN24G.16 transcript levels were increased approximately fivefold (72 h) during incompatible interaction, while no variation in FYN24G.15 mRNA levels was detected (Fig. 4b). For the other tested treatment (compatible interaction after inoculation with C. lindemuthianum) no significant change in FYN24G.15 or FYN24G.16 transcript levels were detected (Fig. 4c). In conclusion, these RT-qPCR experiments revealed that FYN24G.15 and FYN24G.16 are differentially up-regulated in common bean seedlings subjected to biotic (incompatible interaction with C. lindemuthianum) and abiotic (dark stress) stresses.
Expression analysis by Real-Time q-PCR of two common bean FDH (FYN24G.15 and FYN24G.16) after dark stress (a), incompatible interaction with the fungus Colletotrichum lindemuthianum (b) and compatible interaction with C. lindemuthianum (c). Quantification of gene expression was performed using the comparative Ct method (Livak and Schmittgen 2001). Data are presented as induction fold from treated relative to control conditions. The dark stress was performed for 5 days, followed by a dark-to -light shift for 2 days
Discussion
FDH is a low copy gene family in plant genome and three common bean FDH genes are localized in a cluster of CNL sequences
In this paper, we present the results of the sequence annotation of a 105-kb BAC clone from P. vulgaris encompassing a CNL-gene-rich region of the B4 R gene cluster located at the end of linkage group B4 (David et al. 2008; Geffroy et al. 2008; Geffroy et al. 2009). Only ten common bean BAC clones have been annotated so far (D’Ovidio et al. 2004; Melotto et al. 2004; Kami et al. 2006; Innes et al. 2008), and no gene prediction program specifically trained for P. vulgaris is yet available. In the present study, we developed a semi-automatic annotation procedure using a combination of prediction and comparative tools. In particular, we used the information derived from a growing amount of ESTs available for Phaseolus [P. vulgaris (83,348), P. coccineus (20,120), P. acutifolius (739), and P. angustissimus (6,672)] to complement the ab initio gene-model prediction of FGENESH and GenMark.hmm (Table 2). In addition to four CNL-encoding genes, we identified three FDH-encoding genes clustered in less than 13 kb (Fig. 1; Table 2). ESTs were very helpful to update the gene model for these three FDH genes, containing six exons and five introns (Fig. 2). Bioinformatic predictions were confirmed by near-full-length cDNA sequencing. We conducted a comprehensive plant FDH sequence database search. One important aspect of our work was to use EST information to increase the number of complete FDH sequences, especially from non-well sampled genomes. Indeed, out of 36 complete FDH sequences, half came from EST clustering, either from PlantGDB or from manually established EST clusters. Consequently, as already demonstrated in other studies, EST databases provide an essential resource for both genome annotation and gene identification from non- sequenced genomes for phylogenetic or diversity analyses (Zhu et al. 2002; Rossi et al. 2003; Radwan et al. 2008).
The current database searches clearly demonstrated that FDH is a low copy gene in plant genomes, since 33 FDH sequences were identified from 27 plant species. In the soybean genome, we identified two chromosomes (13 and 19) containing FDH genes. G. max diverged from P. vulgaris approximately 20 million years ago (Mya) (Lavin et al. 2005; Shoemaker et al. 2006), and has undergone a subsequent polyploidy event. Since FDH is a low copy gene in plant genomes, these results suggest that these two scaffolds correspond to homoeologous regions of the soybean genome. It also suggests that these two chromosome regions correspond to the orthologous regions of the bean B4 R gene cluster. Concerning the two model legume species (Young et al. 2005; Cannon et al. 2006; Sato et al. 2008), a partial FDH sequence (because of its location at the end of a BAC clone) has been identified in L. japonicus contig “CM0201”, confirming that this contig corresponds to the putative orthologous region of the bean B4 R gene cluster (David et al. 2008). This L. japonicus FDH sequence was not included in the present study based on complete plant FDH sequences. In M. truncatula, we identified a single FDH sequence derived from EST clustering. So far, no M. truncatula BAC clone containing a FDH gene is publicly available, suggesting that in M. truncatula the orthologous region of the B4 R gene cluster has not yet been completely sequenced.
