Abstract
Three-fourths of the recognition-dependent disease resistance genes (R-genes) identified in plants encode nucleotide binding site (NBS) leucine-rich repeat (LRR) proteins. NBS-LRR homologs have only been isolated on a limited scale from sunflower (Helianthus annuus L.), and most of the previously identified homologs are members of two large NBS-LRR clusters harboring downy mildew R-genes. We mined the sunflower EST database and used comparative genomics approaches to develop a deeper understanding of the diversity and distribution of NBS-LRR homologs in the sunflower genome. Collectively, 630 NBS-LRR homologs were identified, 88 by mining a database of 284,241 sunflower ESTs and 542 by sequencing 1,248 genomic DNA amplicons isolated from common and wild sunflower species. DNA markers were developed from 196 unique NBS-LRR sequences and facilitated genetic mapping of 167 NBS-LRR loci. The latter were distributed throughout the sunflower genome in 44 clusters or singletons. Wild species ESTs were a particularly rich source of novel NBS-LRR homologs, many of which were tightly linked to previously mapped downy mildew, rust, and broomrape R-genes. The DNA sequence and mapping resources described here should facilitate the discovery and isolation of recognition-dependent R-genes guarding sunflower from a broad spectrum of economically important diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Many recognition-dependent plant disease resistance (R) genes encode proteins comprised an amino-terminal domain, a nucleotide binding site (NBS) domain, and leucine-rich repeats (LRRs) (Bent 1996; Hammond-Kosack and Jones 1997; Hulbert et al. 2001). NBS-LRR encoding genes are often highly duplicated and evolutionarily diverse, with hundreds of family members in plant genomes (Meyers et al. 1999, 2003; Dangl and Jones 2001; Goff et al. 2002; Richly et al. 2002; McHale et al. 2006; Jones and Dangl 2006; Ameline-Torregrosa et al. 2007; Kohler et al. 2008). NBS-LRR resistance proteins directly or indirectly recognize pathogen avirulence factors, which trigger signal transduction cascades leading to rapid defense responses and frequently to hypersensitive reactions and programmed cell death (Dangl and Jones 2001; Bent 1996; Hammond-Kosack and Jones 1997; Hulbert et al. 2001). Of the 40 or so R-genes known to confer resistance to bacterial, fungal, viral, and nematode pathogens in plants, 75% encode NBS-LRR proteins. LRRs are determinants of response specificity and are typically under strong diversifying selection, whereas NBS domains are not (Parniske et al. 1997; McDowell et al. 1998; Meyers et al. 1998a; Sun et al. 2001; Kuang et al. 2004).
Conserved amino acid sequence motifs in the NBS domain have been widely used to isolate and classify NBS-LRR encoding genes (Meyers et al. 1999, 2002, 2003; Pan et al. 2000; Bai et al. 2002; He et al. 2004; Cannon et al. 2002). Two superfamilies have been described in dicots, one with an N-terminal structural domain homologous to the intracellular signaling domains of the Drosophila Toll and mammalian interleukin1 receptor (TIR) and the other lacking the TIR structural domain (non-TIR) (Meyers et al. 1999; Pan et al. 2000). Coiled-coil (CC) motifs are commonly found in the N-terminal structural domains of genes encoding non-TIR-NBS-LRR proteins, and members of the two superfamilies can be distinguished using conserved amino acid sequence motifs in the NBS domain (Meyers et al. 1999; Pan et al. 2000). Hundreds of NBS-LRR encoding genes have been identified in sequenced plant genomes and create a complex pathogen defense network—the Arabidopsis genome harbors ca. 150, the Populus trichocarpa and Medicago truncatula genomes harbor ca. 400, and the rice (Orzya sativa L.) genome harbors ca. 600 NBS-LRR encoding genes (Meyers et al. 1999, 2003; Dangl and Jones 2001; Cannon et al. 2002; Goff et al. 2002; Richly et al. 2002; Holt et al. 2003; Jones and Dangl 2006; Ameline-Torregrosa et al. 2007; Kohler et al. 2008). The number and diversity of NBS-LRR encoding genes in the sunflower (Helianthus annuus L.; 2n = 2x = 34) genome are not known.
NBS-LRR encoding genes have been isolated from numerous species by cloning and sequencing genomic DNA fragments amplified by degenerate oligonucleotide primers complementary to conserved amino acid sequence motifs in the NBS domain, typically from the P-loop to the GLPALP motif (Kanazin et al. 1996; Leister et al. 1996; Yu et al. 1996; Shen et al. 1998). The spectrum of resistance gene candidates (RGCs) isolated from sunflower using the degenerate primer strategy has been limited, and only a few of the previously identified RGCs have been genetically mapped, primarily using RFLP markers (Gentzbittel et al. 1998; Gedil et al. 2001). The latter facilitated the discovery and cloning of downy mildew (Plasmopara halstedii (Farl.) Berl. & De Toni 1888) R-genes found in two tightly linked families of NBS-LRR encoding genes (Gentzbittel et al. 1998; Bert et al. 2001; Gedil et al. 2001; Bouzidi et al. 2002; Radwan et al. 2003, 2004; Slabaugh et al. 2003).
EST databases are another resource for identifying NBS-LRR encoding genes (Zhu et al. 2002; Rossi et al. 2003). The sunflower genome is physically large (3,500 Mbp; Baack et al. 2005) and has not been sequenced; however, 326,421 ESTs have been produced from H. annuus, Jerusalem artichoke (Helianthus tuberosus L.; 2n = 6x = 102), silverleaf sunflower (Helianthus argophyllus Torr. and Gray; 2n = 2x = 34), and other wild species (http://cgpdb.ucdavis.edu/cgpdb2/). We identified several hundred RGCs by mining the sunflower EST database and sequencing genomic DNA fragments amplified from H. annuus, H. tuberosus, and other wild sunflower species using degenerate primers complementary to conserved amino acid sequence motifs in the NBS domain. The newly isolated RGCs are described here. Nearly one-fourth of the unique NBS-LRR encoding genes identified in the present study were genetically mapped using insertion–deletion (INDEL) or single-strand conformational polymorphism (SSCP) markers (Orita et al. 1989; Kukita et al. 1997; Larsen et al. 1999, 2007). The analyses described here build a more complete picture of the genetic diversity and genomic distribution of NBS-LRR encoding genes in sunflower and supply a genomic framework for identifying and isolating recognition-dependent R-genes conferring resistance to a wide range of pathogens of sunflower.
