Abstract
This study was conducted to identify microsatellite markers (SSR) linked to the adult-plant leaf rust resistance gene Lr22a and examine their cross-applicability for marker-assisted selection in different genetic backgrounds. Lr22a was previously introgressed from Aegilops tauschii Coss. to wheat (Triticum aestivum L.) and located to chromosome 2DS. Comparing SSR alleles from the donor of Lr22a to two backcross lines and their recurrent parents showed that between two and five SSR markers were co-introgressed with Lr22a and the size range of the Ae. tauschii introgression was 9–20 cM. An F2 population from the cross of 98B34-T4B × 98B26-N1C01 confirmed linkage between the introgressed markers and Lr22a on chromosome 2DS. The closest marker, GWM296, was 2.9 cM from Lr22a. One hundred and eighteen cultivars and breeding lines of different geographical origins were tested with GWM296. In total 14 alleles were amplified, however, only those lines predicted or known to carry Lr22a had the unique Ae. tauschii allele at GWM296 with fragments of 121 and 131 bp. Thus, GWM296 is useful for selecting Lr22a in diverse genetic backgrounds. Genotypes carrying Lr22a showed strong resistance to leaf rust in the field from 2002 to 2006. Lr22a is an ideal candidate to be included in a stack of leaf rust resistance genes because of its strong adult-plant resistance, low frequency of commercial deployment, and the availability of a unique marker.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leaf rust (Puccinia triticina Eriks.) of wheat (Triticum aestivum L.) is a widespread foliar disease that causes significant reductions in grain yield and quality (Samborski 1985). Host genetic resistance is a desirable and proven method of leaf rust control. However, deployment of single race-specific genes allows the pathogen to evolve and accumulate new virulence (Dyck and Kerber 1985). This creates the need to identify new effective genes and can lead an ongoing cycle of breeding, deployment and subsequent erosion of resistance.
Over 50 leaf rust resistance (Lr) genes have been identified in wheat. Of these, approximately half were introgressed from related species (McIntosh et al. 1995). Five named Lr genes have been introgressed into common wheat from Aegilops tauschii Coss.; these are Lr21 (Rowland and Kerber 1974), Lr32 (Kerber 1987), Lr41 (Cox et al. 1994; Singh et al. 2004), Lr42 (Cox et al. 1994), and the adult-plant resistance (APR) gene Lr22a (Rowland and Kerber 1974). It should be noted that while Lr22a is expressed only at the adult-plant stage, the degree of resistance conferred is comparable to highly resistant seedling Lr genes in contrast to the slow-rusting type APR conferred by genes like Lr34.
In the search for improved genetic solutions for leaf rust resistance it is widely believed that combinations or stacks of multiple Lr genes would confer a more durable resistance than the same genes deployed individually. Such gene stacks might include race non-specific adult plant resistance genes (e.g. Lr34), “undefeated” genes for which no virulence has been detected such as Lr22a, and partially “defeated” genes, for which virulence already exists in the pathogen population (e.g. Lr16). One advantage of creating gene stacks is the synergistic interaction of leaf rust resistance genes which often confers a higher level of resistance than would be expected from the level of resistance demonstrated by the genes in isolation (Samborski and Dyck 1982). The resistance genes Lr34 (German and Kolmer 1992) and Lr13 (Kolmer 1992) were both demonstrated to enhance the level of resistance synergistically when in combination with other Lr genes. Complex stacks of Lr genes can be difficult to construct using phenotypic selection. For example the Canadian cultivar Pasqua carries five Lr genes including Lr11, Lr13, Lr14b, Lr30 and Lr34, but failed to retain Lr16 and Lr22a from the parental cross (Dyck 1993a). With suitable markers a stack of seven Lr genes could have been stabilized in this cross.
Close molecular markers for all Lr genes can help in the assembly and dissection of complex gene stacks in any cross. Lr22a has been physically mapped to chromosome 2DS using telocentric mapping (Rowland and Kerber 1974). In this paper we report both the genetic location and the identification of a closely linked, unique molecular marker allele for Lr22a that was retained from Ae. tauschii. The application of this marker in different genetic backgrounds was examined on a broad range of wheat germplasm.
