Abstract
Key message
Here, we describe a strategy to improve broad-spectrum leaf rust resistance by marker-assisted combination of two partial resistance genes. One of them represents a novel partial adult plant resistance gene, named Lr75.
Abstract
Leaf rust caused by the fungal pathogen Puccinia triticina is a damaging disease of wheat (Triticum aestivum L.). The combination of several, additively-acting partial disease resistance genes has been proposed as a suitable strategy to breed wheat cultivars with high levels of durable field resistance. The Swiss winter wheat cultivar ‘Forno’ continues to show near-immunity to leaf rust since its release in the 1980s. This resistance is conferred by the presence of at least six quantitative trait loci (QTL), one of which is associated with the morphological trait leaf tip necrosis. Here, we used a marker-informed strategy to introgress two ‘Forno’ QTLs into the leaf rust-susceptible Swiss winter wheat cultivar ‘Arina’. The resulting backcross line ‘ArinaLrFor’ showed markedly increased leaf rust resistance in multiple locations over several years. One of the introgressed QTLs, QLr.sfr-1BS, is located on chromosome 1BS. We developed chromosome 1B-specific microsatellite markers by exploiting the Illumina survey sequences of wheat cv. ‘Chinese Spring’ and mapped QLr.sfr-1BS to a 4.3 cM interval flanked by the SSR markers gwm604 and swm271. QLr.sfr-1BS does not share a genetic location with any of the described leaf rust resistance genes present on chromosome 1B. Therefore, QLr.sfr-1BS is novel and was designated as Lr75. We conclude that marker-assisted combination of partial resistance genes is a feasible strategy to increase broad-spectrum leaf rust resistance. The identification of Lr75 adds a novel and highly useful gene to the small set of known partial, adult plant leaf rust resistance genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hexaploid bread wheat (Triticum aestivum L.) is one of the three most important cereal crops with an annual global production of 713 million tonnes (FAOSTAT 2013 http://faostat3.fao.org). Wheat is attacked by many pathogens of which the fungal rust diseases are the most widespread and devastating. There are three species of wheat rust: leaf or brown rust (Puccinia triticina), stripe or yellow rust (Puccinia striiformis f. sp. tritici) and stem or black rust (Puccinia graminis f. sp. tritici). Leaf rust is the most common and most widespread rust disease (Bolton et al. 2008; Kolmer 2013). Yield losses caused by leaf rust are characterized by reduced kernel weight and a lower number of kernels per spike (Bolton et al. 2008; Huerta-Espino et al. 2011).
The release of crop varieties with high levels of durable disease resistance represents the most sustainable strategy to reduce production losses caused by fungal diseases. To date, 74 leaf rust resistance (Lr) genes have been described (McIntosh et al. 2013). Most Lr genes confer race-specific resistance. Rapid pathogen adaptation often results in the emergence of new virulent leaf rust races and consequently a breakdown of race-specific Lr resistance (Huerta-Espino et al. 2011; McIntosh et al. 2013). Hence, there is a need to identify Lr genes that show a more durable resistance in the field. A particular type of durable disease resistance is referred to as slow-rusting resistance. It is characterized by a partial resistance phenotype that is often only effective at the adult plant stage but not in seedlings (Caldwell 1968). This type of resistance is therefore also referred to as adult plant resistance (APR). Because of their partial nature it is challenging to combine several APR genes in a single genotype through classical breeding. Marker-assisted selection can serve as a suitable approach to track the presence of APR loci in breeding programs. It has been described that the combination of two or more APR loci with additive effects can result in near immune resistance levels (Singh et al. 2000; Lillemo et al. 2011).
Wheat breeders at the International Wheat and Maize Improvement Center (CIMMYT) have exploited the strategy of combining slow-rusting genes in wheat disease resistance breeding. The CIMMYT wheat breeding material has therefore been the most important source for the discovery of slow-rusting genes. Cultivars such as ‘Frontana’, ‘Pavon76’, ‘Parula’, ‘Trap’ or ‘Mango’ have been released during the past decades using CIMMYT bread wheat germplasm and they show near immune resistance responses to wheat leaf rust by the additive effect of 3–4 slow-rusting genes (Singh and Rajaram 1991; Singh et al. 1998, 2005). The main APR genes identified in these cultivars are Lr34/Yr18/Sr57/Pm38, Lr46/Yr29/Pm39 and Lr68. There are also other sources of slow-rusting resistance genes besides CIMMYT germplasm. For example, the gene Lr67/Yr46/Sr55/Pm46 has been identified in the common wheat accession PI250413 which was collected from Pakistan (Dyck and Samborski 1979; Moore et al. 2015). Some of these genes are mapped at high resolution or cloned such as Lr34/Yr18/Sr57/Pm38, Lr67/Yr46/Sr55/Pm46 and Lr68 (Lagudah et al. 2006; Krattinger et al. 2009; Moore et al. 2015). On the other hand, slow-rusting resistance has only been poorly studied in the Central European winter wheat genepool.