Materials and methods
Plant materials
Genomic DNAs were isolated from young leaves harvested from greenhouse grown plants of an elite oilseed inbred line (RHA373) and a wild self-incompatible outbred population (ANN1811; PI 494567) of H. annuus and wild, self-incompatible, outbred populations of H. argophyllus (ARG-1807, PI 494573), Helianthus deserticola (DES-2345, PI 649872), Helianthus paradoxus (PAR-Cibola), and H. tuberosus (TUB-49, PI 650095). Seeds of PAR-Cibola were supplied by Dr. Loren Rieseberg (Indiana University, Bloomington, Indiana). Seeds of the other germplasm accessions were supplied by the United States Department of Agriculture (USDA) Agricultural Research Service (ARS) National Plant Germplasm System (http://www.ars-grin.gov/npgs/) or the USDA-ARS Northern Crop Science Research Laboratory. DNA was isolated from manually ground fresh or frozen leaf tissues using a modified CTAB (cetyltrimethylammonium bromide) method (Murray and Thompson 1980).
NBS fragment isolation by PCR amplification using degenerate primers
Genomic DNA fragments spanning conserved NBS sequences were isolated from common and wild sunflower species using two pairs of degenerate forward and reverse oligonucleotide primers (NBS-F1/NBS-R1 and NBS-F2/NBS-R2) complementary to conserved P-loop and GLPLAL motifs in the NBS domain (Table 1; Leister et al. 1996; He et al. 2004) and were predicted to amplify ca. 500 bp DNA fragments. Standard PCR methods were used, amplicons were separated on 1.5% agarose gels (Sigma, USA), and bands of the predicted length were excised and purified using the QIAquick method (Qiagen USA), cloned into the pCR 4-TOPO vector (Invitrogen, USA), chemically transformed into TOP10 E. coli cells (Invitrogen, USA), and plated on Luria broth agar containing 100 mg of ampicillin/l. We randomly selected 192 clones from the ANN1811, ARG-1807, DES-2345, PAR-Cibola, and TUB-49 libraries and 288 clones from the RHA373 library for DNA sequencing. The selected clones were bidirectionally Sanger sequenced on an ABI3730XL using ABI Big-Dye technology (Applied Biosystems, USA). Sequences of clones harboring putative pseudogenes were substantiated by resequencing.
Mining sunflower and lettuce EST databases for NBS-LRR encoding resistance gene homologs
Sunflower and lettuce transcript assemblies (TAs) in the Compositae Genome Database (CGPdb; http://www.cgpdb.ucdavis.edu/) were screened to identify unigenes homologous to NBS-LRR encoding genes using “NBS”, “LRR”, and “NBS-LRR” keyword searches (TAs were BLAST annotated). Homologs with BLASTX scores ≤e−0.15 were selected, cDNA sequences were translated in six frames using the CGPdb pipeline (http://cgpdb.ucdavis.edu/database/sms/translation.php), and BLASTX analyses were performed against the NCBI Protein Database to substantiate the homology of the selected sunflower and lettuce unigenes to NBS-LRR encoding R-genes identified in other plant species (Altschul et al. 1997; http://www.ncbi.nlm.nih.gov). Sunflower NBS-LRR sequences were used to search for lettuce homologs in the CGPdb EST database.
Sequence and genetic diversity analyses
Nucleotide binding site nucleotide and amino acid sequences were aligned using Clustal_X (Thompson et al. 1997). JALVIEW was used to display and identify redundant sequences (Clamp et al. 2004; http://www.jalview.org/). NBS amino acid sequences were screened for motifs characteristic of proteins encoded by plant disease resistance genes and were classified as non-TIR or TIR using amino acid sequence motifs characteristic of the two superfamilies (Bent 1996; Hammond-Kosack and Jones 1997; Meyers et al. 1999, 2003; Pan et al. 2000). TIR sequences were identified by the presence of the RNBS-A TIR motif (FLENIRExSKKHGLEHLQKKLLSKLL) and aspartic acid (D) as the last amino acid residue in the Kin-2 motif, whereas non-TIR sequences were identified by the presence of the RNBS-A non-TIR motif (FDLxAWVCVSQxF) and tryptophan (W) as the last amino acid residue in the Kin-2 motif (Meyers et al. 1999, 2003; Pan et al. 2000).
Because many of the newly isolated NBS sequences (n = 758) were closely related (>90% amino acid similarity), sequences with <90% amino acid similarity (n = 69) were selected for in-depth analysis. The NBS nucleotide sequence alignment for the newly isolated RGCs (n = 758) and the amino acid sequence alignment for the selected sunflower NBS sequences (n = 69) have been supplied as GeneDoc input files [Appendices (S1) and (S2)]; GeneDoc is freeware and can be downloaded from http://www.nrbsc.org/gfx/genedoc/index.html or http://www.nrbsc.org/downloads/. Neighbor-joining (NJ) trees were produced from analyses of NBS amino acid similarity matrices with bootstrap resampling (k = 1,000 permutations) (Saitou and Nei 1987; Thompson et al. 1997). Two trees were produced, one from an alignment and analysis of the 69 selected sunflower sequences with <90% amino acid similarity and 24 lettuce (Lactuca sativa L.) sequences selected from previously identified families (Meyers et al. 1998b; Kuang et al. 2004) and one from an alignment and analysis of sunflower and lettuce sequences with sequences from diverse NBS-LRR encoding R-genes isolated from other plant species: RPS4 (AJ243468), RPP1 (AF098962), ADR1 (AJ581996), RPS5 (AF074916), RPM1 (X87851), RPP13 (AF209730), RPS2 (ATU14158), RPP5 (ATRPP5LE1), and RPP8 (AF089711) from Arabidopsis; L6 (LUU27081) from flax (Linum usitatissimum L.); N (NGU15605) from tobacco (Nicotiana tabacum L.) and NRG1 (DQ054580) from Nicotiana benthamiana; Mi (AF039681) and Rx (AJ011801) from potato (Solanum tuberosum L.); I2 (AF004878) from tomato (Solanum lycopersicon L.); Xa1 (AB002266), Pi-ta (AF207842), and Pib (AB013449) from rice (Oryza sativa L.); Rp1 (AF107293) from maize (Zea mays L.); and Dm3 (LSRGC2B1) from lettuce (GenBank accession numbers are shown in parentheses).