Materials and methods
Plant material and populations
Lr22a was previously transferred to a synthetic hexaploid RL5404 by crossing tetra-Canthatch (2n = 2x = 28, AABB) with Aegilops tauschii var. strangulata RL5271 (2n = 14, DD; Dyck and Kerber 1970; Rowland and Kerber 1974). This was followed by six backcrosses with Thatcher to produce the line RL6044 (Thatcher*7//tetra-Canthatch/RL5271) which was then backcrossed with AC Domain to produce the leaf rust resistant line 98B34-T4B (AC Domain*6/RL6044) with Lr22a. RL5404 was also crossed with Neepawa (Neepawa*6/RL5404) to create the Lr22a-carrying near-isogenic line (NIL) RL4495.
An F2 population that segregated for Lr22a was created by crossing 98B34-T4B with the leaf rust susceptible line 98B26-N1C01 (AC Domain*2/Sumai3//Grandin*2/AC Domain).
Thatcher NILs carrying single Lr genes (Lr11, Lr13, Lr16, Lr22a and Lr34), cultivars carrying Lr22a (AC Minto) and cultivars believed to carry Lr22a (5500HR and 5600HR) were used in field tests to evaluate leaf rust resistance.
Disease rating
Segregation of Lr22a was followed in the 98B34-T4B × 98B26-N1C01 F2 population by inoculating flag leaves with Puccinia triticina isolate TJBJ-77-2 (Long and Kolmer 1989; McCallum and Seto-Goh 2003) following the procedures of McCallum and Seto-Goh (2003). Plants were grown with 16 h supplemental lighting in a greenhouse at approximately 20°C. Twelve days after inoculation plants were classified as resistant (infection types; to 1), moderately resistant (infection types 1+ to 2) and susceptible (infection types 3–4) as described by Stakman et al. (1962). Parents of the F2 population were tested for leaf rust resistance at the seedling and adult stage.
Thatcher NILs and cultivars carrying Lr22a were grown in field plots at Glenlea, MB, Canada from 2003 to 2006 and 2002 to 2006, respectively, to observe Lr gene effectiveness. Plants were infected with natural leaf rust inoculum and rows of susceptible wheat throughout the nurseries were artificially inoculated with a mixture of virulence phenotypes found in western Canada during the previous year. Adult-plants were rated for leaf rust based on a modified Cobb scale for incidence (Peterson et al. 1948) and severity was scored as R = resistant (flecks and small uredinia with necrosis), MR = moderately resistant (large necrotic flecks and large uredinia), M = moderate (mixture of uredinia sizes), MS = moderately susceptible (moderate to large uredinia with chlorosis), and S = susceptible (large uredinia without chlorosis or necrosis).
Molecular mapping
DNA was extracted from lyophilized leaf tissue using a modified CTAB extraction (Kleinhoffs et al. 1993). PCR was performed with a cycle of 2 min 94°C, then 1 min 95°C, 1 min 50–60°C, and 50 s 73°C for 30 cycles, followed by 5 min 73°C, using the following conditions: PCR buffer 1×, dNTPs 0.2 mM each, MgCl2 1.5 mM, primers 10 pmol each, Taq DNA polymerase 1 U, and approximately 50 ng genomic DNA. PCR products were separated on 5% denaturing polyacrylamide gels in TBE buffer (89 mM tris, 89 mM boric acid, 20 mM EDTA) at 85 W for 2 h, and stained with silver (Promega, Madison, WI, USA). Lr22a has been previously located on chromosome 2DS (Rowland and Kerber 1974). Microsatellite marker (SSR) alleles located on chromosome 2DS were compared between RL5271, RL5404, RL6044, Thatcher, 98B34-T4B, and AC Domain. Polymorphic markers were tested for linkage to Lr22a using 68 of the susceptible individuals from the F2 population of 98B34-T4B × 98B26-N1C01. Susceptible F2 plants were used for mapping because their genotypes at the Lr22a locus were known. These susceptible individuals were selected for pustule uniformity to avoid assigning homozygous susceptible genotypes at the Lr22 locus to heterozygous individuals that would falsely appear to be recombinants. The location of linked SSR alleles was confirmed using nulli/tetra and ditelocentric cytogenetic stocks (Sears 1966; Sears and Sears 1978).