Our phylogenetic results (Fig. 3) are consistent with two duplication events for bean FDH. A first duplication event, leading to FYN24G.15 and (FYN24G.16/FYN24G.17) common ancestor, probably occurred after the M. truncatula/(G. max/P. vulgaris common ancestor) divergence, estimated at ~54 Mya (Cronk et al. 2006). A second more recent duplication event, leading to FYN24G.16 and FYN24G.17, probably occurred after the divergence of P. vulgaris and G. max (~20 Mya). This second duplication event corresponds to an inverted duplication giving rise to FYN24G.16 and FYN24G.17. As mentioned earlier, since G. max is a polyploid species, this scenario implies the loss of one FDH on soybean chromosome 13. Inverted duplication events from a founder gene result either in tail-to-tail or head-to-head (present case for FYN24G.16 and FYN24G.17) orientation. Interestingly, this evolutionary mechanism has been found for gene families other than bean FDHs and has been demonstrated to be involved in micro RNA biogenesis (Allen et al. 2004). For example, in Arabidopsis thaliana, two miRNA genes, MIR161 and MIR163 evolved relatively recently by inverted duplication events from their target genes (Allen et al. 2004).
Prediction programs are in favor of the ‘mitochondrial only’ localization of plant FDH
Sequence comparisons of the 36 FDH sequences identified in our database search revealed that they are well conserved (Supplementary Fig. 2). The occurrence of conserved Thr76 and Thr333 in all the 36 FDH (except Thr333 which is absent in the Neurospora crassa FDH NcrFD01), is striking. In potato tubers, FDH has been shown to be phosphorylated on residues Thr76 and Thr333 (Bykova et al. 2003). This suggests that phosphorylation could be a general mechanism for NAD-dependent FDH protein regulation.
In fact, sequence comparison of the 36 identified FDH revealed that most of the nucleotide variations reside in the amino terminal region of the coding sequence, which corresponds to the predicted signal peptide (Supplementary Fig. 2). Signal peptides, which target a particular subcellular compartment, are not well conserved among proteins and organisms, but have common characteristics (Hurt et al. 1985; von Heijne 1986; Hartl et al. 1989). For example, mitochondrial precursors generally contain their targeting information in a cleavable N-terminal presequence, which is rich in hydroxylated and positively charged residues and generally produces an N-terminal amphiphilic α-helix. In yeasts and fungi, FDH is localized in the cytosol (van Dijken et al. 1976; Yurimoto et al. 2003) and this localization is confirmed by the present in silico predictions (Table 3). In higher plants, FDH is encoded by nuclear genes and imported into the mitochondrion, as demonstrated in S. tuberosum by antibody recognition of the protein in mitochondrial fractions (Colas des Francs-Small et al. 1993) and GFP targeting (Ambard-Bretteville et al. 2003a). In A. thaliana only, a dual localization of FDH in the chloroplast as well as in mitochondria has been proposed (Olson et al. 2000; Herman et al. 2002), but has never been shown by GFP-targeting as seen in the Arabidopsis SUBcellular database [SUBA, (Heazlewood et al. 2007)]. Furthermore, two proteomic studies report identification of FDH as well as other very abundant mitochondrial proteins in chloroplastic fractions of A. thaliana, and stress that they can only be contaminants (Friso et al. 2004; Kleffmann et al. 2004). In the present study, we present the results obtained using three independent prediction programs (Predotar, TargetP and Mitoprot) on a very large subset of 33 plant FDHs. All the predictions are in favor of the ‘mitochondrial only’ localization of FDH, even in A. thaliana, strongly suggesting that the ‘mitochondrial only’ localization is the rule in higher plants. Ambard-Bretteville et al. (2003a) suggested that the first three residues of the presequence are very important for mitochondrial import, since the mutation of a single amino acid in the potato FDH presequence was sufficient to strongly inhibit import, and the deletion of the first two residues totally abolished it. They also observed that the presequence bearing the “MAM” motif in position 1–3 lead to very high scores for mitochondrial import predictions. In the present work, the recurrence of the conserved “MAM” motif at the beginning of most of the plant FDH presequences is striking (found in 25 out of 33 plant FDH sequences).