DNA marker development, genotyping, and genetic mapping
Single-strand conformational polymorphism and INDEL markers were developed for genotyping NBS-LRR loci by designing flanking oligonucleotide primers complementary to cDNA and genomic DNA reference sequences. Whenever possible, nucleotide polymorphisms were used to identify paralog-specific sequences for designing primers for INDEL and SSCP marker genotyping. INDEL markers were manually genotyped on 1.5% NuSieve agarose gels (FMC, USA). SSCP markers were manually genotyped by silver-staining polyacrylamide gels (Bassam et al. 1991; Slabaugh et al. 2003). PCRs were performed using 30 ng of DNA template, 0.65 U Taq polymerase (Qiagen, USA), 1× PCR reaction buffer, 2.5 mM Mgcl2, 0.2 mM dNTPs, 0.16 μM of each primer (one pair/marker), and a ‘touchdown’ PCR protocol (Don et al. 1991; Hecker and Roux 1996) with an initial denaturation temperature of 94 C for 3 min, followed by 1 cycle of 94 C for 30 s, 68 C for 30 s and 72 C for 60 s; annealing temperatures in subsequent cycles were decreased by 1 C until reaching 58 C, products were amplified for 35 cycles at 94 C for 30 s/cycle, 58 C for 30 s/cycle, and 72 C for 60 s/cycle, and products were extended in the final cycle at 72 C for 15 min. Genomic DNA amplicons were separated on agarose to screen for INDEL polymorphisms and, when absent, were separated and genotyped using SSCP analysis. Polymorphic INDEL and SSCP markers were genotyped on 94 RHA280 × RHA801 recombinant inbred lines (RILs) (Tang et al. 2002; Yu et al. 2003) or 94 NMS373 × (NMS373 × ANN1238) BC1 progeny (Gandhi et al. 2005). NBS-LRR INDEL and SSCP marker loci were anchored, grouped, and ordered against a previously mapped genome-wide backbone of SSR and INDEL markers (Tang et al. 2002; Yu et al. 2003; Gandhi et al. 2005). Genetic mapping analyses were done using MAPMAKER (Lander et al. 1987) as previously described (Tang et al. 2002). DNA marker loci were ordered by allowing 0 or 2% genotyping errors (Lincoln and Lander 1992), primarily for comparing likelihoods of locus orders in genomic regions densely populated with NBS-LRR marker loci.
Results
Sunflower RGCs identified by targeting conserved NBS sequence motifs and mining the EST database
Genomic DNA fragments were amplified from H. annuus, H. argophyllus, desert sunflower (H. deserticola Heiser; 2n = 2x = 34), Pecos sunflower (H. paradoxus Heiser; 2n = 2x = 34), and H. tuberosus using two pairs of degenerate oligonucleotide primers complementary to highly conserved NBS sequence motifs (Leister et al. 1996; Meyers et al. 1999; He et al. 2004, Tables 1, 2). Both primer pairs (NBS-F1/R1 and NBS-F2/R2) were predicted to amplify ca. 500 bp fragments from TIR and non-TIR-NBS-LRR encoding genes. Collectively, 1,248 amplicons were cloned and sequenced (196–288/genotype), 1,052 of the 1,248 resequenced amplicons (RSAs) were homologous to NBS sequences in NBS-LRR encoding genes previously isolated from sunflower or other plant species, 784 RSAs had uninterrupted ORFs from the P-loop to the GLPLAL motif, 542 RSAs were unique and lacked stop codons or frame shift mutations, and 242 RSAs harbored stop codons or frame shift mutations and were suspected to be pseudogenes [Table 2; GenBank Acc. No. EF559385-EF560169; Appendix (S3)]. Sequencing errors should have been minimal because each clone was bidirectionally sequenced and putative pseudogenes were resequenced. The 542 unique NBS sequences ranged in length from 164 to 173 amino acids (GenBank Acc. No. ABQ58077-ABQ57529). Three-fourths of the newly isolated NBS sequences lacking stop codons were TIR class (Meyers et al. 1999, 2003; Pan et al. 2000). Nearly half of the NBS sequences isolated from H. annuus were TIR (53.6%) class, whereas 93.9% of the NBS sequences isolated from H. tuberosus were TIR class. Of the NBS sequences harboring stop codons, 95.3% were TIR class [Appendix (S3)].
Eighty-eight unigenes homologous to NBS-LRR encoding R-genes were identified by screening transcript assemblies (TAs) developed from 284,745 Helianthus ESTs in the Compositae Genome Program Database (CGPdb; http://cgpdb.ucdavis.edu/cgpdb2/). The ESTs spanned pre-NBS, NBS, LRR-, or post-LRR domains or combinations thereof [Appendix (S3)]. ESTs spanning NBS domains were aligned with previously and newly isolated NBS domain sequences (Gedil et al. 2001; Radwan et al. 2003; Plocik et al. 2004). Nucleotide identities ranged from 28 to 100%, while amino acid similarities ranged from 16 to 100%.
Diversity of sunflower homologs encoding NBS-LRR resistance proteins
Sunflower NBS sequences (n = 758) spanning the P-loop to the GLPLAL motif were translated and aligned to identify and eliminate redundant and closely related (>90% amino acid similarity) sequences and select a subset of unique sequences (<90% amino acid similarity) for genetic diversity analyses; 69 NBS sequences were selected for cluster analyses [Appendix (S1, S2)]. Of the latter, 15 were previously identified (Gedil et al. 2001; Radwan et al. 2003; Plocik et al. 2004) and belonged to a single TIR family (see group T3 in Fig. 1), eight were ESTs identified in the CGPdb, and 46 were newly isolated genomic DNA sequences [Appendix (S3)]. The 69 sunflower NBS sequences were aligned with 28 lettuce NBS sequences. Of the latter, 21 were previously identified and shared <98% identity (Meyers et al. 1998b; Kuang et al. 2004) and seven were identified by mining the lettuce EST database (http://cgpdb.ucdavis.edu/cgpdb2/). Several other lettuce ESTs homologous to NBS-LRR encoding genes were identified, but spanned non-NBS sequences.
