Abstract
The broadest based resistance to anthracnose of common bean (Phaseolus vulgaris L.) is conferred by the Co-4 locus. We sequenced a bacterial artificial chromosome clone harboring part of the Co-4 locus of the bean genotype Sprite and assembled a single contig of 106.5 kb for functional annotation. This region contained five copies of the COK-4 gene that encodes for a serine threonine kinase protein previously mapped to the Co-4 locus and 19 novel genes with no similarity to any previously identified genes of common bean. Several putative genes of the Co-4 locus seemed to be expressed as they matched perfectly with bean expressed sequence tags. The expression of the COK-4 genes was assessed by reverse transcription (RT)-PCR, and a single 850-bp cDNA fragment was sequenced and compared with the genomic sequences of the COK-4 homologs. Although the COK-4 cDNA was isolated from a different bean cultivar, it showed high similarity (95%) to the exons of genes BA17 and BA21, suggesting that they were expressed. In a phylogenetic tree including all currently available Pto-like sequences from Phaseolus species, the COK-4 homologs formed a single cluster with the Pto gene, whereas two sequences from P. coccineus and all sequences of P. vulgaris formed two closely related clusters. The Co-4 locus was physically mapped to the short arm of bean chromosome 3, which corresponds to linkage group B8. This study represents a first step in gaining an understanding of the genomic organization of an anthracnose resistance locus of common bean and provides molecular data for comparative analysis with other plant species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Independent of the plant taxa or the pathogen involved, resistance gene-encoded proteins have been shown to possess a small number of conserved functional domains, such as leucine-rich repeats (LRR), nucleotide-binding sites (NBS), leucine zippers (LZ), and protein kinases (PK) (Hammond-Kosack and Jones 1997). Each resistance gene may encode one or more domains. For example, the Pto gene encodes for a serine threonine kinase (STK; Martin et al. 1993) protein, whereas RPM1 encodes for a three-domain protein with LZ, NBS, and LRR (Boyes et al. 1998). Some resistance genes that encode for LRR and NBS domains are divided into two classes based on the presence of either a Toll/interleukin 1 cytoplasmic receptor (TIR) or a coiled coil (CC) domain at the amino terminus of the protein (Graham et al. 2002).
Each domain has a specific role in the resistance/defense process. In some instances, LRR proteins appear to be responsible for recognition specificity as they can bind to the corresponding ligands from the pathogen (Jia et al. 2000). In other cases, LRR proteins interact with putative targets of pathogen virulence proteins (Dixon et al. 2000). NBS, LZ, and kinases may be involved in signal transduction pathways and they are usually involved in activating other proteins, which may trigger a chain of events leading to a defense response (Bent 1996; Hammond-Kosack and Jones 1997). Unlike LRR and NBS, the STK domain found in disease resistance proteins may also be involved in other cellular processes, which makes it difficult to isolate an STK-like disease resistance gene in a certain species using the nucleotide information of this domain from another unrelated species. However, the STK domain of resistance genes is evolutionarily different from other kinase domains as it clusters separately in phylogenetic trees (Vallad et al. 2001). Pto-like kinases are unique to plant species, unlike other kinases that are present in most eukaryotes (Vallad et al. 2001). Therefore, disease resistance genes that encode for kinases may be identified in other species based on comparisons with the Pto gene. However, genetic linkage and functionality still have to be demonstrated to assign the role of those kinases in conferring resistance to pathogens.
The ideal approach to finding genes of interest is to have whole genome sequences available. However, due to the large size of many plant genomes, this is not economically feasible for many crop plants. A more cost-effective approach is to sequence relatively short genomic fragments known to carry the target gene. The Co-4 locus that confers resistance to anthracnose in common bean has been tagged with several molecular markers (Young et al. 1998; Melotto and Kelly 2001) and is a good candidate for map-based cloning. This locus is highly valuable for breeding purposes as it controls 97% of all currently identified races of the pathogen Colletotrichum lindemuthianum (Melotto et al. 2000). We recently provided genetic and molecular evidence that Co-4 is a multi-gene family with single nucleotide polymorphisms (SNPs) among homologs of different bean genotypes possessing different alleles at the Co-4 locus. A Pto-like kinase gene, COK-4, was also identified in the Co-4 locus (Melotto and Kelly 2001). However, neither the physical location of this locus in the bean chromosomes nor the genomic content of this locus has been characterized.
In the investigation reported here, we were provided with a bacterial artificial chromosome (BAC) library from the bean cultivar Sprite (Vanhouten and MacKenzie 1999) to further characterize the Co-4 locus. Using the SAS13 marker as a probe, we were able to identify four BAC clones. The largest BAC clone, which contained a 106.5-kb genomic region linked to the Co-4 locus, was completely sequenced and annotated. The expression of the COK-4 gene homologs and several other genes was assessed by comparison with expressed sequence tagged (EST) sequences and/or reverse transcription (RT)-PCR. The bean COK-4 gene homologs were also compared to all previously described Pto-like gene sequences in the genus Phaseolus using phylogenetic analysis. In addition, the BAC clone was physically mapped to the bean chromosome 3 using fluorescent in situ hybridization (FISH). This is the first report on the genomic content of the longest contiguous DNA sequence of common bean.
Materials and methods
Genetic material
The four BAC clones used in this study—78L17, gF11, 53Eg, and 56P12—were selected from a bean library constructed with the snap bean cultivar Sprite (Vanhouten and MacKenzie 1999) using the SAS13 marker previously found to be tightly linked to the Co-4 2 locus (Young et al. 1998). The presence of the SAS13 marker in the four BAC clones was also tested using PCR as described by Young et al. (1998). The FISH experiment was conducted with bean genotype Calima from Colombia, which had been used to establish a reference integrated chromosomal map of common bean (Pedrosa et al. 2003). COK-4 cDNA was isolated from bean genotype SEL 1308 that carries the Co-4 2 locus. SEL 1308 was derived from a backcross between the anthracnose-susceptible cultivar Talamanca from Costa Rica and the resistant parent Colorado de Teopisca (CIAT accession number G2333) from Mexico.