Why is plant FDH localized in mitochondria since formate can be produced in all cell compartments (Hourton-Cabassa et al. 1998; Igamberdiev et al. 1999)? Although formate is beneficial as a C1-unit donor (folate synthesis), it is also known to be toxic to plants at concentrations from 4 mM added to the culture medium (Li et al. 2002). Although 4 mM seems to be a high concentration it is not dissimilar to the Km of plant FDHs for formate, which has been reported to be 0.5–2 mM (Uotila and Koivusalo 1979; Farinelli et al. 1983; Li et al. 2000; Ambard-Bretteville et al. 2003b). This formate toxicity could be crucial to mitochondria since formate has been reported to inhibit cytochrome-c oxidase and respiration (Nicholls 1975). Therefore, we propose that FDH might be localized inside mitochondria to regulate the intra-mitochondrial concentrations of formate and protect the mitochondria from the deleterious effect of high concentrations of this compound which are thought to arise under stress conditions (Ambard-Bretteville et al. 2003b).
Two nearly identical FDH genes are differentially up-regulated in common bean seedlings subjected to biotic and abiotic stresses
In most higher plants, FDH transcripts are scarce in unstressed leaves, but accumulate in response to numerous abiotic stresses. The first evidence for this was described in S. tuberosum leaves submitted to abiotic stresses such as drought, osmotic stress, cold, hypoxia, prolonged darkness, wounding as well as foliar application of metabolites (formate, methanol, ethanol…) (Hourton-Cabassa et al. 1998). Similar observations were made in Arabidopsis thaliana where FDH was shown to be up-regulated after dark, cold, drought, heat treatments, and wounding (Li et al. 2001, 2002). In P. vulgaris, we detected an increase in FYN24G.15 transcript levels, during prolonged darkness (72 h), and an accumulation of FYN24G.16 transcript levels during incompatible interaction with C. lindemuthianum (72 h) (Fig. 4). Interestingly, this time (72 h post inoculation) corresponds to the transition between biotrophic and necrotrophis phases, a key step in the infection process of the hemibiotrophic fungus C. lindemuthianum (Dufresne et al. 2000). Although we know that all three bean FDH genes are transcriptionally active (cDNA analysis), we were not able to test FYN24G.17 in qPCR experiments since no specific primers could be designed for that gene. Our results constitute an example of gene duplication and subsequent divergence, because two genes FYN24G.15 and FYN24G.16, with 87% nucleic acid identity, are differentially up-regulated. During the duplication events, flanking sequences were duplicated together with the FDH-coding sequences (Supplementary Fig. 3). FYN24G.15 and FYN24G.16, which present distinct expression patterns, also show the highest nucleotide variation in their 5′ flanking regions. Our results not only confirm the FDH transcript accumulation in response to abiotic stresses, but constitute a first literature report of FDH transcript accumulation in response to a biotic stress (infection with a fungus). This is backed up by transcriptome data in publicly available databases such as Genevestigator (https://www.genevestigator.ethz.ch), which reveal a high accumulation of FDH transcripts during infection of Arabidopsis thaliana by numerous pathogens (for example Botrytis cinerea, Phytophtora infestans, Pseudomonas syringae). The physiological role of FDH proteins in plants during biotic and abiotic stresses is not fully understood yet. Formate, the substrate for FDH, can arise from various pathways including photorespiration, cell wall degradation or synthesis, and glycolysis (Igamberdiev et al. 1999). In stressed plants, formate biosynthesis may result from enhancement of any of these pathways. As hypothesised by Hourton-Cabassa et al. (1998) formate could be the signal for the accumulation of FDH transcripts. Indeed, since formate accumulation is known to have deleterious effects in mitochondria at high concentrations (Nicholls 1975), FDH might protect mitochondria from these harmful effects. As we detected bean FDH transcript accumulation after both dark treatment and C. lindemuthianum incompatible infection, our study constitutes an additional example of connection between biotic and abiotic stresses (Fujita et al. 2006; Sudo et al. 2008). Mechanisms that underlie signaling pathway connections remain unclear and further work is needed to clarify these connection points, but formate could be a good candidate as a signaling molecule involved in both pathways.