Neighbor-joining tree produced from an analysis of the amino acid similarity matrix for 69 sunflower (Ha prefix) nucleotide binding site (NBS) sequences with <90% amino acid identity and 26 lettuce (Ls prefix) NBS sequences. Seven non-TIR (N1–N7) and eight TIR groups (T1–T8) were identified. Numbers shown on branches are percentages of bootstrap replications supporting nodes
The sunflower NBS sequences clustered into 14 groups with strong bootstrap support, eight solely comprised TIR (T1–T8) and six solely comprised of non-TIR (N1–N6) NBS sequences (Fig. 1). Two groups (T7 and N6) were populated with ESTs only. Nine groups (T1–5, T8, and N3–5) lacked ESTs; however, most of the ESTs spanned non-NBS sequences and could not be classified [Appendix (S3)]. One additional non-TIR group was identified (N7) and was solely populated with lettuce sequences (Fig. 1). The latter belong to a large and diverse cluster of NBS-LRR encoding genes harboring downy mildew R-genes in lettuce (Kuang et al. 2004). The other lettuce RGCs clustered with sunflower RGCs in two of the seven non-TIR groups (N1 and N6) and two of the eight TIR (T4 and T6) groups, with 35–98% identity among sequences within a group. Sunflower RGCs homologous to lettuce RGCs in group N7 have not been identified and lettuce RGCs homologous to sunflower RGCs in several groups (N2–5, T1–3, T5, and T7–8) have not been identified.
Sunflower and lettuce NBS sequences selected to sample diversity across the 15 groups (Fig. 1) were clustered with NBS sequences from 20 previously identified monocot and dicot R-genes (Fig. 2). The selected RGCs were highly similar to one or more known TIR and non-TIR-NBS-LRR encoding R-genes. Two non-TIR groups were identified, one harboring ADR1, NRG1, and a single sunflower (Ha-RGC185) and lettuce (Ls-BQ866268) RGC each (non-TIR1), and the other harboring the other non-TIR R-genes and RGCs (non-TIR2). The latter harbored sunflower and lettuce RGCs from each of the previously identified non-TIR groups other than N3 (Figs. 1, 2). Sunflower and lettuce RGCs in the TIR superfamily were most closely related to N, a TIR-NBS-LRR encoding R-gene isolated from tobacco (N. tabacum L.) (GenBank Acc. No. NGU15605; Whitham et al. 1994; Lawrence et al. 1995).
Genetic mapping and genomic distribution of sunflower homologs encoding NBS-LRR resistance proteins
Single-strand conformational polymorphism and INDEL markers were developed for 196 RGCs, 88 from ESTs and 108 from RSAs [Appendix (S4)]. The parents of the RHA280 × RHA801 RIL (Tang et al. 2002) and NMS373 × ANN1811 BC1 (Gandhi et al. 2005) mapping populations were screened for presence-absence polymorphisms (dominant INDELs), SSCPs, or both (Fig. 3). Because NBS-LRR encoding genes are commonly duplicated, allelic and non-allelic strands were separated by SSCP analysis to facilitate paralog discovery and mapping. The number of loci amplified by a particular SSCP marker was inferred from the number of strands identified, e.g., two loci were inferred when four strands were amplified and three loci were inferred when six strands were amplified from one or both parents, assuming two complementary strands per allele (Bassam et al. 1991). Of the 196 DNA markers developed and screened in the present study, 43 amplified a single locus, 153 amplified two or more loci, and 150 (76.5%) were polymorphic (10 were polymorphic for two or three loci each).
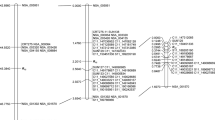
Collectively, 167 NBS-LRR loci were genetically mapped, nine by genotyping INDEL and 158 by genotyping SSCP markers (Figs. 3, 4). INDEL and SSCP markers for NBS-LRR loci were integrated into a framework of previously mapped SSR and INDEL marker loci distributed throughout the sunflower genome (Tang et al. 2002; Yu et al. 2003; Gandhi et al. 2005). One-third of the mapped NBS-LRR markers (58/167) were developed from ESTs, 66.9% of the SSCP markers developed from ESTs were polymorphic (58/88), and 88.9% of the SSCP markers developed from RSAs were polymorphic (96/108). NBS-LRR loci were found throughout the genome—the upper and lower segments of most linkage groups harbored at least one NBS-LRR locus (Fig. 4). The 17 linkage groups (x = 17) displayed in Fig. 4 were developed by integrating NBS-LRR loci mapped in the NMS373 × ANN1811 BC1 population into the framework of DNA marker loci mapped in the RHA280 × RHA801 RIL population by interpolating genetic distances and binning loci within the shortest interval possible flanked by common DNA marker loci–individual linkage groups for the two populations are displayed in a comparative mapping database (http://www.sunflower.uga.edu/CMAP) developed using CMAP (http://www.gmod.org/wiki/index.php/CMAP; Gonzales et al. 2005).
Genomic locations for 167 NBS-LRR loci in sunflower (2n = 2x = 34) mapped using INDEL or SSCP markers. Species sources for DNA markers developed from ESTs are shown as prefixes in locus names (ANN, H. annuus; ARG, H. argophyllus; PET H. petiolaris; TUB, H. tuberosus; LSE, L. serriola). Groups (clusters), when known, are identified by alphanumeric suffixes (T1–T8 for TIR and N1–N7 for non-TIR-NBS-LRR groups) in locus names (groups for SSCP or INDEL markers developed from non-NBS sequences were not known). Putative paralogous loci found on different linkage groups are identified by numerical suffixes, e.g., the RGC131 (a member of group T1) SSCP marker identified paralogous loci on linkage groups 9 and 10 (RGC131-9-T1 and RGC131-10-T1). Putative paralogous loci found on a single linkage group are identified by alphanumeric suffixes, e.g., the RGC145 (a member of group T3) SSCP marker identified paralogous loci (A and B) on linkage group 9 (RGC145-9A-N3 and RGC145-9B-N3)
Wild species ESTs were a particularly rich source of novel RGCs, with 31 originating from H. tuberosus, 20 originating from domesticated and wild H. annuus, three originating from prairie sunflower (H. petiolaris Nutt.; 2n = 2x = 34), one originating from H. argophyllus, and one originating from prickly lettuce (L. serriola L.). ESTs facilitated the discovery of 14 new NBS-LRR clusters or singletons, which were found on the upper and lower segments of linkage group (LG) 1, upper segments of linkage groups 2, 3, 4, 5, 9, 10, and 14, and lower segments of linkage groups 6, 8, 12, 15, 16, and 17 (Fig. 4). Several NBS-LRR homologs identified from ESTs mapped to the two largest and most complex and well known clusters in sunflower, a TIR cluster (T3) on the upper end of LG 8 and a non-TIR cluster (N1) on the lower end of LG 13 (Gentzbittel et al. 1998; Bert et al. 2001; Gedil et al. 2001; Bouzidi et al. 2002; Radwan et al. 2003; Slabaugh et al. 2003). NBS-LRR encoding genes from each of the 14 groups identified from analyses of the NBS amino acid sequence alignment [Fig. 1; Appendix (S2)], other than N7, were genetically mapped, and members of several groups were found on two or more linkage groups. Collectively, 44 NBS-LRR clusters or singletons were identified by genetic mapping (Fig. 4).