Pulse field gel electrophoresis (PFGE)
To determine the size of the BAC clone inserts, we digested the plasmids with NotI in the presence of bovine serum albumin and separated the resulting fragments on a 1% ultra pure agarose gel using a contour-clamped homogeneous electric field unit (CHEF DRII; BioRad, Hercules, Calif.) under the following conditions: ramp time of 1–12 s, 6 V/cm, 0.5× TBE buffer, running time of 15 h at 15°C.
Shotgun sub-cloning and sequencing
The 78L17 BAC plasmid DNA was extracted with the Plasmid Midi kit (Qiagen, Valencia, Calif.) from cells cultured for 16 h in 200 ml of Luria-Bertani (LB) medium at 37°C or until culture absorbance at 600 nm reached 1–1.5. The BAC plasmid (1 μg) was then sonicated once for 10 s using an ultrasonic homogenizer (Cole Parmer) to produce fragments between 700 bp and 1,400 bp. The sonicated sample was checked for size and quantity on an agarose gel. The shotgun sub-cloning library was constructed with 100 ng of sonicated DNA. DNA fragment end repair, dephosphorylation, ligation to the PCR 4Blunt-TOPO vector, and Escherichia coli transformation were performed with the TOPO Shotgun Subcloning kit (Invitrogen, Carlsbad, Calif.) following the manufacturer’s protocol. Recombinant colonies were incubated in LB medium supplemented with 100 μg/ml ampicillin and 8% glycerol for 20 h at 37°C in 96-well microplates.
Plasmid miniprep was conducted in 96-well plates using standard procedures. DNA sequencing was performed using the Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, Calif.) and the M13 reverse or M13 forward primer. The PCR profile consisted of 25 cycles of 10 s at 96°C, 5 s at 50°C, and 4 min at 60°C. Following PCR product precipitation, automated sequencing was carried out in a capillary system (3700 ABI Sequencer, Applied Biosystems). BAC-end sequencing was performed as described above, except that the DNA template used for the PCR reaction was BAC plasmid (500 ng) sonicated into lengths of 2–4 kb to facilitate the sequencing of the DNA downstream of the M13 primer.
Sequence assembly, finishing and annotation
Sequence reads were assembled using the phred/phrap/consed package (http://www.phrap.org Ewing et al. 1998; Ewing and Green 1998; Gordon et al. 1998). To improve the quality of some regions and to confirm the sequence of contiguous repetitive regions, we used flanking primers designed with the primer picking function of consed to amplify those regions. PCR products were purified, both strands were sequenced, and the resulting reads were added to the assembly. Each base was sequenced at least twice with a phred quality higher than 20. The predicted restriction map of the final consensus sequence was compared with EcoRI, EcoRV, and HindIII digests of the BAC plasmid in order to confirm its assemblage (data not shown).
The consensus sequence was analyzed for the presence of coding regions with two different computer programs— genescan (Burge and Karlin 1997; http://genes.mit.edu/GENSCAN.html) and fgenesh (http://www.softberry.com/)—using Arabidopsis as the model organism. The putative function of coding regions was determined by comparison with known genes available at the GenBank database using the blastx algorithm (Altschul et al. 1997). Conserved domains were identified using the motif scan software (http://hits.isb-sib.ch/cgi-bin/PFSCAN Falquet et al. 2002). Predicted genes were also compared to the bean EST database (http://lgm.esalq.usp.br/BEST Melotto et al., unpublished) and to plant unigene databases (Wheeler et al. 2003) of the National Center for Biotechnology Information (NCBI) using blastn to assess the expression of those genes. Sequences of predicted genes with same function were aligned with the clustalw software (Thompson et al. 1994) to assess their similarities. The annotated BAC sequence herein described has been deposited in the NCBI database under accession number AY341443.
Reverse transcriptase-PCR analysis
mRNA samples from SEL 1308 seedlings inoculated with race 73 of Colletotrichum lindemuthianum and non-inoculated control seedlings were used as templates in the reverse transcriptase (RT) reaction using the GeneRacer kit (Invitrogen). cDNA was synthesized using the oligo dT primer and 1 μg of mRNA in a reaction incubated for 1 h at 42°C according to the manufacturer’s instructions. The COK-4 cDNA was PCR-amplified with the COK-4-specific primers described by Melotto and Kelly (2001). The PCR reaction contained 1× PCR buffer, 200 μM dNTP, 1 μl of the cDNA reaction, 10 pmol of each primer, 1 U ThermoZyme DNA polymerase (Invitrogen) in a total volume of 50 μl. Controls to test for DNA contamination of the mRNA samples were included in this analysis where mRNA was used in the PCR reaction instead of cDNA. The amplification profile consisted of 3 min at 94°C, 30 cycles of 30 s at 94°C, 30 s at 70°C, and 2 min at 72°C, and final extension of 7 min at 72°C. To produce a well-defined band, we carried out a second-round of PCR under the same conditions as described above using 1 μl of the first PCR reaction. Although a high-fidelity DNA polymerase was used in the PCR reaction, one cannot discard the possibility of nucleotide substitution during amplification resulting in sequencing errors. This experiment was conducted twice.
COK-4 cDNA fragments isolated from inoculated or non-inoculated bean seedlings were cloned using the TOPO TA Cloning kit (Invitrogen). Both strands of the cloned cDNA fragments were sequenced using a 377 DNA Sequencer (Applied Biosystems) as previously described by Melotto et al. (1996). cDNA sequences were compared to the genomic DNA sequence of the COK-4 gene using the two-sequence alignment software (blast2) available at the NCBI web site and also to the protein database (Pfam at http://www.sanger.ac.uk/Software/Pfam/ ; Bateman et al. 2002) to determine its putative function.