The Swiss winter bread wheat cultivar ‘Forno’ (pedigree: ‘NR72837 × Kormoran’) was released in Switzerland in 1986. ‘Forno’ shows near immune levels of leaf rust resistance in the field against all leaf rust isolates tested so far. At least six QTLs contribute to this remarkable leaf rust resistance of ‘Forno’ (Schnurbusch et al. 2004). Among them is the well-known multi-pathogen resistance gene Lr34 located on chromosome 7D. Lr34 encodes for an ATP-binding cassette transporter (Krattinger et al. 2009) and it explained 33–43 % of the phenotypic variance in the QTL study of Schnurbusch et al. (2004). Lr34 is associated with leaf tip necrosis (LTN), a senescence-like process. LTN is often seen as an unwanted trait in Western European wheat cultivars and Lr34 has therefore only rarely been used in the Western European wheat breeding programs (Kolmer et al. 2008). A second major QTL for leaf rust resistance, QLr.sfr-1BS, was identified on chromosome arm 1BS. QLr.sfr-1BS explained 28–32 % of the phenotypic variance and was not linked with LTN. In the ‘Arina’ × ‘Forno’ recombinant inbred line (RIL) population generated by Schnurbusch et al. (2004), QLr.sfr-1BS was mapped to an interval of 16 cM on chromosome 1BS close to the microsatellite marker gwm604 (Schnurbusch et al. 2004). This locus interacted with four other QTLs: Lr34, two minor QTLs that were contributed by the susceptible parent ‘Arina’ and QLr.sfr-7BL, a minor QTL contributed by ‘Forno’ that was not linked to LTN. In a different mapping population derived from a cross of ‘Forno’ with the spelt wheat cv. ‘Oberkulmer’, Messmer et al. (2000) identified six QTLs for durable leaf rust resistance in cultivar ‘Forno’. Three of these QTLs were not detected by Schnurbusch et al. (2004). The strongest QTL detected in the ‘Forno’ × ‘Oberkulmer’ population was on chromosome 7BL with a phenotypic variance of 35.8 %. This QTL fell into the same genetic interval as QLr.sfr-7BL identified by Schnurbusch et al. (2004). Also, a QTL on chromosome 1BS was identified by Messmer et al. (2000) that explained 10.6 % of the phenotypic variance across four environments. This is most likely be the same QTL as QLr.sfr-1BS because it was identified in the same genetic interval as QLr.sfr-1BS in the ‘Arina’ × ‘Forno’ RIL population.
The objectives of the research presented here were (1) the marker-assisted introgression of the two leaf rust resistance QTLs, QLr.sfr-1BS and QLr.sfr-7BL into the popular but leaf rust-susceptible Swiss winter wheat cultivar ‘Arina’ and (2) the mapping of QLr.sfr-1BS, a yet uncharacterized, partial leaf rust APR gene that was designated as Lr75.
Materials and methods
Plant material
Two different mapping populations were used in this study. The first population consisted of 240 recombinant inbred lines (RIL) generated from a cross of two Swiss winter wheat cultivars, ‘Arina’ and ‘Forno’ (Schnurbusch et al. 2004). A second near isogenic line (NIL) population was developed from a cross of ‘Arina’ and a back-cross line ‘ArinaLrFor’ (Arina*3/Forno). The recombinants of the ‘Arina’ × ‘ArinaLrFor’ NIL population were phenotyped qualitatively as F3, F4 and F5 rows in the field in Reckenholz, Switzerland during 2013, 2014 and 2015.
Selection of ‘Forno’ leaf rust QTLs for backcrossing
For re-evaluation of the original phenotypic data of the ‘Arina’ × ‘Forno’ RIL population developed by Schnurbusch et al. (2004), we grouped a subset of 117 RILs into different classes based on the availability of unambiguous marker information for the target regions of the three QTLs on chromosomes 1BS, 7DS and 7BL. These groups were made based on the following markers: gwm604 and gwm131 for Lr75; cssfr1 and cssfr2 for Lr34 (Lagudah et al. 2009) and ksuD2 and gbxGb218 for QLr.sfr-7BL.
Development of near isogenic lines, ‘ArinaLr75’, ‘ArinaQLr.sfr-7BL’ and ‘ArinaLrFor’
In order to introgress both Lr75 and QLr.sfr-7BL into the susceptible cv. ‘Arina’, 101 BC2F5 back-cross lines (Arina*3/Forno) were generated as described by Krattinger et al. (2009). The back-cross lines were screened with the flanking markers of the Lr75 target region (barc128–gwm131). Lines that had the ‘Forno’ alleles for both markers were further screened with the microsatellite markers gwm146 and gwm344 for presence of the QLr.sfr-7BL region and for absence of Lr34 with the diagnostic markers cssfr1 and cssfr2 (Lagudah et al. 2009). The resulting backcross line carrying Lr75 and QLr.sfr-7BL was named ‘ArinaLrFor’. We developed two additional near isogenic lines, ‘ArinaLr75’ and ‘ArinaQLr.sfr-7BL’, that carry individual gene introgressions. For this, ‘ArinaLrFor’ was crossed with ‘Arina’ and segregating progeny homozygous for either Lr75 or QLr.sfr-7BL were selected based on the markers described above. In addition, a near isogenic line ‘ArinaLr34’ with Lr34 in the genetic background of susceptible cv. ‘Arina’ was also generated (Arina*4/Forno). The presence of Lr34 was confirmed with the Lr34 diagnostic markers cssfr1 and cssfr2 (Lagudah et al. 2009).
Characterization of leaf rust resistance in the field
For field trials in Switzerland, the parental lines, ‘Arina’, ‘Forno, ‘ArinaLr75’, ‘ArinaQLr.sfr-7BL’, ‘ArinaLrFor’, ‘ArinaLr34’ and the recombinants were sown in randomized 5-row ×1.3 m plots in two replications with 40 seeds per row. Parental lines were replicated after 20 plots and one parental line was sown per plot. The first and the last row in each plot consisted of spreader rows containing a 1:1:1 mixture of highly susceptible wheat lines ‘Morocco’, ‘Bernina’ and ‘Arina’ to facilitate high and uniform pathogen density in the field. All field trials were inoculated with a mixture of 16 Swiss leaf rust isolates as described in Messmer et al. (2000). Inoculation was started by planting artificially infected plants into the spreader rows. Repeated leaf rust observations were made throughout the growing season. The final leaf rust severity on the flag leaves of the parental lines and the population was recorded when the susceptible cv. ‘Arina’ displayed leaf rust infection levels of 60 % or more.
‘Arina’, ‘Forno’, ‘ArinaLrFor’ and ‘ArinLr34’ were also tested in the fields in Cobbitty, Australia in 2014. The lines were planted at the Karalee site of the Plant Breeding Institute of the University of Sydney. Leaf rust-susceptible genotype ‘Sonora’ was used in spreader rows. Urediniospores of P. triticina (Pt) pathotype 76-1, 3, 5, 7, 9, 10, 12 +Lr37 (Plant Breeding Institute Culture Number = 621) and 10-1, 3, 9, 10, 11, 12 (Plant Breeding Institute Culture Number = 592) were used for infections. The virulence/avirulence formulae of Pt pathotypes which were used to test the parental lines is provided in Online Resource 1. Disease severity to leaf rust on the flag leaves of adult plants in the field were recorded when ‘Arina’ showed an infection level of 60 %.