SSR markers on chromosome 2DS were compared between RL5271, RL5404, RL6044, Thatcher, 98B34-T4B, AC Domain, RL4495, Neepawa, BW63, AC Minto, 5500HR, and 5600HR to determine which markers were co-introgressed with Lr22a from Ae. tauschii into Canadian wheat.
New SSR markers were added to the Agriculture and Agri-Food Canada Cereal Research Centre (AAFC-CRC) International Triticeae Mapping Initiative (ITMI) genetic map (Opata85/Synthetic W-7984; Somers et al. unpublished, ITMI-CRC). A newly developed Opata85/Synthetic doubled haploid (DH) population was tested to confirm the marker order on 2DS found in the ITMI-CRC population. The ITMI-CRC map was used to align the Lr22a linkage group with the genetic map of 2DS. Genetic distances were calculated using the Kosambi mapping function (Kosambi 1944). All maps were constructed using MapMaker version 3.0 (Lander et al. 1987).
To asses the potential of GMW296, the closest marker to Lr22a, as a selection tool in varying genetic backgrounds 118 different cultivars and breeding lines were surveyed (Table 2). Alleles from these genotypes were compared to the allele amplified from RL5271, the Ae. tauschii donor of Lr22a.
Results
Fourteen polymorphic microsatellite markers on chromosome 2DS were evaluated in Thatcher, AC Domain, the Aegilops tauschii donor of Lr22a (RL5271), the original synthetic (RL5404 = Tetra-Canthatch/RL5271) and two backcross lines (RL6044 = Thatcher*7/RL5404 and 98B34-T4B = AC Domain*6/RL6044). Microsatellite alleles from GWM296 and GWM455 were introgressed from RL5271 via RL5404 to these two resistant backcross lines (Table 1; Fig. 1). Thus GWM296 and GWM455 alleles from RL5271 have remained associated with Lr22a through a minimum of 13 cycles of recombination in common wheat.
Alleles of GWM296 found in RL5271 (Lr22a donor), RL6044 (Thatcher*6/RL5271; carries Lr22a), Thatcher (recurrent wheat parent), Chinese Spring, nulli-2D/tetra-2B, nulli-2A/tetra-2B, ditelo 2DS, and ditelo 2DL on a silver-stained polyacrylamide gel. The allele associated with Lr22a has DNA fragments that are 131 and 121 bp in size. A common chromosome 2DS allele in the North American wheat lines tested has the 167 and 135 bp fragments. The 176 bp fragment is from the chromosome 2A locus
The same 14 SSR markers were used to assess the introgression of Lr22a into RL4495 (Neepawa*6/RL5404) and its derivatives which included BW63, AC Minto, 5500HR and 5600HR. RL4495 retained Ae. tauschii alleles from GWM455, GWM296, GWM261, WMC25, and WMC503, and transmitted them to its derivatives (Table 1). Two other introgression sizes were detected in RL4495, one with GWM455, GWM296, and GWM261, and one with GWM455 and GWM296. Since these introgressions are smaller than the one found in BW63 they cannot be in the line of descent to AC Minto, 5500HR and 5600HR as these cultivars have retained the largest introgression found in RL4495.