Phylogenetic analysis of bean serine/threonine kinases
The COK-4 gene homologs were aligned to previously cloned STKs of common bean, OG5, OG8, OG9, SG1, and SG5 (accession nos. gi|14010506, gi|14010500, gi|140104898, gi|14010488, gi|14010482, respectively; Vallad et al. 2001), two Pto-like ESTs from Phaseolus coccineus (gi|27395850 and gi|27395849), and to the Pto protein from tomato (gi|1809257) using the clustalw software. The phylogenetic trees of amino acid sequences were constructed using parsimony analysis, and the robustness of the final topology was tested with the tree-building method PAUP (Swofford 1998). An STK sequence of Arabidopsis (Swiss Prot accession no. Q06548) was used as the outgroup.
FISH mapping
The BAC clone 56P12 was used to physically map the Co-4 locus on the common bean chromosomes. Assignment of this clone to a specific chromosomal region was confirmed by re-hybridization with a pool of [restriction fragment length polymorphism (RFLP)] clones from linkage group F (RFLP pool F′: Bng 62, Bng 69, Bng 125, Bng 186, Bng 214) corresponding to linkage group B8 of the bean core map (Freyre et al. 1998). This pool of clones can be used as a marker for the short arm of chromosome 3 (Vallejos et al. 1992; Pedrosa et al. 2003). Both probes were labeled by nick translation (Roche Diagnostics, Indianapolis, Ind.) with Cy3-dUTP (Amersham Pharmacia Biotech, Piscataway, N.J.).
Chromosome preparation was as described in Pedrosa et al. (2003) and slide selection and pre-treatment as in Pedrosa et al. (2001). Chromosome and probe denaturation and post-hybridization washes were performed according to Heslop-Harrison et al. (1991), with the modifications described in Pedrosa et al. (2002) for hybridization without blocking DNA. Preparations were counterstained and mounted with 2 μg/ml DAPI in Vectashield (Vector). Re-probing of slides for confirmation of chromosome identity was performed according to Heslop-Harrison et al. (1992).
Photographs were taken on a Zeiss Axioplan (Carl Zeiss) equipped with a mono cool view CCD camera (Photometrics). Images from the camera were combined and pseudo-colored using the IPLab spectrum software (iplab). The distance of FISH signals to the closest telomere and total chromosome length were measured using the analyze-measure length function of the same software. The position of the BAC clone was based on measurements of 20 chromosomes and was defined as a proportion of the total chromosome length (telomere of the short arm is indicated as zero and telomere of the long arm as one). This position was integrated into the map of chromosome 3 established by Pedrosa et al. (2003). Digital images were imported into adobe photoshop ver. 6 for final processing.
Results
Selection of bean BAC clones and shotgun library preparation
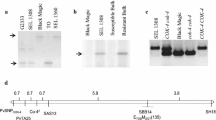
A bean BAC library of the cultivar Sprite that covers three- to five fold the bean genome (Vanhouten and Mackenzie 1999) was screened with the SAS13 marker that is tightly linked to the Co-4 locus that confers resistance to anthracnose in common bean (Young et al. 1998). This marker contains part of an open-reading frame (ORF) that encodes for an STK similar to the Pto gene of tomato. Four BAC clones that strongly hybridized with the marker were selected for further analysis. Of these four BAC clones, we chose to sequence 78L17 because it exhibited the SAS13 banding pattern (Fig. 1a) similar to that of the anthracnose-resistant genotype SEL 1308 and carried the longest insert as determined by PFGE (greater than 100 kb; Fig. 1b). The SAS13 primers also amplified other bands from the 78L17 clone, suggesting the presence of repetitive sequences. The BAC-end sequencing of the 56P12 clone indicated that it does not extend the 78L17 clone and carries a genomic fragment smaller than that of 78L17 (Fig. 1b). The genome region covered by the 56P12 insert is shown in Fig. 2.
a DNA amplification using the SAS13 primers. Lanes: 1 100-bp ladder (Gibco, Gaithersburg, Md.), 2 anthracnose resistance bean genotype SEL 1308, 3 anthracnose-susceptible bean genotype Black Magic, 4 56P12, 5 gF11, 6 78L17, 7 53Eg. b Pulse field agarose gel showing the approximate insert size of the BAC clones. Lanes: 1 78L17, 2 gF11, 3 53Eg, 4 56P12, 5 low-range molecular-weight marker (New England Biolabs, Beverly, Mass.). Numbers on the left or right of each panel indicate molecular weight in kilobases
The predicted genes found in BAC clone 78L17. Genes that could not be assigned with a putative function based on the blastx results are classified as “hypothetical protein (HP)”. Exons are indicated by rectangles and introns as the thin lines between them. The coding strand is indicated by the arrowhead of each gene. The three segments with high homology to known genes but with no identified ORFs are indicated as thin-interrupted arrows. The position of the reads corresponding to the end-sequence of BAC clone 56P12 is shown as a small gray arrow. Thin arrows under some genes indicate the alignment of the gene exons with corresponding bean ESTs. Numbers to the left are the positions in kilobases of the 78L17 BAC insert
A shotgun library of the 78L17 clone was prepared, and 2,400 sub-clones were picked for sequencing. A sample of 20 sub-clones was used to calculate the average insert size present in the library by PCR amplification with the M13R and M13F primers. The average insert size was 780 bp and ranged from 400 bp to 1,600 bp (data not shown). These 2,400 clones were sequenced in one direction, and their sequences were trimmed for vector sequences and low-quality bases, which sometimes resulted in entire reads being removed before assembly. Trimming, assembling, and analyses were performed with the phred/phrap/consed software package. In this first analysis 20 contigs were formed, and although some of these contained highly similar sequences these contigs did not align completely. To close the gaps and to assemble the repeated regions correctly, we sequenced the opposite DNA strand of the clones placed at the end of each contig. Several primer sets were also designed based on the flanking sequences of the repetitive regions of the BAC insert. Single fragments amplified by each primer set were sequenced in both directions, and the consensus sequence was used as an anchor to help in the assembly of the highly similar repeats. A total of 2,165 reads was used to produce a single contig of 106,592 bp with an average phred quality of 86.5 per base and a minimum quality of 20. This contig size is equivalent to the size of the BAC insert as determined by PFGE.