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- FDH:
-
Formate dehydrogenase
- Mya:
-
Million years ago
References
Agarwal PK, Agarwal P, Reddy MK, Sopory SK (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25:1263–1274
Allen E, Xie ZX, Gustafson AM, Sung GH, Spatafora JW, Carrington JC (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36:1282–1290
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ambard-Bretteville F, Small I, Grandjean O, Colas des Francs-Small C (2003a) Discrete mutations in the presequence of potato formate dehydrogenase inhibit the in vivo targeting of GFP fusions into mitochondria. Biochem Biophys Res Commun 311:966–971
Ambard-Bretteville F, Sorin C, Rebeille F, Hourton-Cabassa C, Colas des Francs-Small C (2003b) Repression of formate dehydrogenase in Solanum tuberosum increases steady-state levels of formate and accelerates the accumulation of proline in response to osmotic stress. Plant Mol Biol 52:1153–1168
Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, Studholme DJ, Yeats C, Eddy SR (2004) The Pfam protein families database. Nucleic Acids Res 32:D138–D141
Belkhadir Y, Subramaniam R, Dangl JL (2004) Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr Opin Plant Biol 7:391–399
Burset M, Guigo R (1996) Evaluation of gene structure prediction programs. Genomics 34:353–367
Bykova NV, Stensballe A, Egsgaard H, Jensen ON, Moller IM (2003) Phosphorylation of formate dehydrogenase in potato tuber mitochondria. J Biol Chem 278:26021–26030
Cannon SB, Sterck L, Rombauts S, Sato S, Cheung F, Gouzy J, Wang XH, Mudge J, Vasdewani J, Scheix T, Spannagl M, Monaghan E, Nicholson C, Humphray SJ, Schoof H, Mayer KFX, Rogers J, Quetier F, Oldroyd GE, Debelle F, Cook DR, Retzel EF, Roe BA, Town CD, Tabata S, Van de Peer Y, Young ND (2006) Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc Natl Acad Sci USA 103:14959–14964
Cao YF, Song FM, Goodman RM, Zheng Z (2006) Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J Plant Physiol 163:1167–1178
Chini A, Grant JJ, Seki M, Shinozaki K, Loake GJ (2004) Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J 38:810–822
Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host–microbe interactions: Shaping the evolution of the plant immune response. Cell 124:803–814
Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786
Colas des Francs-Small C, Ambard-Bretteville F, Small ID, Remy R (1993) Identification of a major soluble-protein in mitochondria from nonphotosynthetic tissues as NAD-dependent formate dehydrogenase. Plant Physiol 102:1171–1177
Cronk Q, Ojeda I, Pennington RT (2006) Legume comparative genomics: progress in phylogenetics and phylogenomics. Curr Opin Plant Biol 9:99–103
D’Ovidio R, Raiola A, Capodicasa C, Devoto A, Pontiggia D, Roberti S, Galletti R, Conti E, O’Sullivan D, De Lorenzo G (2004) Characterization of the complex locus of bean encoding polygalacturonase-inhibiting proteins reveals subfunctionalization for defense against fungi and insects. Plant Physiol 135:2424–2435
Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
David P, Sévignac M, Thareau V, Catillon Y, Kami J, Gepts P, Langin T, Geffroy V (2008) BAC end sequences corresponding to the B4 resistance gene cluster in common bean: a resource for markers and synteny analyses. Mol Genet Genomics 280:521–533
Davison D (1949) The distribution of formic and alcohol dehydrogenases in the higher plants, with particular reference to their variation in the pea plant during its life cycle. Proc Linn Soc NSW 74:26–36
Desveaux D, Singer AU, Dangl JL (2006) Type III effector proteins: doppelgangers of bacterial virulence. Curr Opin Plant Biol 9:376–382
Dong QF, Schlueter SD, Brendel V (2004) PlantGDB, plant genome database and analysis tools. Nucleic Acids Res 32:D354–D359
Doyle JJ, Luckow MA (2003) The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol 131:900–910