Characterization of leaf rust resistance at the seedling stage
The parents ‘Arina’, ‘Forno’, ‘ArinaLr75’, ‘ArinaQLr.sfr-7BL’ and ‘ArinaLrFor’ along with ‘ArinaLr34’ were characterized at the seedling stage in the greenhouse with the seven Swiss isolates 91,047, 96,002, 95,219, 93,012, 96,209, 95,001 and 90,035 that were also used in the isolate mixture for field infections. For each line, 20–25 seeds were sown in two replicates in soil (Rasenerde [20 % org. matter, pH (CaCl2) 6.5, 1.4 g/L salt content (KCl), filler (DIN EN 12580)], ökohum GmbH, Herrenhof, Switzerland) in pots with a diameter of 13 cm. After treatment with growth inhibitor (Cycocel® Extra (4 mL/L), Omya AG, AGRO, Oftringen, Switzerland) and fertilizer (Wuxal® Profi (2–3 mL/L), Maag Garden, Syngenta, Düsseldorf, Germany), they were grown for 10 days under diurnal conditions (16 h light/20 °C, 8 h dark/16 °C, 70 % humidity). At the two-to-three-leaf stage (approx. after 10 days) the plants were inoculated with urediniospores suspended in oil (3 M™ Fluorinert™ FC-43, 3 M Electronics, Zwijndrecht, Belgium). After inoculation, the plants were allowed to air-dry for 30 min before they were placed in darkness for 24 h at 16 °C with 95 % humidity. Afterwards, the plants were transferred to growth chambers providing 16 h light/20 °C, 8 h dark/16 °C, 70 % humidity. The disease was assessed 10 days after inoculation (dai) using the 0–4 infection type scoring system described by Roelfs et al. (1992).
Measurement of pustule density of urediniospores
Pustule density of the uredinial infection was measured on field-infected flag leaves of ‘Arina’, ‘Forno’, ‘ArinaLrFor’ and ‘ArinaLr34’ when the plants were between the Zadoks growth stages 55–69 i.e. from half ear emergence to complete anthesis (Zadoks et al. 1974). For each line, leaves from two different plots were sampled and nine flag leaves per plot were randomly selected. For each leaf, a surface of 4 cm in the middle of the leaf was marked. The marked area was photographed at 3 different time points (84, 87 and 93 days after inoculating the spreader rows) with a Nikon camera using a macro objective lens. Subsequently, the images were analyzed with the image analysis software ImageJ 1.48 (Abràmoff et al. 2004). The area of interest (4 cm × width of the leaf) was calculated using the polygon selection tool and the pustules were counted using the oval selection tool. Not fully visible pustules at the edge of the leaf or the marked area were ignored. The average pustule density (number of pustules per area) was calculated for each of the four lines at different time points. Differences in pustule density among lines and time points were assessed using ANOVA. The distribution of the residual was tested with the Shapiro test and pustule density data were cube root transformed to fit normal distribution of the residuals. Then, a full model that included the line, time, plots effect and their potential interactions was used. As only marginally significant (p = 0.051), the plot effect was dropped from the full ANOVA. Only the line, time effects and their interaction were kept for further analysis. Pair-wise p value between lines and time points were computed with the Tukey’s HSD (Honestly Significant Difference) test. All analyses were performed using R v.3.1.3 (R Core Team 2016).
Marker analysis and genetic linkage mapping
The parents and BC3F2 recombinants developed from the cross of ‘Arina’ × ‘ArinaLrFor’ were grown in the greenhouse and leaf tissue was harvested from the seedlings (8–10 days old). DNA was extracted with a CTAB (cetyl trimethyl ammonium bromide) protocol as described by Stein et al. (2001). The quantity and concentration of DNA was measured using a NanoDrop spectrophotometer (Witec AG, Lucerne, Switzerland). The final concentration was standardized to 650 ng/µL. Dilutions of 13 and 65 ng/µL were used in PCR reaction for 6 % LiCOR gel (LiCOR DNA Sequencer 4200) and agarose gel electrophoresis, respectively. The simple sequence repeat identification tool, SSRIT (http://www.archive.gramene.org/db/markers/ssrtool) was used to identify the repeat motifs and SSR primers were designed using the software program Primer3 (v. 0.4.0). The PCR products of SSR primers were resolved on 6 % LiCOR gel. SSR markers were named as ‘swm’ (swiss wheat microsatellites). Primer sequences along with their repeat motifs are given in Online Resource 2. The genetic linkage map was constructed on a subset of F2-derived F3 lines (lines with missing phenotypic data were excluded) by calculating the recombination frequency between the markers. MapChart 2.3 (Voorrips 2002) was used to draw the linkage map.
To further saturate the 8 cM target region between wmc230 and gwm18 with additional markers, we exploited the flow-sorted Illumina survey sequences of chromosome 1BS of wheat cv. ‘Chinese Spring’ (http://wheat-urgi.versailles.inra.fr/Seq-Repository), the 1BS physical map generated by Raats et al. (2013) and information obtained by comparative genetics from a 1BS reference zipper based on synteny information of Brachypodium, rice and sorghum. The 1BS reference zipper was constructed in a similar manner as described by Breen et al. (2013). The gene-containing 1BS wheat Illumina sequences were physically anchored to BAC end sequences of wheat chromosome 1BS. These Illumina sequence contigs were further anchored to the reference zipper. The sequences of the flanking markers wmc230 and gwm18 were aligned against the integrated model of Illumina sequence contigs and reference zipper and the target region was defined. Then, we searched the Illumina sequences for microsatellite motifs within this target region and designed primers flanking the repeat motifs.