Functional annotation of the BAC clone 78L17
Analysis of the 106,592-bp sequence located at the Co-4 region revealed the presence of 24 putative genes (Table 1, Fig. 2), denoted BA1 to BA24, where BA stands for bean anthracnose. The predicted gene sequences were compared with the GenBank database using the blastx algorithm (Altschul et al. 1997). It was possible to identify seven novel putative genes with no similarities to any other genes currently described, five copies of the COK-4 gene, and genes that encoded for retrotransposon elements, anthocyanin acyltransferase, mitochondrial carrier protein, phytochelatin synthetases, reversibly glycosylated protein, cytochrome P450, and B12D protein ortholog.
Three regions with no predicted genes were found to have similarities to previously described genes and were considered to be gene segments (Fig. 2). Segment one, 851 bp, had similarities with disease resistance genes, such as the Cf-4 of tomato, Xa21 of rice, and other genes containing LRR domains. The largest ORF identified in this region (444 bp) encoded for a LRR domain (E-value of 3.6−25), as indicated by a scan of this sequence against the HMM library (Gough et al. 2001; http://supfam.org/Superfamily). Segment two had similarities with RTs associated with transposable elements. This segment was found inside an intron but was coded by the opposite strand of the predicted gene BA18, close to a predicted retrotransposon (gene BA19). The third segment was highly similar to the COK-4 gene (Melotto and Kelly 2001) and was located between two copies of the COK-4 gene.
Amino acid sequences of the predicted genes were compared to the protein database Pfam to determine the presence of conserved domains (Table 1 in electronic supplementary material). The predicted genes BA5, BA14, BA15, BA16, BA18, and BA20 showed no match with conserved domains available in the database. All of these genes had no match with genes available in the GenBank, except for gene BA16 that encoded for a B12D protein homolog. The retrotransposon elements, genes BA1, BA4, BA19, and BA23, differed at the nucleotide level, however, all four encoded for retroviral Pol protein and the last three also encoded for GAG protein. Genes BA1, BA4, and BA19 are copia-type retrotransposons, whereas gene BA23 is of a gypsy type. The COK-4 homologs, genes BA3, BA6, BA17, BA21, and BA22, encoded for PK with one ATP-binding region signature or tyrosine PK-specific active-site signature. The anthocyanin 5-aromatic acyltransferase (gene BA7) encoded for a transferase, indicating that this gene may be involved in anthocyanin biosynthesis. The conserved domains encoded by gene BA8 (mitochondrial carrier protein, PF00153), genes BA9, BA10, and BA11, (phytochelatin synthetases, PF04833), and gene BA13 (cytochrome P450) agree with the results obtained with blastx. The reversibly glycosylated protein (gene BA12) encodes for a glycosyl transferase (PF00535) that may be involved in the biosynthesis of polysaccharides. Finally, genes BA2 and BA24, which showed no homology to any known gene, encoded for conserved sequences. BA2 had two phosphorylation sites, and BA24 had a lectin domain of a ricin B chain profile.
The consensus sequence of the bean BAC clone 78L17 was compared to the genomes of Zea mays, Brassica oleracea, Medicago truncatula, and Arabidopsis thaliana through the megablast search tool, however no synteny was observed. Only three of the predicted genes that encode for mitochondrial carrier protein (BA8), phytochelatin synthetase (BA11), and reversibly glycosylated protein (BA12) were found to be putative orthologs (E-value=0.0) of the Arabidopsis genes At5 g15640, At5 g15630, At5 g15650, respectively. These genes are clustered in the Arabidopsis chromosome V as determined by the sequencing and mapping of the BAC clone F14F8 (http://www.tigr.org/ ).
The contig was also compared to bean ESTs and plant unigenes databases to determine whether the predicted genes are expressed at the mRNA level. All significant alignments occurred with regions of the contig containing the predicted genes (Table 2 in electronic supplementary material). Although the phytochelatin synthetases, genes BA9, BA10, and BA11, are very similar to each other, a nearly perfect match was only observed between the bean EST PVEPSE2034B07.g (CB541439) and gene BA11 (99%), suggesting that this is the only detectably expressed copy among the three herein described. Another bean EST, PVEPSE2004A02.g (CB539337), showed 71% similarity to gene BA10 and 87% similarity to gene BA11, indicating that other expressed copies of the phytochelatin synthetase gene exist in other regions of the bean genome. The gene BA12, which encodes for a reversibly glycosylated protein, was similar (80% identity) to the bean EST PVEPSE2027E09.g (CB541006), the cytochrome P450 gene (BA13) showed 100% nucleotide match to the bean EST PVEPSE2012F04.g (CB539942), and the retrotransposon (gene BA23) was similar (77% identity) to the bean EST PVEPSE3030B07.g (CB543509).
Perfect alignment was observed between the mitochondrial carrier protein (BA8) and the phytochelatin synthetase (gene BA10) with ESTs sequenced from P. coccineus (CA907529 and CA910091, respectively). These two genes along with reversibly glycosylated protein (BA12) and cytochrome P450 (BA13) appeared to be the most conserved genes among the plant species compared. The disease resistance gene candidates did not align with any bean EST. However, high similarities (76–80%) were observed between the COK-4 homologs and soybean ESTs (Table 2 in electronic supplementary material).