Discussion
The NBS-LRR markers mapped in the present study supply landmarks for identifying and isolating R-genes conferring resistance to diverse pathogens in sunflower [Fig. 4; Appendix (S3, S4)]. Of the 14 NBS-LRR groups identified by cluster analysis (Figs. 1, 2), only four had previously been identified (Gedil et al. 2001; Radwan et al. 2003; Plocik et al. 2004). Moreover, of the 44 NBS-LRR clusters and singletons mapped in the present study (Fig. 4), only four had previously been mapped and only two were linked to previously identified R-genes, the T3 cluster on LG 8 and the N1 cluster on LG 13 harboring downy mildew R-genes (Gentzbittel et al. 1998; Bert et al. 2001; Gedil et al. 2001; Bouzidi et al. 2002; Radwan et al. 2003; Slabaugh et al. 2003). Previously, RGCs had not been identified for downy mildew R-genes on LG 1 (Pl ARG), black rust (Puccinia helianthi Schw.) R-genes on LGs 8 (R 1) and 13 (R ADV), a broomrape [Orobanche cernua var. cumana (Wallr.) G. Beck] R-gene on LG 3 (Or 5), and a chlorotic mottle virus R-gene on LG 14 (Rcmo-1) (Gentzbittel et al. 1998; Bert et al. 2001; Gedil et al. 2001; Bouzidi et al. 2002; Radwan et al. 2003, 2004; Slabaugh et al. 2003; Tang et al. 2003; Yu et al. 2003; Dußle et al. 2004; Pérez-Vich et al. 2004; Lenardon et al. 2005); the locations of R 1 and R ADV were inferred from linkages of both loci to SCAR markers (Lawson et al. 1998) mapped by Yu et al. (2003) and cross-referenced in the present study. Other than Rcmo-1 (Lenardon et al. 2005), newly identified and mapped NBS-LRR loci were linked to each of the previously mapped recognition-dependent R-genes in sunflower (Pl ARG, Pl 8, R 1, R ADV, and Or 5). RGCs linked to Pl 8, a downy mildew resistance gene on LG 13, were previously identified by mapping RFLP markers (Radwan et al. 2003, 2004). The DNA sequences and markers described here [Appendix (S1–S4)] should greatly facilitate the isolation of Pl 8 and other R-genes in sunflower.
While 167 NBS-LRR loci were mapped in the present study, including 100% of the polymorphic INDEL or SSCP markers developed from ESTs and members of each of the 14 sunflower NBS-LRR groups (Fig. 1), the genomic locations of several hundred of the newly identified RGCs are not known, although most of the unmapped RGCs were highly similar (>91% amino acid similarity) to one or more of the mapped RGCs [Appendix (S3); Fig. 4]. The development and genetic mapping of additional NBS-LRR markers could be accelerated using multiplex capillary SSCP genotyping (Larsen et al. 1999, 2007) or other high-throughput DNA fragment analysis methods (Slabaugh et al. 2003) and might identify unique loci or clusters of loci, although one or more members of nearly every group have been mapped. Until the sunflower genome has been sequenced, the discovery of RGCs will be limited to the sorts of forward genetic and comparative genomic approaches described here, as has been done in many other plant species (Kanazin et al. 1996; Leister et al. 1996; Yu et al. 1996; Collins et al. 1998; Shen et al. 1998; Pan et al. 2000; Wang et al. 2001; Wei et al. 2002; Zhu et al. 2002; Lee et al. 2003; Madsen et al. 2003; Meyers et al. 2003; He et al. 2004; Rossi et al. 2003). On the basis of analyses of sequenced plant genomes (Meyers et al. 2003; Ameline-Torregrosa et al. 2007; Kohler et al. 2008), the full complement of NBS-LRR encoding genes in the sunflower genome is undoubtedly larger than the complement isolated so far [Appendix (S1–S3)].
Wild species are rich sources of genetic diversity for disease resistance and other traits in domesticated plants (Tanksley and McCouch 1997; Michelmore 2003) and have been an important source of R-genes in sunflower (Pustovoit and Kroknin 1978; Škoric 1985; Miller and Gulya 1988, 1991; Seiler 1991a, b; Tan et al. 1992; Quresh et al. 1993; Degener et al. 1999; Viguié et al. 2000; Langar et al. 2002; Miller et al. 2002; Vear et al. 2003; Slabaugh et al. 2003; Dußle et al. 2004). Several novel sunflower NBS-LRR loci were identified in the present study by mining wild species ESTs and isolating NBS sequences from wild species. We sequenced more deeply than had been done in earlier studies and targeted wild species to increase the depth and breath of sequences discovered by the degenerate primer approach, particularly since wild species have played an important role in disease resistance breeding in sunflower, e.g., the downy mildew R-genes found on three chromosomes (Pl ARG on LG 1, Pl 7 on LG 8, and Pl 8 on LG 13) were introgressed from wild species (Miller et al. 2002; Slabaugh et al. 2003; Dußle et al. 2004).
Between ESTs and RSAs, at least 918 unique NBS-LRR sequences have been identified in sunflower so far [Gentzbittel et al. 1998; Bert et al. 2001; Gedil et al. 2001; Bouzidi et al. 2002; Radwan et al. 2003, 2004; Slabaugh et al. 2003; Plocik et al. 2004; Appendix (S3)]. Wild species ESTs were a particularly rich source of novel RGCs, were the sole source of NBS-LRR encoding genes in four of 10 newly identified groups (T2, T4, T8, and N5), and were the sole source of RGCs linked to previously mapped R-genes for which RGCs have not been identified (Figs. 1, 4). H. tuberosus ESTs supplied the largest number of novel RGCs, several of which were linked to R-genes previously identified from resistance phenotypes. H. tuberosus has been a historically important source of R-genes transferred across the diploid × hexaploid bridge, e.g., the first dominant race-specific broomrape R-gene (Or 1) was discovered in H. tuberosus, perhaps as early as 1916, and was introgressed into H. annuus through backcross breeding (Vranceanu et al.1980; Parker and Riches 1993). Several clusters of NBS-LRR loci identified from ESTs and RSAs are not linked to R-genes identified from resistance phenotypes, but could be the source of as yet undiscovered R-genes.