Deletion bin mapping
Chromosome 1B specificity of the SSR markers was confirmed by the absence of amplification in nulli-tetrasomic lines of cultivar ‘Chinese Spring’. Further, to determine the bin localization of all SSR markers, a set of 11 deletion lines for chromosome 1B was used along with two ditelosomic lines of wheat cv. ‘Chinese Spring’. Six deletion lines for the short arm (1BS4-sat-0.52, 1BS18-sat-0.50, 1BS2-sat-1.06, 1BS9-0.84, 1BS10-0.50 and 1BS1-0.35) and five deletion lines for the long arm (1BL11-0.23, 1BL6-0.32, 1BL1-0.47, 1BL2-0.69 and 1BL3-0.85) were used (Fig. 4b). Two ditelosomic lines DT1BS where the 1BL arm is missing and DT1BL where the 1BS arm is absent were also used. The fraction length (FL) value of each deletion line depicts the length of the remaining chromosome arm from the centromere after deletion relative to the length of the complete arm (Endo and Gill 1996). All the cytogenetic stocks were kindly provided by J. Raupp, Wheat Genetic Resource Centre, Department of Plant Pathology, Kansas State University, USA.
Results
Selection of ‘Forno’ leaf rust QTLs for backcrossing
Schnurbusch et al. (2004) identified QLr.sfr-1BS (subsequently referred to as Lr75) as the strongest leaf rust resistance QTL that was not associated with LTN in the ‘Arina’ × ‘Forno’ RIL population. Lr75 interacted with the minor QTL, QLr.sfr-7BL. Based on this information from the QTL study by Schnurbusch et al. (2004) we selected these two loci as candidates for backcrossing into the leaf rust-susceptible cultivar ‘Arina’. To validate our selection, we first re-evaluated the original phenotypic data of the ‘Arina’ × ‘Forno’ RIL population in order to estimate the phenotypic effect of this gene combination. For this, we grouped RIL lines based on marker information and compared the phenotypes of different groups. Lr34 was also included for comparison.
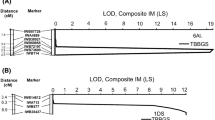
The RIL group that only contained Lr34 showed the strongest leaf rust resistance provided by a single QTL. The AUDPC value for the group with Lr75 alone (238.0) was lower than for the group with no resistance QTL (388.6) although the difference was not significant (p = 0.16). No significant difference in AUDPC values was observed between the group with only QLr.sfr-7BL and the group with no QTL. However, the combination of Lr75 and QLr.sfr-7BL resulted in leaf rust resistance levels comparable to Lr34, confirming the original finding that Lr75 and QLr.sfr-7BL are additive (Fig. 1). All gene combinations with Lr34 were associated with LTN whereas the ones without Lr34 were not. Based on the re-evaluation of these original RIL data we expected that the combination of Lr75 and QLr.sfr-7BL would result in partial leaf rust resistance levels similar to Lr34 but without LTN. Lr75 and QLr.sfr-7BL were therefore co-introduced into the genetic background of ‘Arina’ through marker-assisted backcrossing.
Phenotypic effect of different leaf rust resistance QTL combinations. The phenotypic data of Schnurbusch et al. (2004) were re-evaluated for area under disease progress curve (AUDPC_ %) (top graph) and leaf tip necrosis (LTN) in millimeter (mm) (bottom graph) on groups of RIL lines. The group of RILs with both Lr75 and QLr.sfr-7BL is highlighted in black. Numbers in brackets indicate the number of RIL lines present in each class. Letters indicate lines with equivalent resistance levels (p > 0.05, Tukey’s HSD test) and error bars represent standard errors of the mean
Evaluation of near isogenic lines ‘ArinaLr75’, ‘ArinaQLr.sfr-7BL’ and ‘ArinaLrFor’ for field resistance
In order to introgress both Lr75 and QLr.sfr-7BL into the susceptible cv. ‘Arina’, 101 BC2F5 back-cross lines (Arina*3/Forno) were screened with the flanking markers of the Lr75 region (barc128–gwm131). Twelve of the 101 backcross lines showed the ‘Forno’ alleles for both the flanking markers in the Lr75 region. By screening these twelve lines with Lr34 diagnostic markers, two lines were positive for Lr34 and were therefore excluded. From the remaining 10 lines, one line, ‘ArinaLrFor’, showed the ‘Forno’ alleles for the flanking markers of both Lr75 and QLr.sfr-7BL region.
The lines, ‘ArinaLrFor’ and ‘ArinaLr34’ were evaluated together with ‘Arina’ and ‘Forno’ for leaf rust resistance in Switzerland and Australia (Table 1). In addition, the lines ‘ArinaLr75’ and ‘ArinaQLr.sfr-7BL’ were evaluated for leaf rust resistance only in Switzerland in 2016. ‘ArinaLr75’ and ‘ArinaQLr.sfr-7BL’ both showed a weak partial resistance response with a final disease severity of 40–60 % and 50–70 %, respectively (Table 1; Fig. 2). The susceptible control ‘Arina’ had leaf rust infection levels of 60–100 % except for 2014 crop season where ‘Arina’ had leaf rust infection levels of 50–80 %. This was due to the emergence of stripe rust and low temperature at the time of rust development which resulted in the minimum infection level of 50 % in ‘Arina’. Despite the relatively weak contributions towards resistance of Lr75 and QLr.sfr-7BL alone, the gene combination in ‘ArinaLrFor’ resulted in good levels of partial leaf rust resistance comparable or even stronger than Lr34. (Table 1; Fig. 2). ‘ArinaLrFor’ displayed a slow-rusting response with a final leaf area coverage ranging from 14 to 40 % in comparison to ‘ArinaLr34’ which had final leaf area coverage of 5–56 %. ‘Forno’ displayed a near-immune response which is due to the combination of Lr75, Lr34, QLr.sfr-7BL and several minor QTLs.
The lines ‘ArinaLrFor’ and ‘ArinaLr34’ along with ‘Arina’ and ‘Forno’ were also tested for leaf rust resistance in Australia. Similar to the results obtained for the Swiss environment, ‘ArinaLrFor’ showed increased leaf rust resistance in the field in Australia (Table 1; Online Resource 3). Hence, ‘ArinaLrFor’ showed good levels of partial leaf rust resistance in two environments.