Isolation of the COK-4 cDNA
In two independent hybridization experiments that were carried out to detect the transcript of the COK-4 gene in bean genotypes, we were unable to detect any with either [32P]- or digoxigenin (DIG)-labeling and detection system on the X-ray film, except for the control band (data not shown). Sensitivity of the COK-4 probe was tested, and as little as 100 pg of plasmid DNA could be detected on the membrane (data not shown). The COK-4 transcript could only be detected by RT-PCR due to its low abundance. An 850-bp cDNA band was amplified from RNA samples collected from either inoculated or non-inoculated seedlings of SEL 1308 (Fig. 3), indicating that the COK-4 gene is not induced upon pathogen infection. cDNA from both samples were cloned and sequenced, and they proved to be identical, indicating that they were transcripts of the same gene; the protocol for its amplification was reproducible.
Agarose gel showing the RT-PCR analysis of the COK-4 gene using mRNA collected from SEL 1308 seedlings (lanes 2, 3) or SEL 1308 seedlings inoculated with anthracnose (lanes 4, 5). Lanes: 1 100-bp DNA ladder (Gibco), 2, 4 PCR reaction using cDNA as template, 3, 5 control PCR reaction using mRNA as a template to test for DNA contamination of the mRNA samples. a Primary PCR, b Secondary PCR. Molecular weight in kilobases is indicated by the numbers on the left
Sequence comparison and phylogenetic analysis of the COK-4 homologs
The predicted mRNA sequences of the five COK-4 homologs found in BAC clone 78L17, the previously cloned COK-4 gene (gb|AAF98554.1; Melotto and Kelly 2001), and the COK-4 cDNA were aligned to determine their similarities (Fig. 1 in electronic supplementary material) using the clustalw software. The SEL 1308 COK-4 gene homolog possibly has an intron between positions 817 and 894 of the alignment, even though a single ORF spanning the intron has been identified. A second intron that appears as a gap in the COK-4 cDNA sequence may also exist in all genomic sequences of COK-4 homologs between positions 996 and 1,016. This intron was not detected by the gene prediction softwares used in this study. Two COK-4 homologs (gene BA17 and BA21), the previously cloned COK-4 gene (Melotto and Kelly 2001), and the COK-4 cDNA proved to be very similar (95% identity), an observation also supported by the phylogenetic analysis presented below.
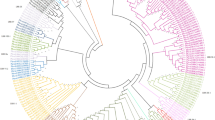
To determine the relationship among the COK-4 homologs obtained in this study and previously described Pto-like sequences of Phaseolus sp. available in the GenBank database, we constructed a phylogenetic tree was constructed using parsimony analysis (Fig. 4). Two well-supported groups were formed. The COK-4 homologs formed a single cluster with the Pto gene, whereas the two ESTs from P. coccineus, and the P. vulgaris STKs (Vallad et al. 2001) formed a separate cluster. Sub-groups within clusters were observed, and this analysis supported the close proximity of the genes BA17, BA21 and the COK-4 homolog present in the bean genotype SEL 1308. Interestingly, predicted COK-4 genes with two and three exons (BA3, BA6, and BA22) fell in one sub-group, and genes with four exons (BA17 and BA21) fell in the other sub-group.
Phylogenetic tree based on the amino acid sequences of the COK-4 homologs, five bean serine/threonine kinases (gi|14010506, gi|14010500, gi|140104898, gi|14010488, gi|14010482), two ESTs isolated from Phaseolus coccineus (gi|27395850 and gi|27395849), the Pto protein of tomato (gi|1809257), and a serine/threonine kinase from Arabidopsis (Q06548). Sequences identified in this study are in bold. An unrooted tree was constructed using parsimony analysis, and the sequence of Arabidopsis was treated as outgroup. Numbers at the nodes represent percentage bootstrap values of 1,000 re-samplings
Chromosomal localization of the Co-4 locus
We chose the BAC clone 56P12 to localize the Co-4 locus by FISH because its insert covers a region that is included in BAC 78L17, but it is shorter and contains less repetitive sequences than 78L17. BAC clone 56P12 hybridized clearly and reproducibly to a chromosome pair that was tentatively identified as bean chromosome 3 (Fig. 5a). To confirm this location, we re-probed the same preparation with the RFLP pool F′, which is a marker for the short arm of chromosome 3. Signals from this probe partially co-localized with signals from BAC 56P12 (Fig. 5b), thereby confirming the assignment of the Co-4 locus to the short arm of chromosome 3 and indicating that this locus is closely linked to the markers included in the RFLP pool F’. This result is in agreement with the mapping of Co-4 to linkage group B8 of the bean core linkage map (Kelly et al. 2003). Linkage group F of the University of Florida map was previously aligned with the B8 of the bean core map (Freyre et al. 1998). Measurements placed the Co-4 locus at position 0.17 (SD=0.07, n=25) of that chromosome. Figure 5c shows a schematic representation of the position of this locus on chromosome 3 and the correspondence to linkage groups B8 and F.
Localization of BAC 56P12 on common bean chromosomes. a BAC 56P12 (visualized in green) hybridized to a single locus on a chromosome pair tentatively identified as chromosome 3. b RFLP pool F′ (red) confirmed the localization of BAC 56P12 (green) in the distal region of the short arm of chromosome 3. c BAC 56P12 can be assigned to linkage group F (University of Florida map) or linkage group B8 of the bean core map. Bar (a): 2.5 μm
Discussion
Previous studies established that the Co-4 locus that confers resistance to anthracnose of common bean is complex and presumably the site of a multi-allelic series (Young et al. 1998). Genetic characterization of this locus resulted in the isolation of the COK-4 gene (Melotto and Kelly 2001), which is similar to the Pto gene of tomato (Martin et al. 1993). Southern analysis indicated that several COK-4 homologs are clustered in the bean genome, but the exact number of copies, the size of the genomic region spanning these copies, and the expression of this gene were not determined. To address these questions, we selected a bean BAC clone, then fully sequenced, annotated, and compared it to plant unigene databases in order to assess the expression of the COK-4 gene and other genes that might be present in that region. In addition, we physically located the Co-4 locus on bean chromosome 3.