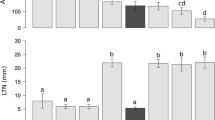
In the F2 of 98B34-T4B × 98B26-N1C01, 88 plants were resistant, 216 plants had a moderately resistant infection type and 102 plants were susceptible. These numbers fit both a 3:1 ratio (resistant plus moderate resistant: susceptible; χ2 3:1 = 0.02, P = 0.87), and a 1:2:1 ratio (resistant: moderate resistant: susceptible; χ2 1:2:1 = 2.64, P = 0.27). As adult plants 98B34-T4B was resistant to P. triticina and 98B26-N1C01 was susceptible, while as seedlings both lines were susceptible. This confirms that the F2 population segregated for the APR conferred by Lr22a. The Ae. tauschii alleles of GWM296 and GWM455 (both present in 98B34-T4B) were both polymorphic with respect to 98B26-N1C01. Based on 68 susceptible F2 plants representing 136 gametes, GWM296 was 2.9 cM and GWM455 was 4.4 cM from the Lr22a locus (Fig. 2). Testing with nulli-2D/tetra-2B, nulli-2A/tetra-2B, ditelo 2DS, and ditelo 2DL confirmed that the locus of GWM296 shown to be linked to the APR of Lr22a was on chromosome 2DS (Fig. 1).
Alignment of the Lr22a and ITMI-CRC maps with two previously published maps of chromosome 2DS. The map of 98B34-T4B/98B26-N1C01 was based on 68 F2 plants while the ITMI-CRC map was derived from 68 recombinant inbred lines from the cross of Opata85/W-7984 (W-7984 = Ae. tauscii/Altar84 durum; this cross is commonly called Opata/Synthetic). The map of Raupp et al. (2001) reports the mapping of Lr39, however, Singh et al. (2004) found that Lr39 = Lr41
A minor conflict exists between our proposed order for markers close to Lr22a and those published elsewhere (Raupp et al. 2001). The marker order was clarified by re-genotyping lines recombinant in the region and adding additional markers to the ITMI-CRC map. An alternate DH population from the ITMI cross was also used to make a comparative map. The DH map agreed with the ITMI-CRC map (data not shown). Based on these results, we concluded that the marker order on 2DS is as shown (Table 1; Fig. 2) with the GWM296 locus 2.9 cM distal to Lr22a.
One hundred and five North American wheat cultivars and breeding lines, four Asian lines, eight European cultivars and one South American wheat cultivar were surveyed for alleles of GWM296 (Table 2). In total 14 alleles were detected, with the most common being that found in Thatcher (Fig. 1). Most Canadian hard-red spring wheat cultivars are derived from Thatcher (B. McCallum and R. DePauw unpublished). In this collection of wheat, the allele from Ae. tauschii RL5271 was present in those lines and cultivars that were expected to carry Lr22a and was absent from all others (Table 2). BW63 was heterogeneous for GWM296 with half the individuals tested carrying the RL5271 allele. Two products were amplified from RL5271 that were 131 and 121 bp in size (Fig. 1). The size range of all other chromosome 2DS alleles found were between 167 and 135 bp.
Cultivars expected to carry Lr22a, including AC Minto, 5500HR, and 5600HR, showed strong resistance to P. triticina in the field from 2002 to 2006 (Table 3). Furthermore, no isolates of P. triticina that are virulent to Lr22a have been found in Canadian virulence surveys between 2002 and 2005 (McCallum and Seto-Goh 2005; McCallum and Seto-Goh unpublished data; Table 4). The Thatcher NILs tested in the field showed that Lr22a provided good resistance singly, while Lr34 showed moderate resistance, Lr16 provided moderate to poor resistance, and Lr11 and Lr13 showed no improved resistance compared to Thatcher (Table 3).
Discussion
Wheat SSR maps reveal that coverage by polymorphic markers is non-uniform (Somers et al. 2004). Of the three genomes of common wheat, the D genome was added most recently and has the shortest evolutionary life span at the hexaploid level. Therefore, the degree of polymorphism is lowest for the D genome chromosomes. By contrast, Ae. tauschii is an older, polymorphic, broadly distributed diploid that has contributed the D genome to many polyploids in the Triticeae. These differences in polymorphism were evident in this study as all 14 SSR markers tested on chromosome 2DS were polymorphic between the Ae. tauschii accession RL5271 and the two recurrent parents, Thatcher and AC Domain. Thatcher and AC Domain were monomorphic for 13 of the 14 SSR markers (Table 1).