Slow-rusting resistance genes are generally associated with a longer latency period, lower uredinial density and smaller uredinial size (Das et al. 1993). The line ‘ArinaLrFor’ has a slow-rusting phenotype as shown in Fig. 2. Measurement of pustule density on the flag leaves of ‘Arina’, ‘Forno’, ‘ArinaLrFor’ and ‘ArinaLr34’ showed that ‘Forno’ displayed a significantly lower number of pustules than the other three lines and, in agreement with the near-immune phenotype, no significant increase in the pustule number was observed over time (84, 87 and 93 days after inoculating the spreader rows) (Table 2). On the other hand, ‘Arina’ showed a constant increase in the number of pustules from day 84 to 93 after inoculating the spreader rows. Both ‘ArinaLrFor’ and ‘ArinaLr34’ showed an intermediate response with a slower increase of pustule density observed in ‘ArinaLrFor’ compared to ‘ArinaLr34’ (Table 2).
‘ArinaLrFor’ shows race-specific resistance at seedling stage
Partial adult plant resistance genes often do not confer seedling resistance. In order to determine the seedling responses of Lr75 and QLr.sfr-7BL individually or in combination we infected ‘ArinaLrFor’, ‘ArinaLr75’ and ‘ArinaQLr.sfr-7BL’ along with ‘Arina’ and ‘Forno’ at the seedling stage in the greenhouse. For the two isolates 91,047 and 95,219 ‘ArinaLrFor’ was as susceptible as ‘Arina’ and showed a moderate infection type (IT = 3+) with medium sized uredia with or without chlorosis. Surprisingly, for some isolates (90,035, 96,002, 93,012, 95,001 and 96,209), ArinaLrFor showed a stronger resistance reaction (;2) than either of its two parents ‘Forno’ and ‘Arina’ (Fig. 3). For these isolates ‘Forno’ showed a mesothetic infection type with various pustule sizes and hypersensitive flecks (X). A similar IT was previously reported for the leaf rust resistance gene Lr14a located on chromosome 7BL (Mcintosh et al. 1995). The differential line ‘Thatcher Lr14a’ showed a similar albeit slightly stronger mesothetic resistance reaction than ‘Forno’. Hence, it is likely that QLr.sfr-7BL in ‘Forno’ is Lr14a and that this gene interacts with another resistance gene in ‘Arina’, which most likely is Lr13 resulting in the strong resistance response of ‘ArinaLrFor’. ‘ArinaLr75’ showed an IT similar to the susceptible cv. ‘Arina’, indicating that Lr75 is ineffective at the seedling stage. The seedling reaction of ‘ArinaQLr.sfr-7BL’ was comparable to ‘ArinaLrFor’, supporting the hypothesis that the seedling resistance in ‘ArinaLrFor’ is most likely due to the interaction of Lr13 gene present in ‘Arina’ background with QLr.sfr-7BL (Fig. 3).
Seedling infection assay on a ‘Arina’, ‘Forno’, ‘ThatcherLr14a’ (ThLr14a) and ‘ArinaLrFor’ (ArLrFor) and on b ‘Arina’, 'Thatcher', ‘ThatcherLr14a’, ‘ArinaLrFor’ (ArLrFor), ‘ArinaLr75’ (ArLr75) and ‘ArinaQLr.sfr-7BL’ (ArQLr.sfr-7BL) using isolate 96,209. Infection type response was scored based on a 0–4 scale (Roelfs et al. 1992). Two images represent results from two independent infection experiments with the same isolate 96,209
Genetic mapping of Lr75
Based on our results, Lr75 can be considered as a partial leaf rust resistance QTL. We therefore decided to further narrow down the Lr75 interval. To define an Lr75 target interval we tested 63 publically available, 1BS specific SSR markers (http://wheat.pw.usda.gov/GG2/index.html). Of the 63 SSRs, nine were 1B-specific and polymorphic (barc128, cwem6c, cfa2158, gpw4069, wmc230, gwm11, gwm18, wmc277 and wmc156) between ‘Arina’ and ‘Forno’ and were added to the genetic map of the ‘Arina’ × ‘Forno’ RIL population (Schnurbusch et al. 2004). This resulted in the establishment of an 8 cM target region spanning the Lr75 gene with wmc230 and gwm18 as the distal and proximal flanking markers, respectively. For precise genetic mapping and phenotypic analysis of Lr75, a near isogenic line (NIL) population consisting of 2067 F2 individuals from a cross of ‘ArinaLrFor’ and ‘Arina’ (Arina*4/Forno) was used. Out of these, 234 lines showed a recombination between the two flanking markers wmc230 and gwm18. These recombinants were further screened with two QLr.sfr-7BL-associated SSR markers (gwm344 and gwm146). Only recombinants without the QLr.sfr-7BL QTL were selected for further mapping in order to avoid interference from the 7BL QTL during phenotyping. This resulted in 65 BC3F2 recombinants that were phenotyped qualitatively as BC3F3–BC3F5 families in comparison to the parental lines.
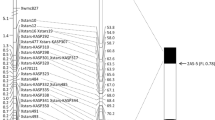
Using the available sequence information of ‘Chinese Spring’, the 1BS physical map and synteny information of Brachypodium, rice and sorghum, 98 SSR primers were designed (Online Resource 2), out of which 8 were polymorphic between the parents. Of these 8 markers, six (swm271, swm275, swm276, swm278, swm281 and swm294) were mapped in the target interval in the BC3F2 fine mapping population (Fig. 4a). The other two markers, swm216 and swm247 were mapped at a distance of 0.16 and 0.31 cM proximal to gwm18, respectively. The addition of the 8 new SSR markers placed Lr75 between the distal marker gwm604 and proximal marker swm271 at a distance of 1.6 and 2.7 cM, respectively (Fig. 4a).