Our analysis of this bean genomic sequence revealed the presence of five copies of the COK-4 gene that had been previously identified in bean genotype SEL 1308 (gb|AAF98554.1), which agrees with the previous observation that this is a complex locus (Melotto and Kelly 2001). We identified 19 other genes clustered with the COK-4 genes, none of which had similarity with any of the Phaseolus genes currently deposited in the GenBank database. Therefore, these 19 are novel genes of common bean. To assess the expression of these genes, we compared their putative coding sequences to plant unigene databases, including the bean EST database (http://lgm.esalq.usp.br/BEST ) recently developed by our group (Melotto et al., unpublished). The phytochelatin synthetases, mitochondrial carrier protein, and reversibly glycosylated protein have the most conserved sequences among the plant unigene databases used for comparison. The transcripts of their genes were detected several times in the unigene databases, suggesting that they are expressed at high levels.
Transcripts of the COK-4 gene were not identified in the currently available unigene databases. The expression of the COK-4 gene was assessed by RT-PCR as different methods of probe labeling and high amounts of RNA in membranes (10–15 μg of total RNA or up to 5 μg of mRNA) were not sensitive enough to detect the COK-4 transcript. These results indicated that COK-4 is expressed at low levels. Notwithstanding, RT-PCR analysis showed that the COK-4 gene is expressed in common bean seedlings as its cDNA was isolated from both non-inoculated and inoculated plants. Our analysis of the COK-4 cDNA indicated that it contains a conserved domain similar to the STK that is present in the Pto gene of tomato that is known to confer resistance to Pseudomonas syringae (Martin et al. 1993). The cDNA sequence and genomic sequences of three COK-4 homologs (SEL 1308, BA17, and BA21) share 95% identity. The COK-4 cDNA was isolated from the bean genotype SEL 1308 that carries at least two copies of the COK-4 gene (Melotto and Kelly 2001), and only one copy has been isolated. The BA17 and BA21 genes were sequenced from the snap bean genotype Sprite, which could explain the small differences between the cDNA and genomic sequences. Since the BA17 and BA21 genes are highly similar to the COK-4 cDNA sequence they are most likely to be functional genes. In soybean, 16 resistance-like genes spanning 118.7 kb were identified, but not all copies seemed to be functional. In addition, their expression varied among different plant tissues examined (Graham et al. 2002).
Interestingly, genes that may be involved in stress and plant defense response were also identified in the BAC sequence. For example, reversibly glycosylated proteins have been implicated in cell-wall biosynthesis in several plants, including Arabidopsis (Delgado et al. 1998) and pea (Dhugga et al. 1997), and cell-wall deposition may be induced by pathogen attacks during the incompatible interaction (Greenberg 1997). Cytochrome P450 has been indicated to be involved in the metabolic pathways of defense mechanisms (Kessmann et al. 1990). A protein homologous to cytochrome P450 is induced in pepper after inoculation with C. gloeosporioides (Oh et al. 1999). A cytochrome P450 gene was also found clustered with disease resistance domains in melon (Wang et al. 2002). However, the exact role of this gene in controlling diseases is not well understood. Phytochelatin synthetases, which are found in plants and yeast, are responsible for the production of phytochelatins in response to cadmium stress (Toppi and Gabbrielli 1999). Phytochelatin forms complexes with cadmium, consequently playing an important role in cadmium detoxification as demonstrated in experiments using Cd-sensitive mutants of Arabidopsis (Howden et al. 1995).
An interesting feature of this region of the Co-4 locus is the presence of retroelements clustered with putative resistance- and defense-related genes. Retroelements have been used to study the evolutionary mechanisms of resistance loci (Graham et al. 2002) as insertions of retroelements may be involved in the appearance of new resistance specificities (Ronald 1998). DNA sequencing of the disease resistance loci of other plant species such as citrus (Yang et al. 2003), barley (Wei et al. 2002), and melon (Wang et al. 2002) also revealed the presence of retroelements and multiple copies of disease resistance-related domains. COK-4 appeared as multiple copies all oriented in the same direction, indicating that this locus may be evolving due to intergenic crossing-over similar to the Cf-4/9 locus of tomato (Parniske et al. 1997) followed by transposon insertion between homologs. One COK-4 segment was also found between the BA21 and BA22 genes, indicating that an intragenic unequal crossing-over may have occurred at this region.
The identification of only four clones that strongly hybridized with the COK-4 gene from a bean library with three- to five fold coverage (Vanhouten and Mackenzie 1999) and the results of the FISH experiments proved that Co-4 is a single locus. Moreover, the physical location of the Co-4 region on bean chromosome 3 is in agreement with its earlier assignment to the linkage group B8 (Kelly et al. 2003) of the bean core map (Freyre et al. 1998) that corresponds to chromosome 3 (Pedrosa et al. 2003). The physical localization of such an economically important trait in common bean is unique. Attempts have been made to localize two low-copy genes, polygalacturonase-inhibiting protein and leghemoglobin, on common bean chromosomes, but the exact chromosomes and the respective linkage groups were not identified (Frediani et al. 1993; Uchiumi et al. 1998). The relationship between genetic and physical distances in common bean has not yet been completely determined, however Llaca and Gepts (1996) estimated a 500 kb/cM ratio based on the region surrounding the phaseolin gene, Phs. Using this estimate as a reference, the sequence reported here spans 0.2 cM around the Co-4 locus. Since it is known that this ratio varies throughout the bean genome (Pedrosa et al. 2003), the exact estimation has yet to be determined and awaits the availability of additional markers that fall on the other side of the Co-4 locus.