Dufresne M, Perfect S, Pellier AL, Bailey JA, Langin I (2000) A GAL4-like protein is involved in the switch between biotrophic and necrotrophic phases of the infection process of Colletotrichum lindemuthianum on common bean. Plant Cell 12:1579–1589
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953–971
Farinelli MP, Fry DW, Richardson KE (1983) Isolation, purification and partial characterization of formate dehydrogenase from soybean seed. Plant Physiol 73:858–859
Ferrier Cana E, Geffroy V, Macadre C, Creusot F, Imbert Bollore P, Sevignac M, Langin T (2003) Characterization of expressed NBS-LRR resistance gene candidates from common bean. Theor Appl Genet 106:251–261
Ferrier Cana E, Macadre C, Sevignac M, David P, Langin T, Geffroy V (2005) Distinct post-transcriptional modifications result into seven alternative transcripts of the CC-NBS-LRR gene JA1tr of Phaseolus vulgaris. Theor Appl Genet 110:895–905
Flor HH (1955) Host–parasite interaction in flax rust. Its genetics and other implications. Phytopathol 45:680–685
Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res 8:967–974
Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, van Wijk KJ (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 16:478–499
Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9:436–442
Geffroy V, Sicard D, de Oliveira JCF, Sevignac M, Cohen S, Gepts P, Neema C, Langin T, Dron M (1999) Identification of an ancestral resistance gene cluster involved in the coevolution process between Phaseolus vulgaris and its fungal pathogen Colletotrichum lindemuthianum. Mol Plant Microbe Interact 12:774–784
Geffroy V, Sevignac M, De Oliveira JCF, Fouilloux G, Skroch P, Thoquet P, Gepts P, Langin T, Dron M (2000) Inheritance of partial resistance against Colletotrichum lindemuthianum in Phaseolus vulgaris and co-localization of quantitative trait loci with genes involved in specific resistance. Mol Plant Microbe Interact 13:287–296
Geffroy V, Sevignac M, Billant P, Dron M, Langin T (2008) Resistance to Colletotrichum lindemuthianum in Phaseolus vulgaris: a case study for mapping two independent genes. Theor Appl Genet 116:407–415
Geffroy V, Macadre C, David P, Pedrosa-Harand A, Sevignac M, Dauga C, Langin T (2009) Molecular analysis of a large subtelomeric NBS-LRR family in two representative genotypes of the major gene pools of Phaseolus vulgaris. Genetics 181:405–419
Grant JJ, Chini A, Basu D, Loake GJ (2003) Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol Plant Microbe Interact 16:669–680
Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL (2006) Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol 60:425–449
Hammond-Kosack KE, Jones JDG (1996) Resistance gene-dependent plant defense responses. Plant Cell 8:1773–1791
Hammond-Kosack KE, Parker JE (2003) Deciphering plant–pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol 14:177–193
Hartl F-U, Pfanner N, Nicholson DW, Neupert W (1989) Mitochondrial protein import. Biochim Biophys Acta 988:1–45
Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH (2007) SUBA: the Arabidopsis subcellular database. Nucleic Acids Res 35:D213–D218
Herman PL, Ramberg H, Baack RD, Markwell J, Osterman JC (2002) Formate dehydrogenase in Arabidopsis thaliana: overexpression and subcellular localization in leaves. Plant Sci 163:1137–1145
Hourton-Cabassa C, Ambard-Bretteville F, Moreau F, de Virville JD, Remy R, Colas des Francs-Small C (1998) Stress induction of mitochondrial formate dehydrogenase in potato leaves. Plant Physiol 116:627–635
Huang XQ, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Hurt EC, Muller U, Schatz G (1985) The 1st 12 amino-acids of a yeast mitochondrial outer-membrane protein can direct a nuclear-encoded cytochrome-oxidase subunit to the mitochondrial inner membrane. EMBO J 4:3509–3518
Igamberdiev AU, Bykova NV, Kleczkowski LA (1999) Origins and metabolism of formate in higher plants. Plant Physiol Biochem 37:503–513