Genetic linkage and physical deletion bin mapping of leaf rust resistance gene Lr75. a Genetic linkage map of the short arm of chromosome 1B of the ‘Arina’ × ‘ArinaLrFor’ mapping population. Marker positions are shown in cM on the left side of the linkage map. b Deletion bin map of the short arm of chromosome 1B of wheat cv. ‘Chinese Spring’. Physical bin localization of the markers wmc230 and swm271 is shown by arrowheads. c Deletion bin mapping of wmc230. d Deletion bin mapping of swm271 on Chinese Spring, ArinaLrFor, water, 1BS4-sat-0.52, 1BS18-sat-0.50, 1BS2-sat-1.06, 1BS9-0.84, 1BS10-0.50, 1BS1-0.35, 1BL11-0.23, 1BL6-0.32, 1BL1-0.47, 1BL2-0.69, 1BL3-0.85, DT1BS, DT1BL (lanes 1–16)
Deletion bin mapping
In order to physically map Lr75, we used the cytogenetic stocks of chromosome 1B of wheat cv. ‘Chinese Spring’. Marker wmc230 amplified on none of the 1BS deletion lines but amplified in all lines with a deletion on the long arm. The marker swm271 on the other hand did not amplify on deletion lines 1BS1-0.35 and 1BS10-0.50 (Fig. 4b–d). Hence, the use of the cytogenetic stocks physically placed the markers wmc230 and swm271 and the gene towards the distal end of chromosome 1BS (Fig. 4). Singh et al. (2013b) mapped Lr71 close to the centromere in the deletion bins 1BS10-0.50 and 1BL6-0.32 respectively (Fig. 4).
Discussion
In this study we introgressed two leaf rust resistance QTLs, Lr75 and QLr.sfr-7BL, from ‘Forno’ into the leaf rust susceptible Swiss winter wheat cv. ‘Arina’. Marker-assisted introgression of these two QTLs resulted in high levels of partial adult plant leaf rust resistance in the field. Further, genetic mapping of Lr75 with SSR markers placed this gene towards the distal end of chromosome 1BS. The only reported leaf rust resistance gene present on chromosome 1BS in the close proximity of Lr75 is Lr71 (Singh et al. 2013b). Lr75 can be distinguished from Lr71 by the marker gwm18 which was reported to be the distal flanking marker of Lr71 (Singh et al. 2013b), whereas gwm18 mapped proximal to the Lr75 gene. In addition, deletion bin mapping also physically separates Lr75 from Lr71. The deletion bin mapping of the SSR markers wmc230 and swm271 mapped Lr75 towards the distal end whereas deletion bin mapping mapped Lr71 towards the centromere on chromosome 1BS as reported by Singh et al. (2013b). Hence, both genetic and physical mapping of Lr75 on chromosome 1BS with SSR markers showed that it is a novel gene as no other leaf rust resistance gene has been described in the target region of Lr75.
Breeding for slow-rusting resistance in European wheat germplasm
All the adult plant slow-rusting genes (Lr34/Yr18/Sr57/Pm38, Lr46/Yr29/Pm39 and Lr68) that were obtained from the CIMMYT wheat germplasm are associated with LTN which is considered undesirable in European wheat breeding programme (Singh et al. 1998; Krattinger et al. 2009; Hiebert et al. 2010; Herrera-Foessel et al. 2012). Due to this reason, the wheat cultivars possessing these genes have not been widely grown and accepted in European wheat breeding. It has therefore become essential to identify additional sources of durable rust resistance in European wheat germplasm without LTN. In our study we described a novel slow-rusting gene, Lr75 present on wheat chromosome 1BS. Lr75 has shown to provide an additive effect when combined with another slow-rusting QTL, QLr.sfr-7BL. Both these QTLs are present in Swiss winter wheat cv. ‘Forno’ and are not associated with LTN. Another example of a non-LTN broad-spectrum APR gene is Lr22a which was introgressed from an Aegilops tauschii accession into cultivated wheat (Hiebert et al. 2007).
Very little information is available about leaf rust APRs in European wheat breeding material. Only a few studies looked at the existence of APR genes in ~100 European wheat lines (Winzeler et al. 2000; Park et al. 2001; Pathan and Park 2006). All studies reported the frequent occurrence of the Lr13 APR gene in European wheat cultivars. Winzeler et al. (2000), detected varying levels of resistance shown by the cultivars carrying Lr13 across Europe which indicates that virulence for Lr13 exist. ‘Arina’ for example is known to possess Lr13 but is susceptible to leaf rust throughout Europe (Pathan and Park 2006).
To our knowledge, only four studies reported on the identification of leaf rust APRs on chromosome 1BS in European wheat breeding material (Messmer et al. 2000; Schnurbusch et al. 2004; Singh et al. 2009; Buerstmayr et al. 2014). Other than ‘Forno’, cultivars ‘Beaver’ and ‘Capo’ possess 1BS QTLs. ‘Beaver’ has the 1BL/1RS translocation and the QTL identified in ‘Beaver’ can therefore not be Lr75. The QTL in ‘Capo’, QLr.ifa-1B mapped close to the centromere but since no detailed study has been available on this QTL, it is not clear whether the genomic locations of Lr75 and QLr.ifa-1B are identical. So far, ‘Forno’ seems to be the only source of Lr75 and interestingly this gene has not been described in any other European wheat cultivar. However, apart from European wheat lines, two CIMMYT wheat lines, ‘Pastor’ and ‘Parula’ also possess a leaf rust resistance QTL on chromosome 1BS (William et al. 1997; Rosewarne et al. 2012). According to the study conducted by Singh and Rajaram (1992), the high level of resistance in ‘Parula’ is due to the combination of three slow-rusting APR genes, Lr34, Lr46 and Lr68 plus some minor genes. William et al. (1997) by using a RIL population developed from a cross of resistant cv. ‘Parula’ and moderately susceptible cv. ‘Siete Cerros’ identified a minor QTL on chromosome 1BS in ‘Parula’ which explained a phenotypic variance of 7–10 %. Another QTL on chromosome 1BS was detected in cv. ‘Pastor’ by Rosewarne et al. (2012). They reported this QTL to be present in the same genomic region as Lr75. Both these QTLs have not been characterized in detail and their precise genetic location is not available. Therefore, it is impossible to conclude if these QTLs are Lr75 or not.