The NBS-LRR copy numbers often vary widely within species, and NBS-LRR loci frequently rearrange and evolve through recombination, unequal crossing-over, gene conversion, insertions–deletions, and point mutations (Leister et al. 1998; Michelmore and Meyers 1998; Meyers et al. 1998a; Noel et al. 1999; Chin et al. 2001; Noir et al. 2001; Zhu et al. 2002; Ashfield et al. 2004; Kuang et al. 2004). The copy number in a large cluster of NBS-LRR loci harboring Pl 1 and other downy mildew R-genes on linkage group 8 is highly variable in sunflower, e.g., 2 to 24 copies/individual have been identified among elite and exotic individuals using INDEL fingerprinting (Slabaugh et al. 2003). The LG 8 cluster is the largest and most complex discovered so far in sunflower, with 54 NBS-LRR loci spanning a 36 cM segment in three subclusters (Fig. 4). One subcluster spans 12 cM, is primarily populated with T3 group members, has two T4 group members in the middle of the cluster (RGC138 and RGC139), and is demarcated on the upper end by two apparently telomeric or near-telomeric loci (RGC230 and RGC237) and lower end by additional T3 group NBS-LRR loci (e.g., RGC244). The second subcluster is slightly downstream, spans 7 cM, and harbors four T3 group members. Finally, additional unclassified NBS-LRR loci were found on the lower end of LG 8, two of which were identified from ESTs and could not be classified (ANN-RGC198 and PET-RGC221).
The second largest NBS-LRR cluster identified so far in sunflower is found on the lower end of LG 13, with 27 loci in two subclusters and a few singletons spanning a 31 cM segment (Fig. 4). This cluster harbors TIR- and non-TIR-NBS-LRR encoding genes. Other than three TIR-NBS-LRR loci, two from the T3 group at the upper end of the segment and one from the T1 group, loci in this cluster belong to a single non-TIR group (N1). This segment harbors downy mildew (Pl 5 and Pl 8) and black rust (R ADV) R-genes (Lawson et al. 1998; Radwan et al. 2003; Slabaugh et al. 2003; Yu et al. 2003), and is now much more densely populated with RGCs, which should facilitate the identification and cloning of R-genes for Pl 8 and R ADV (Fig. 4). The presence of complex mixed clusters of NBS-LRR loci, such as the TIR and non-TIR cluster found on LG 13, is common in the genomes of higher plants (Meyers et al. 1999; Zhu et al. 2002).
While an unknown number of genes encoding NBS-LRR proteins have presumably not yet been discovered in sunflower, a significant number have and supply resources for cataloging and cross-referencing phenotypic and quantitative trait loci conferring resistance to diverse pathogens in sunflower and, when coupled with the steadily growing collection of NBS-LRR sequences and mapped NBS-LRR loci, should greatly facilitate the discovery of additional R-genes and a greater understanding of R-gene diversity in domesticated and wild sunflower species (Michelmore and Meyers 1998; Noel et al. 1999; Ellis et al. 2000; Noir et al. 2001; Cannon et al. 2002; Michelmore 2003). Wild species have been an important source of R-genes in sunflower and, not coincidentally, RGCs identified from wild species ESTs were linked to several important R-genes in sunflower (Pl ARG, Or 5, Pl 8, R 1, and R ADV). Genetic analyses of RGCs for many of these R-genes are underway and should lead to the discovery of functionally important determinants of resistance and the development of additional high-throughput DNA markers for accelerating disease resistance breeding in sunflower through marker-assisted selection.
References
Altschul SF, Madden TL, Schaffer AA, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ameline-Torregrosa C, Wang BB, O’Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB (2007) Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol 146:5–21
Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes R (2004) Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 16:309–318
Baack EJ, Whitney KD, Rieseberg LH (2005) Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytol 167:623–630
Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE, Hulbert SH (2002) Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res 12:1871–1884
Bassam BJ, Caetano-Anollés G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Bent AF (1996) Plant disease resistance genes: function meets structure. Plant Cell 8:1757–1771
Bert PF, Labrouhe DTD, Philippon J, Mouzeyar S, Jouan I, Nicolas P, Vear F (2001) Identification of a second linkage group carrying genes controlling resistance to downy mildew (Plasmopara halstedii) in sunflower (Helianthus annuus L.). Theor Appl Genet 103:992–997
Bouzidi MF, Badaoui S, Cambon F, Vear F, De Labrouhe DT, Nicolas P, Mouzeyar S (2002) Molecular analysis of a major locus for resistance to downy mildew in sunflower with specific PCR-based markers. Theor Appl Genet 104:592–600
Cannon SB, Shu H, Baumgarten AM, Spangler R, May G, Cook DR, Young ND (2002) Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol 54:548–562
Chin DB, Arroyo-Garcia R, Ochoa OE, Kesseli RV, Lavelle DO, Michelmore RW (2001) Recombination and spontaneous mutation at the major cluster of resistance genes in lettuce (Lactuca sativa). Genetics 157:831–849
Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20:426–427
Collins NC, Webb CA, Seah S, Ellis JG, Hulbert SH, Pryor A (1998) The isolation and mapping of disease resistance gene analogs in maize. Mol Plant Microbe Interact 11:968–978
Dangl JL, Jones JDG (2001) Plant pathogens and integrated defense responses to infection. Nature 411:826–833
Degener J, Melchinger AE, Hahn V (1999) Interspecific hybrids as source of resistance to Sclerotinia and Phomopsis in sunflower breeding. Helia 22:49–60
Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS (1991) ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19:4008
Dußle CM, Hahn V, Knapp SJ, Bauer E (2004) Pl Arg from Helianthus argophyllus is unlinked to other known downy mildew resistance genes in sunflower. Theor Appl Genet 109:1083–1086
Ellis J, Dodds P, Pryor T (2000) The generation of plant disease resistance gene specificities. Trends Plant Sci 5:373–379
Gandhi SD, Heesacker AF, Freeman CA, Argyris J, Bradford K, Knapp SJ (2005) The self-incompatibility locus (S) and quantitative trait loci for self-pollination and seed dormancy in sunflower. Theor Appl Genet 111:619–629