In this study SSR markers previously mapped to 2DS were compared between backcross lines and the recurrent parents in order to identify candidate markers that might be linked to Lr22a. Out of the 14 loci investigated, two polymorphic markers were transferred through thirteen rounds of backcrossing and both were then shown to be closely linked to the target gene. This screening strategy was successful because of the high marker density and high degree of polymorphism between chromosome 2DS of Ae. tauschii and the corresponding chromosome of common wheat.
Based on recombination in the F2 population, the minimum size of the segment introgressed into RL6044 and 98B34-T4B was 4.4 cM and the probable size was 9 cM (assuming the crossovers occurred at the midpoint of the flanking intervals; Fig. 2). In contrast, the Lr22a introgression from RL5271, via RL5405, into RL4495 included up to five SSR markers. The largest introgression represents a minimum of 17 cM and a probable size of 20 cM as estimated from the ITMI-CRC map (Table 1; Fig. 2). This larger introgression has persisted from RL4495 into three Canadian cultivars, AC Minto, 5500HR, and 5600HR.
Three-point linkage values observed in the F2 population (98B34-T4B/98B26-N1C01) confirmed that Lr22a, GWM296 and GWM455 are closely linked (Fig. 2). In a study of durable leaf rust resistance in Swiss winter wheat, QTL analysis revealed a significant narrow QTL close to the GWM296 locus (Schnurbusch et al. 2004). Schnurbusch et al. (2004) speculated that this QTL on 2DS could represent Lr22 and our data supports this hypothesis. GWM455 was previously reported to be polymorphic between Thatcher and RL6044 in an effort to exclude Lr22a as an allele of other Lr genes introgressed from Ae. tauschii, but linkage experiments between Lr22a and GWM455 were not performed (Raupp et al. 2001).
Other genes of interest are found near the Lr22 locus on chromosome 2DS. Lr41 (Lr41 = Lr39; Singh et al. 2004) is distal to GWM210 (Raupp et al. 2001) whereas Lr22a is proximal. The order of loci near Lr22 is inconsistent between our map (ITMI-CRC) and the map of Raupp et al. (2001). After re-genotyping lines recombinant on 2DS, the marker order as presented in the ITMI-CRC map is the most likely (Fig. 2). However, the marker orders in the ITMI-CRC map and the map presented by Raupp et al. (2001) both place Lr22a and Lr41 on opposite sides of GWM210 (Fig. 2). Both introgressions of Lr22a into wheat, RL6044 and RL4495, retained SSR markers proximal to GWM210. Furthermore, Lr41 is a seedling resistance gene and Lr22a is an adult plant resistance gene. McIntosh et al. (1995) recorded no instances of an adult-plant resistance gene with an allele that confers seedling resistance for any of the three rusts of wheat. In the particular case of Lr22 there are two alleles that have been reported and both provide adult-plant resistance (Rowland and Kerber 1974; Dyck 1979). This data agrees with the conclusion of Raupp et al. (2001) that Lr41 is not allelic to Lr22a. The dwarfing gene Rht8 is closely linked with GWM261 (Korzun et al. 1998) and is proximal to the Lr22 locus (Fig. 2).
Even though the D genome of wheat has reduced polymorphism, 14 alleles for GWM296 were identified on 2DS. Although both of the 2DS alleles illustrated in Fig. 1 have two fragments, other genotypes tested exhibited a single fragment. For example, the 2DS allele of GWM296 found in AC Splendor has a single fragment of 167 bp while Superb has fragments of 149 and 137 bp (not shown). Thus heterozygotes may have two, three or four fragments from chromosome 2DS while homozygotes may have one or two. Therefore caution is required when classifying an individual’s genotype without prior knowledge of parental haplotypes and the degree of homozygosity. It should be noted that GWM296 also amplified a locus on chromosome 2A; however, this larger fragment (176 ± 4 bp) did not overlap with the 2DS fragments in any of the genotypes tested. Despite the allele diversity of GWM296 on chromosome 2D, the allele that was co-introgressed with Lr22a from Ae. tauschii was not found in any common wheat lines tested except for those believed to carry Lr22a (Table 2). The uniqueness of the GWM296 allele, ease of scoring, and its close association with Lr22a provides a means to include Lr22a in complex gene stacks and to classify existing lines for the distribution of Lr22a provisionally.