Slow-rusting APR genes are influenced by environment
The knowledge of an environmental influence on resistance genes allows wheat breeders to deploy resistance gene combinations most effectively in different regions. Our study showed that the combination of Lr75 and QLr.sfr-7BL provided partial resistance in Switzerland and Australia. Similarly, Lr34 also showed partial resistance at the adult plant stage in Switzerland and Australia. However, the level of resistance shown by these genes varied at the two locations. In Australia, the resistance provided by Lr34 was stronger than that provided by the gene combination of Lr75 and QLr.sfr-7BL, whereas in Switzerland, except for one crop season (2016), Lr34 alone showed a weaker resistance response. Similar findings have also been reported in the literature where the environment plays a role in modifying the resistance response of slow-rusting genes. Herrera-Foessel et al. (2012) compared the leaf rust resistance response of Lr68, Lr34, Lr46 and Lr67 during three crop seasons (2008–2009, 2009–2010, 2010–2011) in the field at Ciudad Obregon, Mexico. They reported that except for one crop season (2010–2011), the effect of Lr68 was smaller as compared to Lr34, Lr46 and Lr67. In 2010–2011 however, Lr68 showed a stronger resistance response than Lr46. Similar results were observed by Lillemo et al. (2011) while studying the additive effect of three APR genes, Lr34/Yr18/Sr57/Pm38, Lr46/Yr29/Pm39 and Lr68 in ‘Avocet-YrA × Parula’ F6 RIL mapping populations across nine different environments. In agreement with Herrera-Foessel et al. (2012), they also observed a smaller resistance response of Lr68 than Lr34 and Lr46 in Mexico. On the other hand, a stronger resistance response of Lr68 as compared to Lr34 was seen in Argentina and Uruguay (Lillemo et al. 2011). Interestingly, the combination of Lr68 and Lr34 showed stronger resistance than either gene alone in all the tested environments which suggests an additive effect of these two genes. In contrast, Silva et al. (2015) while studying the effect and interaction of Lr68, Lr34 and Sr2 genes in two wheat populations derived from ‘Parula’ at sites in Uruguay did not observe an additive effect of the combination of Lr68 and Lr34. Instead, the effect of the combination of Lr68 and Lr34 was comparable to the effect of Lr68 alone. These studies clearly showed that resistance gene combinations do not necessarily behave in the same manner in all environments. Stem rust resistance gene, Sr2 is known to be tightly linked to seedling resistance gene Lr27 (Mago et al. 2011) and Lr27 was also reported to be responsible for reducing leaf rust severity (Bariana et al. 2007). Silva et al. (2015) studied the effect of the Sr2 gene in reducing leaf rust in Uruguay. They observed that the stem rust resistance gene Sr2 does not have any effect on leaf rust resistance when present alone but a significant increase in resistance level was seen when Sr2 was present in combination with Lr68. From their study it was clear that Sr2 alone is not strong enough to provide resistance and rather it enhances the effect of Lr68.
QLr.sfr-7BL in ‘Forno’ is most likely the leaf rust resistance gene Lr14a
In a survey of wheat leaf rust in Western Europe, Park et al. (2001) reported the presence of Lr14a in ‘Forno’. This gene was mapped to the distal end of chromosome 7BL in a wheat consensus map (Gale et al. 1995). A major leaf rust resistance QTL, (QLr.ubo-7B.2) was also identified by Maccaferri et al. (2008) on chromosome 7BL within an 8.2 cM region in durum wheat cv. ‘Creso’. Their study also postulated that this QTL is effective at both seedling and adult plant stages. The two microsatellite markers gwm146 and gwm344 were reported to be tightly linked to this QTL on chromosome 7BL. The same SSR markers were also reported to be closely linked to another gene, LrLla which was more precisely mapped on chromosome 7BL in a population of 98 F3 lines derived from Chilean durum cv. Llareta INIA by Herrera-Foessel et al. (2008). They postulated this gene to be Lr14a based on the resistance response and chromosomal location. In addition, (Singh et al. 2013a) also identified a major QTL for leaf rust resistance on chromosome 7BL close to marker gwm146 in French durum wheat cv. ‘Sachem’. They reported that this QTL is effective at both seedling and adult plant stages when tested in different environments in Mexico. The APR gene Lr68 was also mapped on chromosome 7BL in the same genomic region as QLr.sfr-7BL by Herrera-Foessel et al. (2012). Like all the QTLs mentioned above, QLr.sfr-7BL identified from ‘Forno’ also shared the same genomic region on chromosome 7BL close to the markers gwm344 and gwm146. In addition, seedling infection data have also shown that the QTL, QLr.sfr-7BL in ‘Forno’ is most likely Lr14a because of the mesothetic resistance response shown by both ‘Forno’ and Lr14a differential line, ‘ThatcherLr14a’. Therefore, it is likely that this QTL is actually the Lr14a gene. Lr14a was transferred from emmer wheat to the bread wheat cv. ‘Hope’ and H-44. Park et al. (2001) reported the frequent occurrence of Lr14a in Europe and that 33.5 % of the area in France has been occupied by Lr14a alone or in combination with Lr13. Virulence against Lr14a was reported in Europe by Goyeau et al. (2010) and Park et al. (2001).
In this research we have successfully shown that marker-assisted introgression of two partial, non-LTN leaf rust resistance genes results in slow-rusting resistance in the susceptible Swiss winter wheat cv. ‘Arina’. Introgression of this slow-rusting gene combination in different cultivars will be useful in improving leaf rust resistance in near future and impedes a greater value in breeding. For the optimal use of Lr75, this gene has to be tested with the local pathotypes of the region and if effective, can be combined with other effective rust resistance genes to minimize the spread of emerging virulent pathotypes.
Author contribution statement
JS, LL, TW, SGK and BK planned and conceptualized the experiments. JS, LL SGK and UB performed field leaf rust infection experiments. JS, LL and SGK generated the mapping population. JS and LL performed the molecular work. JS and TW developed the strategy for marker development. JS, SGK and BK wrote the manuscript.