Gedil MA, Slabaugh MB, Berry S, Johnson R, Michelmore R, Miller J, Gulya T, Knapp SJ (2001) Candidate disease resistance genes in sunflower cloned using conserved nucleotide-binding site motifs: genetic mapping and linkage to the downy mildew resistance gene Pl1. Genome 44:205–212
Gentzbittel L, Mouzeyar S, Badaoui S, Mestries E, Vear F, Tourvieille de Labrouhe D, Nicolas P (1998) Cloning of molecular markers for disease resistance in sunflower, Helianthus annuus L. Theor Appl Genet 96:519–525
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:79–92
Gonzales MD, Archuleta E, Farmer A, Gajendran K, Grant D, Shoemaker R, Beavis WD, Waugh ME (2005) The Legume Information System (LIS): an integrated information resource for comparative legume biology. Nucleic Acids Res 33:D660–D665
Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Ann Rev Plant Phys Plant Mol Biol 48:575–607
He L, Du C, Covaleda L, Xu Z, Robinson AF, Yu JZ, Kohel RJ, Zhang HB (2004) Cloning, characterization, and evolution of the NBS-LRR-encoding resistance gene analogue family in polyploid cotton (Gossypium hirsutum L.). Mol Plant Microbe Interact 17:1234–1241
Hecker KH, Roux KH (1996) High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. Biotechniques 20:478–485
Holt BF, Hubert DA, Dangl JL (2003) Resistance gene signaling in plants: complex similarities to animal innate immunity. Curr Opin Immunol 15:20–25
Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Ann Rev Phytopathol 39:285–312
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329
Kanazin V, Marek LF, Shoemaker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA 93:11746–11750
Kohler A, Rinaldi C, Duplessis S, Baucher M, Geelen D, Duchaussoy F, Meyers BC, Boerjan W, Martin F (2008) Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol Biol 66:619–636
Kuang H, Woo SS, Meyers BC, Nevo E, Michelmore RW (2004) Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16:2870–2894
Kukita Y, Tahira T, Sommer SS, Hayashi K (1997) SSCP analysis of long DNA fragments in low pH gel. Hum Mutat 10:400–407
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 25:511–557
Langar K, Griveau Y, Kaan F, Serieys H, Varès D, Bervillé A (2002) Evaluation of parameters accounting for Phomopsis resistance using natural infection and artificial inoculation on recombinant inbred lines from a cross between susceptible and resistant sunflower. Eur J Plant Path 108:307–315
Larsen LA, Christiansen M, Vuust J, Andersen PS (1999) High-throughput single-strand conformation polymorphism analysis by automated capillary electrophoresis: robust multiplex analysis and pattern-based identification of allelic variants. Hum Mutat 13:318–327
Larsen LA, Jespersgaard C, Andersen PS (2007) Single-strand conformation polymorphism analysis using capillary array electrophoresis for large-scale mutation detection. Nat Protoc 2:1458–1466
Lawrence GJ, Finnegan EJ, Ayliffe MA, Ellis JG (1995) The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 7:1195–1206
Lawson RR, Goulter KC, Henry RJ, Kong GA, Kochman JK (1998) Marker-assisted selection for two rust resistance genes in sunflower. Mol Breed 4:227–234
Lee SY, Seo JS, Rodriguez-Lanetty M (2003) Comprative analysis of superfamilies of NBS-encoding disease resistance gene analogs in cultivated and wild apple species. Mol Gen Genomics 269:101–108
Leister D, Ballvora A, Salamini F, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421–429
Leister D, Kurth J, Laurie DA, Yano M, Sasaki T, Devos K, Graner A, Schulze-Lefert P (1998) Rapid reorganization of resistance gene homologues in cereal genomes. Proc Natl Acad Sci USA 95:370–375
Lenardon SL, Bazzalo ME, Abratti G, Cimmino CJ, Galella MT, Grondona M, Giolitti F, Leon AJ (2005) Screening sunflower for resistance to sunflower chlorotic mottle virus and mapping the Rcmo-1 resistance gene. Crop Sci 45:735–739
Lincoln SE, Lander ES (1992) Systematic detection of errors in genetic linkage data. Genomics 14:604–610
Madsen LH, Collins NC, Rakwalska M, Backes G, Sandal N, Krusell L, Jensen J, Waterman EH, Jahoor A, Ayliffe M, Pryor AJ, Langridge P, Schulze-Lefert P, Stougaard J (2003) Barley disease resistance gene analogs of the NBS-LRR class: identification and mapping. Mol Genet Genomics 269:150–161
McDowell JM, Dhandaydham M, Long TA, Aarts MG, Goff S, Holub EB, Dangl JL (1998) Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10:1861–1874
McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7:212
Meyers BC, Shen KA, Rohani P, Gaut BS, Michelmore RW (1998a) Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10:1833–1846
Meyers BC, Chin DB, Shen KA, Sivaramakrishnan S, Lavelle DO, Zhang Z, Michelmore RW (1998b) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10:1817–1832
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Meyers BC, Morgante M, Michelmore RW (2002) TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J 32:77–92
Meyers BC, Kozik A, Griego A, Kuang H, Michelomore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Michelmore RW (2003) The impact zone: genomics and breeding for durable disease resistance. Curr Opin Plant Biol 6:397–404
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth- and- death process. Genome Res 8:1113–1130
Miller JF, Gulya TJ (1988) Registration of six downy mildew resistant sunflower germplasm lines. Crop Sci 28:1040–1041
Miller JF, Gulya TJ (1991) Inheritance of resistance to race 4 of downy mildew derived from interspecific cross in sunflower. Crop Sci 31:40–43
Miller JF, Gulya TJ, Seiler GJ (2002) Registration of five fertility restorer sunflower germplasms. Crop Sci 42:989
Murray MG, Thompson WR (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Noel L, Moores TL, van Der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JD (1999) Pronounced intraspecific haplotype divergence at RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11:2099–2112
Noir S, Combes MC, Anthony F, Lashermes P (2001) Origin, diversity and evolution of NBS-type disease-resistance gene homologues in coffee trees (Coffea L.). Mol Gent Genomics 265:654–662
Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA 86:2766–2770
Pan Q, Liu YS, Budai-Hadrian O, Sela M, Carmel-Goren L, Zamir D, Fluhr R (2000) Comparative genetics of nucleotide binding site-leucine rich repeat resistance gene homologues in the genomes of two dicotyledons: tomato and arabidopsis. Genetics 155:309–222
Parker C, Riches CR (1993) Parasitic weeds of the world: biology and control. CAB Int, Wallingford
Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD (1997) Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91:821–832
Pérez-Vich B, Akhtouch B, Knapp SJ, Leon AJ, Velasco L, Fernández-Martínez JM, Berry ST (2004) Quantitative trait loci for broomrape (Orobanche cumana Wallr.) resistance in sunflower. Theor Appl Genet 109:92–102
Plocik A, Layden J, Kesseli R (2004) Comparative analysis of NBS domain sequences of NBS-LRR disease resistance genes from sunflower, lettuce, and chicory. Mol Phylogen Evol 31:153–163
Pustovoit GV, Kroknin EY (1978) Inheritance of resistance in interspecific hybrids of sunflower to downy mildew. Rev Plant Pathol 57:209
Quresh Z, Jan CC, Gulya TJ (1993) Resistance of sunflower rust and its inheritance in wild sunflower species. Plant Breed 110:297–306
Radwan O, Bouzidi MF, Vear F, Philippon J, Tourvieille de Labrouhe D, Nicolas P, Mouzeyar S (2003) Identification of non-TIR-NBS-LRR markers linked to PL5/PL8 locus for resistance to downy mildew in sunflower. Theor Appl Genet 106:1438–1446
Radwan O, Bouzidi MF, Nicolas P, Mouzeyar S (2004) Development of PCR markers for the Pl5/ Pl8 locus for resistance to Plasmopara halstedii in sunflower, Helianthus annuus L. from complete CC-NBS-LRR sequences. Theor Appl Genet 109:176–185
Richly E, Kurth J, Leister D (2002) Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol 19:76–84
Rossi M, Araujo PG, Paulet F, Garsmeur O, Dias VM, Chen H, Van Sluys MA, D’Hont A (2003) Genomic distribution and characterization of EST-derived resistance gene analogs (RGAs) in sugarcane. Mol Genet Genomics 269:406–419
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Seiler GJ (1991a) Registration of 15 interspecific sunflower germplasm lines derived from wild annual species. Crop Sci 31:1389–1390
Seiler GJ (1991b) Registration of 13 downy mildew tolerant interspecific sunflower germplasm lines derived from wild annual species. Crop Sci 31:1714–1716
Shen KA, Keyers BC, Islam-Faridi MN, Chin DB, Stelly DM, Michelmore RW (1998) Resistance gene candidate identified by PCR with degenerate oligonucleotide primers map to clusters of resistance genes in lettuce. Mol Plant Microbe Interact 11:815–823
Škoric D (1985) Sunflower breeding for resistance to Diaporthe/Phomopsis helianthi. Helia 8:21–23
Slabaugh MB, Yu JK, Tang S, Heesacker A, Hu X, Lu G, Han F, Bidney D, Knapp SJ (2003) Haplotyping and mapping a large cluster of resistance gene candidates in sunflower using multilocus intron fragment length polymorphisms. Plant Biotech J 1:167–185
Sun Q, Collins NC, Ayliffe M, Smith SM, Drake J, Pryor T, Hulbert SH (2001) Recombination between paralogues at the Rp1 rust resistance locus in maize. Genetics 158:423–438
Tan AS, Jan CC, Gulya TJ (1992) Inheritance of resistance to race 4 of sunflower downy mildew in wild sunflower accessions. Crop Sci 32:949–952
Tang S, Yu JK, Slabaugh MB, Shintani DK, Knapp SJ (2002) Simple sequence repeat map of the sunflower genome. Theor Appl Genet 105:1124–1136
Tang S, Heesacker A, Kishore VK, Fernandez A, Sadik E, Cole G, Knapp SJ (2003) Genetic mapping of the Or5 gene for resistance to Orobanche race E in sunflower. Crop Sci 43:1021–1028
Tanksley SD, McCouch S (1997) Seed banks and molecular maps: unlocking the genetic potential from the wild. Science 277:1063–1066
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgens DG (1997) The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vear F, De Labrouhe DT, Miller JF (2003) Inheritance of the wide-range downy mildew resistance in the sunflower line RHA 419. Helia 26:19–24
Vranceanu AV, Tudor VA, Stoenescu FM, Pirvu N (1980) Virulence groups of Orobanche cumana Wallr., different hosts and resistance sources and genes in sunflower. In: Proceedings of the 9th International Sunflower Conference, Torremolinos, Spain, pp 74–82
Viguié A, Labrouhe DTD, Vear F (2000) Inheritance of several sources of resistance to Phomopsis stem canker (Diaporthe helianthi Munt.-Cvet.) in sunflower (Helianthus annuus L.). Euphytica 116:167–179
Wang Z, Taramino G, Yang D, Liu G, Tingey SV, Miao GH, Wang GL (2001) Rice ESTs with disease-resistance gene or defense-response gene-like sequences mapped to regions containing major resistance genes or QTLs. Mol Gen Genet 265:302–310
Wei FS, Wong RA, Wise RP (2002) Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14:1903–1917
Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B (1994) The product of tobacco mosaic virus resistance gene N: similarity to Toll and interleukin-1 receptor. Cell 78:1101–1115
Yu T, Buss GR, Saghai-Maroof MA (1996) Isolation of the super family of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc Natl Acad Sci USA 93:11751–11756
Yu JK, Tang S, Slabaugh MB, Heesacker AF, Cole G, Herring M, Soper J, Han F, Chu WC, Webb DM, Thompson L, Edwards KJ, Berry ST, León AJ, Olungu C, Maes N, Knapp SJ (2003) Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci 43:367–387
Zhu H, Cannon SB, Young ND, Cook DR (2002) Phylogeny and genomic organization of the TIR and non-TIR NBS-LRR resistance gene family in Medicago truncatula. Mol Plant Microbe Interact 15:529–539
Acknowledgments
This research was supported by grants from the United States Department of Agriculture National Research Initiative Plant Genome Program (No. 2000-04292) and National Science Foundation Plant Genome Program (No. 0421630).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Shirasu.
DDBJ/EMBL/GenBank sequence accession numbers. Sunflower nucleotide and amino acid sequences have been deposited in DDBJ/EMBL/GenBank under accession numbers EF560168-EF559378 and ABQ58077-ABQ57529.
Appendix
Appendix
S1. GeneDoc (http://www.nrbsc.org/gfx/genedoc/index.html) input file displaying a CLUSTAL-X alignment of 785 sunflower NBS nucleotide sequences.
S2. GeneDoc (http://www.nrbsc.org/gfx/genedoc/index.html) input file displaying a CLUSTAL-X alignment of 69 sunflower NBS amino acid sequences sharing less than 90% similarity.
S3. Sunflower NBS-LRR sequence database.
S4. Sunflower NBS-LRR SSCP and INDEL marker database.
Rights and permissions
About this article
Cite this article
Radwan, O., Gandhi, S., Heesacker, A. et al. Genetic diversity and genomic distribution of homologs encoding NBS-LRR disease resistance proteins in sunflower. Mol Genet Genomics 280, 111–125 (2008). https://doi.org/10.1007/s00438-008-0346-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-008-0346-1