In the survey of wheat breeding lines and cultivars GWM296 was useful in tracking the transmission of Lr22a through the genealogy of Canadian wheat. BW63 is an important source of leaf rust resistance in the pedigree of several Canadian bread wheat varieties containing Lr11, Lr14b, Lr22a, Lr30 and Lr34 (Dyck 1993a). The data indicated that BW63 was heterogeneous for the presence of Lr22a and this could explain why AC Minto (Columbus/BW63//Katepwa/BW552; Townley-Smith et al. 1993) inherited Lr22a from BW63 while Pasqua (BW63*2/Columbus) did not (Dyck 1993a). Since BW63 would have demonstrated a high degree of adult-plant resistance it would have been difficult to track the incorporation Lr22a during the development of Pasqua. All of the Canadian cultivars (Table 2) and breeding lines tested that inherited the RL5271 allele of GWM296, and presumably Lr22a, were derived from AC Minto except for 98B34-T4B (AC Domain*6/3/Thatcher*7//tetra-Canthatch/Ae. tauschii RL5271).
In Canada Lr22a is deployed in AC Minto (registered in 1991) and putatively in 5500HR (2000) and 5600HR (1999). There are no reports of US cultivars that carry Lr22a (JA Kolmer, personal communication). The Canadian cultivars that carry Lr22a have only occupied small percentages (0.13–0.95%) of the wheat production area in Canada from 1998 to 2006 (Canadian Wheat Board, http://www.cwb.ca). Thus Lr22a has had relatively low exposure to P. triticina in Canada and the US. The detection of an ineffective race-specific “b” allele of Lr22 (Dyck 1979) implies that Lr22a will also prove to be race specific. The absence of virulence on Lr22a could be explained in part by its lack of exposure to P. triticina.
AC Minto carries Lr11, Lr13 and Lr22a (Kolmer 1997). Presense of Lr22a in AC Minto is confirmed by presense of the five Ae. tauschii alleles found in BW63 (Table 1). Each year AC Minto showed field resistance that was better than the Lr22a NIL, indicating that Lr13 and/or Lr11 had the ability to interact synergistically with Lr22a even though they were ineffective independently (Table 3). Lr13 is known to interact synergistically with some other Lr genes (Kolmer 1992), however, no data has been reported on the potential interaction between Lr13 and Lr22a. Cultivars 5600HR (AC Minto//Columbus/Roblin) and 5500HR (AC Minto/3/MN72506/Columbus//RL4473) both received the Ae. tauschii alleles found in BW63, and presumably Lr22a, via AC Minto (Table 1). In addition to Lr22a, 5600HR and 5500HR may also carry Lr11, Lr13, and Lr16, and Lr11 and Lr16, respectively, based on pedigree and their reactions to different P. triticina virulence phenotypes (Samborski and Dyck 1982; Dyck 1993b; B. McCallum unpublished). In the field all three Canadian cultivars that carry Lr22a showed leaf rust resistance that is better than the individual genetic components of their resistance (Table 3). Although Lr22a appeared to be the key component of the leaf rust resistance in these cultivars, the undesirable scenario where Lr22a is deployed as a single resistance gene has not occurred.
It would be desirable to stack Lr22a with other broadly effective resistance genes such as Lr21 (Huang and Gill 2001; Huang et al. 2003) or Lr34 (Lagudah et al. 2006) that have been characterized with reliable DNA markers. While the markers and mapping we have described can facilitate the stacking of Lr22a with Lr21, in fact markers are not needed to stack these two genes together. Lr21, or other seedling Lr genes, could be detected in the presence of Lr22a with seedling tests while Lr22a could be fixed by top crossing or back crossing with one or more “single gene” lines. Markers may prove more beneficial in complex crosses with several Lr genes segregating.