A putative gene that encodes for an anthocyanin 5-aromatic acyltransferase involved in the biosynthesis of anthocyanin (Fujiwara et al. 1998) was found in the reported sequence. Tight genetic linkage was observed previously between the Co-4 locus, formerly known as Mex2 (Kelly et al. 2003), and the Anp gene that is responsible for pod anthocyanin accumulation (Gantet et al. 1991). The Anp gene is believed to be the same as the Prp (purple pod) locus (M. Bassett, personal communication) that maps to a cluster of seed color genes (C, R, Prp) in the linkage group B8 (McClean et al. 2002) in the vicinity of the Co-4 gene. The color of bean plant organs is determined by the combination of alleles of each gene, and in certain combinations the anthocyanin intensification syndrome is observed when Prp is present (Bassett 1994). However, whether the anthocyanin acyltransferase described herein is one of the color genes and plays a role in the seed-coat color pattern remains to be determined. Similar tight linkages have been observed between other color genes (B) involved in anthocyanin production in common bean (Beninger et al. 2000) and the virus resistance I gene (Temple and Morales 1986).
We report here the gene content and location of a BAC clone spanning at least part of the Co-4 locus in common bean. A cluster of novel genes of common bean that might be involved in abiotic and biotic stress responses has been identified as well as retrotransposons and resistance gene candidates. The presence of genes clusters favors the occurrence of unequal cross-over with further gene duplication that would subsequently result in the evolution of new resistance specificities. In addition, the presence of alleles conferring susceptibility to certain pathogen races in a cluster hinders the development of broad-resistance genotypes due to the difficulty of breaking the linkage. This study represents a first step in gaining an understanding of the genomic organization of an anthracnose resistance locus of common bean and provides molecular data for comparative analysis with other plant species.
References
Altschul SF, Madden TL, Schäffer A, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bassett MJ (1994) Tight linkage of purple pod character and the complex C locus in common bean. J Hered 85:288–290
Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein family database. Nucleic Acids Res 30:276–280
Beninger CW, Bassett MJ, Owens S, Hosfield GL (2000) Chemical and morphological expression of the B and Asp seedcoat genes in Phaseolus vulgaris. J Am Soc Hortic Sci 125:52–58
Bent AF (1996) Plant disease resistance genes: function meets structure. Plant Cell 8:1757–1771
Boyes DC, Nam J, Dangl JL (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA 95:15849–15854
Burge C, Karlin S (1997) Prediction of complete gene structure in human genomic DNA. J Mol Biol 268:78–94
Delgado IJ, Wang Z, Rocher A de, Keegstra K, Raikhel NV (1998) Cloning and characterization of AtRGP1, a reversibly autoglycosylated Arabidopsis protein implicated in cell wall biosynthesis. Plant Physiol 116:1339–1349
Dhugga KS, Tiwari SC, Ray PM (1997) A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: purification, gene cloning and trans Golgi localization. Proc Natl Acad Sci USA 94:7679–7684
Dixon MS, Golstein C, Thomas CM, van der Biezen EA, Jones JDG (2000) Genetic complexity of pathogen perception by plants: The example of Rcr3, a tomato gene required specifically by Cf-2. Proc Natl Acad Sci USA 97:8807–8814
Ewing B, Green P (1998) Base calling of automated sequencer traces using Phred. II error probabilities. Genome Res 8:186–194
Ewing B, Hillier L, Wendl M, Green P (1998) Base calling of automated sequencer traces using Phred. I accuracy assessment. Genome Res 8:175–185
Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A (2002) The prosite database, its status in 2002. Nucleic Acids Res 30:235–238
Frediani M, Cremonini R, Salvi G, Caprari C, Desiderio A, D’Ovidio R, Cervone F, De Lorenzo G (1993) Cytological localization of the PGIP genes in the embryo suspensor cells of Phaseolus vulgaris L. Theor Appl Genet 87:369–373
Freyre R, Skroch PW, Geffroy V, Adam-Blondon A-F, Shirmohamadali A, Johnson WC, Llaca V, Nodari RO, Pereira PA, Tsai S-M, Tohme J, Dron M, Nienhuis J, Vallejos CE, Gepts P (1998) Towards an integrated linkage map of common bean 4. Development of a core linkage map and alignment of RFLP maps. Theor Appl Genet 97:847–856
Fujiwara H, Tanaka Y, Yonekura-Sakakibara K, Fukuchi-Mizutani M, Nakao M, Fukui Y, Yamaguchi M, Ashikari T, Kusumi T (1998) cDNA cloning, gene expression and subcellular localization of anthocyanin 5-aromatic acyltransferase from Gentiana triflora. Plant J 16:421–431
Gantet P, Bettini P, Grisvard J, Dron M (1991) Genetic linkage between Mex2, a specific resistance gene to anthracnose, and Anp, a gene involved in pod anthocyanin accumulation in bean. Plant Dis 75:941–942
Gordon D, Abajian C, Green P (1998) consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Gough J, Karplus K, Hughey R, Chothia C (2001) Assignment of homology to genome sequences using a library of Hidden Markov Models that represent all proteins of known structure. J Mol Biol 313:903–919
Graham MA, Marek LF, Shoemaker RC (2002) Organization, expression and evolution of a disease resistance gene cluster in soybean. Genetics 162:1961–1977
Greenberg JT (1997) Programmed cell death in plant pathogen interactions. Annu Rev Plant Physiol Plant Mol Biol 48:525–545
Hammond-Kosack KE, Jones JD (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48:575–607
Heslop-Harrison JS, Schwarzacher T, Anamthawat-Jónsson K, Leitch AR, Shi M, Leitch IJ (1991) In situ hybridization with automated chromosome denaturation. Technique 3:109–115
Heslop-Harrison JS, Harrison GE, Leitch IJ (1992) Reprobing of DNA:DNA in situ hybridization preparations. Trends Genet 8:372–373
Howden R, Goldsbrough PB, Andersen CS, Cobbett CS (1995) Cadmium-sensitive, cadl mutants of Arabidopsis thaliana are phytochelatin deficient. Plant Physiol 107:1059–1066