Innes RW, Ameline-Torregrosa C, Ashfield T, Cannon E, Cannon S, Chacko B, Chen NWG, Couloux A, Dalwani A, Denny R, Deshpande S, Egan AN, Glover N, Hans CS, Howell S, Ilut D, Jackson S, Lai H, Mammadov J, Martin del Campo S, Metcalf M, Nguyen A, O’Bleness M, Pfeil BE, Podicheti R, Ratnaparkhe MB, Samain S, Sanders I, Segurens B, Sevignac M, Sherman-Broyles S, Thareau V, Tucker DM, Walling J, Wawrzynski A, Yi J, Doyle JJ, Geffroy V, Roe BA, Saghai Maroof MA, Young ND (2008) Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol 148:1740–1759
Jiang SY, Ramamoorthy R, Bhalla R, Luan HF, Venkatesh PN, Cai M, Ramachandran S (2008) Genome-wide survey of the RIP domain family in Oryza sativa and their expression profiles under various abiotic and biotic stresses. Plant Mol Biol 67:603–614
Kami J, Poncet V, Geffroy V, Gepts P (2006) Development of four phylogenetically-arrayed BAC libraries and sequence of the APA locus in Phaseolus vulgaris. Theor Appl Genet 112:987–998
Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjolander K, Gruissem W, Baginsky S (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol 14:354–362
Langlois-Meurinne M, Gachon CMM, Saindrenan P (2005) Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol 139:1890–1901
Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54:575–594
Li R, Ziola B, King J (2000) Purification and characterization of formate dehydrogenase from Arabidopsis thaliana. J Plant Physiol 157:161–167
Li R, Bonham-Smith PC, King J (2001) Molecular characterization and regulation of formate dehydrogenase in Arabidopsis thaliana. Can J Bot 79:796–804
Li R, Moore M, Bonham-Smith PC, King J (2002) Overexpression of formate dehydrogenase in Arabidopsis thaliana resulted in plants tolerant to high concentrations of formate. J Plant Physiol 159:1069–1076
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Lopez CE, Acosta IF, Jara C, Pedraza F, Gaitan-Solis E, Gallego G, Beebe S, Tohme J (2003) Identifying resistance gene analogs associated with resistances to different pathogens in common bean. Phytopathology 93:88–95
Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115
Martin GB, Bogdanove AJ, Sessa G (2003) Understanding the functions of plant disease resistance proteins. Ann Rev Plant Biol 54:23–61
McHale L, Tan XP, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7:212
Melotto M, Coelho MF, Pedrosa Harand A, Kelly JD, Camargo LEA (2004) The anthracnose resistance locus Co-4 of common bean is located on chromosome 3 and contains putative disease resistance-related genes. Theor Appl Genet 109:690–699
Nicholls P (1975) Formate as an inhibitor of cytochrome-C oxidase. Biochem Biophys Res Commun 67:610–616
Olson B, Skavdahl M, Ramberg H, Osterman JC, Markwell J (2000) Formate dehydrogenase in Arabidopsis thaliana: characterization and possible targeting to the chloroplast. Plant Sci 159:205–212
Popov VO, Lamzin VS (1994) NAD(+)-Dependent formate dehydrogenase. Biochem J 301:625–643
Radwan O, Gandhi S, Heesacker A, Whitaker B, Taylor C, Plocik A, Kesseli R, Kozik A, Michelmore RW, Knapp SJ (2008) Genetic diversity and genomic distribution of homologs encoding NBS-LRR disease resistance proteins in sunflower. Mol Genet Genomics 280:111–125
Rairdan G, Moffett P (2007) Brothers in arms? Common and contrasting themes in pathogen perception by plant NB-LRR and animal NACHT-LRR proteins. Microbes Infect 9:677–686
Ramirez M, Graham MA, Blanco-Lopez L, Silvente S, Medrano-Soto A, Blair MW, Hernandez G, Vance CP, Lara M (2005) Sequencing and analysis of common bean ESTs. Building a foundation for functional genomics. Plant Physiol 137:1211–1227
Robert-Seilaniantz A, Navarro L, Bari R, Jones JD (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10:372–379
Rossi M, Araujo PG, Paulet F, Garsmeur O, Dias VM, Chen H, Van Sluys MA, D’Hont A (2003) Genomic distribution and characterization of EST-derived resistance gene analogs (RGAs) in sugarcane. Mol Genet Genomics 269:406–419
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics 16:944–945
Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, Fujishiro T, Katoh M, Kohara M, Kishida Y, Minami C, Nakayama S, Nakazaki N, Shimizu Y, Shinpo S, Takahashi C, Wada T, Yamada M, Ohmido N, Hayashi M, Fukui K, Baba T, Nakamichi T, Mori H, Tabata S (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15:227–239
Sessa G, Yang XQ, Rat V, Eyal Y, Fluhr R (1995) Dark induction and subcellular-localization of the pathogenesis-related PRB1-1b protein. Plant Mol Biol 28:537–547
Shoemaker RC, Schlueter J, Doyle JJ (2006) Paleopolyploidy and gene duplication in soybean and other legumes. Curr Opin Plant Biol 9:104–109
Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4:1581–1590
Sudo E, Itouga M, Yoshida-Hatanaka K, Ono Y, Sakakibara H (2008) Gene expression and sensitivity in response to copper stress in rice leaves. J Exp Bot 59:3465–3474
Suzuki K, Itai R, Suzuki K, Nakanishi H, Nishizawa NK, Yoshimura E, Mori S (1998) Formate dehydrogenase, an enzyme of anaerobic metabolism, is induced by iron deficiency in barley roots. Plant Physiol 116:725–732
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Thunberg T (1921) Sur la presence de certains ferments oxydants dans les grains de Phaseolus vulgaris. Arch Int Physiol 18:601–606
Uotila L, Koivusalo M (1979) Purification of formaldehyde and formate dehydrogenases from pea seeds by affinity chromatography and S-formylglutathione as the intermediate of formaldehyde metabolism. Arch Biochem Biophys 196:33–45
van Dijken JP, Oostra-Demkes GT, Otto R, Harder W (1976) S-formylglutathione: the substrate for formate dehydrogenase in methanol-utilizing yeasts. Arch Microbiol 111:77–83
van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162
von Heijne G (1986) Mitochondrial targeting sequences may form amphiphilic helices. EMBO J 5:1335–1342
Young ND, Cannon SB, Sato S, Kim D, Cook DR, Town CD, Roe BA, Tabata S (2005) Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol 137:1174–1181
Yurimoto H, Lee B, Yano T, Sakai Y, Kato N (2003) Physiological role of S-formylglutathione hydrolase in C-1 metabolism of the methylotrophic yeast Candida boidinii. Microbiology Sgm 149:1971–1979
Zhu HY, Cannon SB, Young ND, Cook DR (2002) Phylogeny and genomic organization of the TIR and non-TIR NBS-LRR resistance gene family in Medicago truncatula. Mol Plant Microbe Interact 15:529–539
Acknowledgments
The authors are grateful to Steven B. Cannon for critical reading of the manuscript. They also thank Mathilde Langlois-Meurinne and Claire Gachon for helpful discussions and Jim Kami and Paul Gepts for providing us the Phaseolus vulgaris BAT93 FY-N24 BAC clone. Special thanks to Georgina Hernández for sharing ESTs data prior to publication. The research was supported by INRA-DGAP, CNRS and the French Ministère de la Recherche.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. T. Nguyen.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2010_1293_MOESM3_ESM.ppt
Multiple alignment performed on 36 FDH deduced amino acid sequences using the Clustal X software and edited in GENEDOC for manual adjustments. Details concerning the origin of the 36 FDH sequences are given in Supplementary Table 1. Peptide signal, as defined on the potato Z21493 sequence (Colas des Francs et al. 2003), is indicated. The multiple alignment used for phylogenetic analyses is based on the region between the two black arrows (PPT 233 kb)
122_2010_1293_MOESM4_ESM.ppt
Analysis of the flanking sequences of the three FDH genes. Hatched areas designate regions presenting > 38% nucleotide identity. Size in base pair of each considered region is indicated in brackets (PPT 165 kb)
Rights and permissions
About this article
Cite this article
David, P., Colas des Francs-Small, C., Sévignac, M. et al. Three highly similar formate dehydrogenase genes located in the vicinity of the B4 resistance gene cluster are differentially expressed under biotic and abiotic stresses in Phaseolus vulgaris . Theor Appl Genet 121, 87–103 (2010). https://doi.org/10.1007/s00122-010-1293-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1293-x