The low level of deployment in commercial cultivars and the lack of reported virulence to Lr22a (Table 3; Park and McIntosh 1994; McCallum and Seto-Goh 2005; Kolmer et al. 2005) make it a good candidate for use in Lr gene stacks. While various strategies can be used to include Lr22a in complex gene combinations, the marker reported here will facilitate gene stacking without the need to backcross. With careful management Lr22a could be widely deployed while maintaining its status as a broad-spectrum and effective Lr gene.
References
Cox TS, Raupp WJ, Gill BS (1994) Leaf rust-resistance genes Lr41, Lr42, and Lr43 transferred from Triticum tauschii to common wheat. Crop Sci 34:339–343
Dyck PL (1979) Identification of the gene for adult-plant leaf rust resistance in Thatcher. Can J Plant Sci 59:499–501
Dyck PL (1993a) The inheritance of leaf rust resistance in the wheat cultivar Pasqua. Can J Plant Sci 73:903–906
Dyck PL (1993b) Inheritance of leaf rust and stem rust resistance in ‘Roblin’ wheat. Genome 36:289–293
Dyck PL, Kerber ER (1970) Inheritance in hexaploid wheat of adult-plant leaf rust resistance derived from Aegilops squarrosa. Can J Genet Cytol 12:175–180
Dyck PL, Kerber ER (1985) Resistance of the race-specific type. In: Roelfs AP, Bushnell WR (eds) The cereal rusts, vol II. Academic Press Inc., Orlando, FL, pp 469–500
German SE, Kolmer JA (1992) Effect of the gene Lr34 in the enhancement of resistance to leaf rust of wheat. Theor Appl Genet 84:97–105
Huang L, Gill BS (2001) An RGA—like marker detects all known Lr21 leaf rust resistance gene family members in Aegilops tauschii and wheat. Theor Appl Genet 103:1007–1013
Huang L, Brooks SA, Wanlong L, Fellers JP, Trick H, Gill BS (2003) Map based cloning of leaf rust resistance gene Lr21 from the large and polyploidy genome of bread wheat. Genetics 164:655–664
Kerber ER (1987) Resistance to leaf rust in hexaploid wheat, Lr32, a third gene derived from Triticum tauschii. Crop Sci 27:204–206
Kleinhoffs A, Kilian A, Saghai MA, Biyashev RM, Hayes P, Chen FQ, Lapitan N, Fenwick A, Blake TK, Kanazin V (1993) A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor Appl Genet 86:705–712
Kolmer JA (1992) Enhanced leaf rust resistance in wheat conditioned by resistance gene pairs with Lr13. Euphytica 61:123–130
Kolmer JA (1997) Virulence in Puccinia recondita f. sp. tritici isolates from Canada to genes for adult-plant resistance to wheat leaf rust. Plant Dis 81(3):267–271
Kolmer JA, Long DL, Hugher ME (2005) Physiological specialization of Puccinia triticina on wheat in the United States in 2003. Plant Dis 89(11):1201–1206
Korzun V, Röder MS, Ganal MW, Worland AJ, Law CN (1998) Genetic analysis of the dwarfing gene (Rht8) in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L.). Theor Appl Genet 96:1104–1109
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lagudah, ES, McFadden H, Singh RP, Huerta-Espino J, Bariana HS, and Spielmeyer W (2006) Molecular genetic characterization of the Lr34/Yr18 slow rusting resistance gene region in wheat. Theor Appl Genet (online first)
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Long DL, Kolmer JA (1989) A North American system of nomenclature for Puccinia recondita f. sp. tritici. Phytopathology 79:525–529
McCallum BD, Seto-Goh P (2003) Physiologic specialization of wheat leaf rust (Puccinia triticina) in Canada in 2000. Can J Plant Pathol 25:91–97
McCallum BD, Seto-Goh P (2005) Physiologic specialization of wheat leaf rust (Puccinia triticina) in Canada in 2002. Can J Plant Pathol 27:90–95