Jia Y, MacAdams SA, Brian GT, Hershey HP, Valent B (2000) Direct interaction between resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19:4004–4014
Kelly JD, Gepts P, Miklas PN, Coyne DP (2003) Tagging and mapping of genes and QTL and molecular marker-assisted selection for traits of economic importance in bean and cowpea. Field Crops Res 82:135–154
Kessmann H, Choudhary AD, Dixon RA (1990) Stress response to alfalfa (Medicago sativa L.) III. Induction of medicarpin and cytochrome P450 enzyme activities in elicitor-treated cell suspension and protoplast. Plant Cell Rep 9:38–41
Llaca V, Gepts P (1996) Pulse field gel electrophoresis analysis of the phaseolin locus region in Phaseolus vulgaris. Genome 39:722–729
Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262:1432–1436
McClean P, Lee R, Otto C, Gepts P, Bassett M (2002) Molecular and phenotypic mapping of genes controlling seed coat pattern and color in common bean (Phaseolus vulgaris L.). J Hered 93:148–152
Melotto M, Kelly JD (2001) Fine mapping of the Co-4 locus reveals a resistance gene candidate, COK-4, that encodes for a protein kinase. Theor Appl Genet 103:508–517
Melotto M, Afanador L, Kelly JD (1996) Development of a SCAR marker linked to the I gene in common bean. Genome 39:1216–1219
Melotto M, Balardin RS, Kelly JD (2000) Host-pathogen interaction and variability of Colletotrichum lindemuthianum. In: Prusky D, Freeman S, Dickman MB (eds) Colletotrichum host specificity, pathology, and host-pathogen interaction. APS Press, St Paul, Minn., pp 346–361
Oh B-J, Ko MK, Kim YS, Kim KS, Kostenyuk I, Kee HK (1999) A cytochrome P450 gene is differentially expressed in compatible and incompatible interactions between pepper (Capsicum annum) and anthracnose fungus, Colletotrichum gloeosporioides. Mol Plant Microbe Interact 12:1044–1052
Parniske M, Hammond-Kosaki KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD (1997) Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf4/9 locus of tomato. Cell 91:821–832
Pedrosa A, Jantsch MF, Moscone EA, Ambros PF, Schweizer D (2001) Characterization of pericentromeric and sticky intercalary heterochromatin in Ornithogalum longibracteatum (Hyacinthaceae). Chromosoma 110:203–213
Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A (2002) Chromosomal map of the model legume Lotus japonicus. Genetics 161:1661–1672
Pedrosa A, Vallejos CE, Bachmair A, Schweizer D (2003) Integration of common bean (Phaseolus vulgaris L.) linkage and chromosomal maps. Theor Appl Genet 106:205–212
Ronald PC (1998) Resistance gene evolution. Curr Opin Plant Biol 1:294–298
Swofford DL (1998) PAUP* Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer, Sunderland, Mass.
Temple SR, Morales FJ (1986) Linkage of dominant hypersensitive resistance to bean common mosaic virus to seed color in Phaseolus vulgaris L. Euphytica 35:331–333
Thompson JD, Higgins DG, Gibson TJ (1994) clustalw: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Toppi LS di, Gabbrielli R (1999) Response to cadmium in higher plants. Environ Exp Bot 41:105–130
Uchiumi T, Kuwashiro R, Miyamoto J, Abe M, Higashi S (1998) Detection of the leghemoglobin gene on two chromosomes of Phaseolus vulgaris by in situ PCR linked-fluorescent in situ hybridization (FISH). Plant Cell Physiol 39:790–794
Vallad G, Rivkin M, Vallejos C, McClean P (2001) Cloning and homology modeling of a Pto-like protein kinase family of common bean (Phaseolus vulgaris L.). Theor Appl Genet 103:1046–1058
Vallejos CE, Sakiyama NS, Chase CD (1992) A molecular marker-based linkage map of Phaseolus vulgaris L. Genetics 131:733–740
Vanhouten W, MacKenzie S (1999) Construction and characterization of a common bean bacterial artificial chromosome library. Plant Mol Biol 40:977–983
Wang Y-H, Choi W, Thomas CE, Dean RA (2002) Cloning of disease-resistance homologues in end sequences of BAC clones linked to Fom-2, a gene conferring resistance to Fusarium wilt in melon (Cucumis melo L.). Genome 45:473–480
Wei F, Wing RA, Wise RP (2002) Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14:1903–1917
Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schrimi LM, Sequeira E, Tatusove TA, Wagner L (2003) Database resources of the national center for biotechnology. Nucleic Acids Res 31:28–33
Yang Z-N, Ye X-R, Molina J, Roose ML, Mirkov TE (2003) Sequence analysis of a 282-kilobase region surrounding the citrus tristeza virus resistance gene (Ctv) locus in Poncirus tifoliata L. Raf. Plant Physiol 131:482–492
Young RA, Melotto M, Nodari RO, Kelly JD (1998) Marker-assisted dissection of the oligogenic anthracnose resistance in the common bean cultivar, ‘G2333’. Theor Appl Genet 96:87–94
Acknowledgements
We thank Dr. Sally MacKenzie (University of Nebraska) for screening her BAC library with the SAS13 marker and kindly donating the four homologous BAC clones, Dr. Carlos Eduardo Vallejos (University of Florida) for providing seeds of the cultivar Calima and the RFLP clones from the University of Florida linkage map, and Dr. Dieter Schweizer (University of Vienna) for supporting the participation of A.P.-H in this project. We thank Dr. Claudia B. Monteiro-Vitorello for assisting with the assembly of the DNA contig, Dr. Jorge L.M. Rodrigues and Dr. Sheng Yang He for helpful suggestions and critical review of the manuscript, and Daniela Truffi and Maria Cristina R. Rangel for assisting with DNA sequencing. M.M. acknowledges financial support for this project from FAPESP—Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil (Grant 00/09049-2). Research at L.E.A. Camargo’s lab is supported by funds from FAPESP (Grant 00/09059-8).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Möllers
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Melotto, M., Coelho, M.F., Pedrosa-Harand, A. et al. The anthracnose resistance locus Co-4 of common bean is located on chromosome 3 and contains putative disease resistance-related genes. Theor Appl Genet 109, 690–699 (2004). https://doi.org/10.1007/s00122-004-1697-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1697-6