McIntosh RA, Wellings CR, Park RF (1995) Wheat rusts: an atlas of resistance genes. CSIRO Publications, East Melbourne, Australia, pp 29–82
Park RF, McIntosh RA (1994) Adult plant resistances to Puccinia recondita f. sp. tritici in wheat. NZ J Crop Hort Sci 22:151–158
Peterson RF, Campbell AB, Hannah AE (1948) A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can J Res 26:496–500
Raupp WJ, Singh S, Brown-Guedira GL, Gill BS (2001) Cytogenetic and molecular mapping of the leaf rust resistance gene Lr39 in wheat. Theor Appl Genet 102:347–352
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier M, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Rowland GG, Kerber ER (1974) Telocentric mapping in hexaploid wheat of genes for leaf rust resistance and other characters derived from Aegilops squarrosa. Can J Genet Cytol 16:137–144
Samborski DJ (1985) Wheat leaf rust. In: Roelfs AP, Bushnell WR (eds) The cereal rusts, vol 2. Academic, Orlando, pp 39–59
Samborski DJ, Dyck PL (1982) Enhancement of resistance to Puccinia recondite by interaction of resistance genes in wheat. Can J Plant Pathol 4:152–156
Schnurbusch T, Paillard S, Schori A, Messmer M, Scharchermayr G, Winzeler M, Keller B (2004) Dissection of quantitative and durable leaf rust resistance in Swiss winter wheat reveals a major resistance QTL in the Lr34 chromosomal region. Theor Appl Genet 108:477–484
Sears ER (1966) Nullisomic-tetrasomic combinations in hexaploid wheat. In: Lewis DR (ed) Chromosome manipulation and plant genetics. Oliver and Boyd, London, pp 29–47
Sears ER, Sears LMS (1978) The telocentic chromosomes of common wheat, In: Ramanujan S (ed) In: Proceedings of the fifth international wheat genetics symposium, New Delhi, India, 23–28 February 1978. Indian Society of Genetics and Plant Breeding, New Delhi, pp 389–407
Singh S, Franks CD, Huang L, Brown-Guedira GL, Marshall DS, Gill BS, Fritz A (2004) Lr41, Lr39, and a leaf rust resistance gene from Aegilops cylindrica may be allelic and are located on wheat chromosome 2DS. Theor Appl Genet 108:586–591
Somers DJ, Isaac P, Edwards K (2004) A high density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Song QJ, Shi JR, Singh S, Fickus EW, Costa JM, Lewis J, Gill BS, Ward R, Cregan PB (2005) Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet 110:550–560
Stakman EC, Stewart DM, Loegering WQ (1962) Identification of physiologic races of Puccinia graminis var. tritici. Agric Res Serv E617
Townley-Smith TF, Czarnecki EM, Campbell AB, Dyck PL, Samborski DJ (1993) AC Minto hard red spring wheat. Can J Plant Sci 73:1091–1094
Acknowledgments
C. Hiebert thanks the Natural Sciences and Engineering Research Council of Canada (NSERC) for the NSERC Post-Graduate Scholarship. Additional financial support was received from the Agriculture and Agri-Food Canada Matching Investment Initiative and the Western Grains Research Foundation Check-off. Inoculum was provided by Pat Seto-Goh. Figure formatting assistance was provided by Mike Shillinglaw.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Keller.
An erratum to this article can be found at http://dx.doi.org/10.1007/s00122-007-0623-0
Rights and permissions
About this article
Cite this article
Hiebert, C.W., Thomas, J.B., Somers, D.J. et al. Microsatellite mapping of adult-plant leaf rust resistance gene Lr22a in wheat. Theor Appl Genet 115, 877–884 (2007). https://doi.org/10.1007/s00122-007-0604-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-007-0604-3