References
Abràmoff MD, Magalhaes PJ, Ram SJ (2004) Image processing with imagej. Biophotonics Int 11:36–42
Bariana H, Miah H, Brown G, Willey N, Lehmensiek A (2007) Molecular mapping of durable rust resistance in wheat and its implication in breeding. In: Wheat production in stressed environments: Proceedings of the 7th international wheat Conf, Mar del Plata, p 723–728
Bolton MD, Kolmer JA, Garvin DF (2008) Wheat leaf rust caused by Puccinia triticina. Mol Plant Pathol 9:563–575. doi:10.1111/j.1364-3703.2008.00487.x
Breen J, Wicker T, Shatalina M et al (2013) A physical map of the short arm of wheat chromosome 1A. PLoS ONE. doi:10.1371/journal.pone.0080272
Buerstmayr M, Matiasch L, Mascher F, Vida G, Ittu M, Robert O, Holdgate S, Flath K, Neumayer A, Buerstmayr H (2014) Mapping of quantitative adult plant field resistance to leaf rust and stripe rust in two European winter wheat populations reveals co-location of three QTL conferring resistance to both rust pathogens. Theor Appl Genet 127:2011–2028. doi:10.1007/s00122-014-2357-0
Caldwell R (1968) Breeding for general and/or specific plant disease resistance. In: Finley KW, Shepherd KW (ed) Proceeding International Wheat Genetic Symposium 3rd, Canberra, pp 263–272
Das M, Rajaram S, Kronstad W, Mundt C, Singh R (1993) Associations and genetics of three components of slow rusting in leaf rust of wheat. Euphytica 68:99–109
Dyck P, Samborski D (1979) Adult-plant leaf rust resistance in PI 250413, an introduction of common wheat. Can J Plant Sci 59:329–332
Endo TR, Gill BS (1996) The Deletion Stocks of Common Wheat. J Hered 87:295–307. doi:10.1093/oxfordjournals.jhered.a023003
Gale M, Atkinson M, Chinoy C, Harcourt R, Jia J, Li Q, Devos K (1995) Genetic maps of hexaploid wheat. In: Li ZS, Xin ZY (eds) Proceedings 8th International Wheat Genet Symp. China Agricultural Scientech Press, Beijing, pp 29–40
Goyeau H, Ammar K, Berder J (2010) Virulence in Puccinia triticina for durum wheat cultivar Creso and other durum wheat cultivars carrying resistance gene Lr14a in France. Plant Dis 94:1068. doi:10.1094/PDIS-94-8-1068A
Herrera-Foessel SA, Singh RP, Huerta-Espino J, Crossa J, Djurle A, Yuen J (2008) Genetic analysis of slow rusting resistance to leaf rust in durum wheat. Crop Sci 48:2132–2140
Herrera-Foessel SA, Singh RP, Huerta-Espino J, Rosewarne GM, Periyannan SK, Viccars L, Calvo-Salazar V, Lan C, Lagudah ES (2012) Lr68: a new gene conferring slow rusting resistance to leaf rust in wheat. Theor Appl Genet 124:1475–1486. doi:10.1007/s00122-012-1802-1
Hiebert CW, Thomas JB, Somers DJ, McCallum BD, Fox SL (2007) Microsatellite mapping of adult-plant leaf rust resistance gene Lr22a in wheat. Theor Appl Genet 115:877–884. doi:10.1007/s00122-007-0604-3
Hiebert CW, Thomas JB, McCallum BD, Humphreys DG, DePauw RM, Hayden MJ, Mago R, Schnippenkoetter W, Spielmeyer W (2010) An introgression on wheat chromosome 4DL in RL6077 (Thatcher*6/PI 250413) confers adult plant resistance to stripe rust and leaf rust (Lr67). Theor Appl Genet 121:1083–1091. doi:10.1007/s00122-010-1373-y
Huerta-Espino J, Singh RP, Germán S, McCallum BD, Park RF, Chen WQ, Bhardwaj SC, Goyeau H (2011) Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 179:143–160. doi:10.1007/s10681-011-0361-x
Kolmer J (2013) Leaf rust of wheat: pathogen biology, variation and host resistance. Forests 4:70–84. doi:10.3390/f4010070
Kolmer JA, Singh RP, Garvin DF, Viccars L, William HM, Huerta-Espino J, Ogbonnaya FC, Raman H, Orford S, Bariana HS, Lagudah ES (2008) Analysis of the rust resistance region in wheat germplasm. Crop Sci 48:1841–1852. doi:10.2135/cropsci2007.08.0474
Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323:1360–1363. doi:10.1126/science.1166453
Lagudah ES, McFadden H, Singh RP, Huerta-Espino J, Bariana HS, Spielmeyer W (2006) Molecular genetic characterization of the Lr34/Yr18 slow rusting resistance gene region in wheat. Theor Appl Genet 114:21–30. doi:10.1007/s00122-006-0406-z
Lagudah ES, Krattinger SG, Herrera-Foessel S, Singh RP, Huerta-Espino J, Spielmeyer W, Brown-Guedira G, Selter LL, Keller B (2009) Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor Appl Genet 119:889–898. doi:10.1007/s00122-009-1097-z
Lillemo M, Singh R, William M, Herrera-Foessel S, Huerta- Espino J, German S, Campos P, Chaves M, Madriaga R, Xia X, Liang S, Liu D, Li Z, Lagudah E (2011) Multiple rust resistance and gene additivity in wheat: lessons from multi-location case studies in the cultivars Parula and Saar. Global Rust Initiative Meeting, St. Paul, pp 111–120
Maccaferri M, Mantovani P, Tuberosa R, Deambrogio E, Giuliani S, Demontis A, Massi A, Sanguineti MC (2008) A major QTL for durable leaf rust resistance widely exploited in durum wheat breeding programs maps on the distal region of chromosome arm 7BL. Theor Appl Genet 117:1225–1240. doi:10.1007/s00122-008-0857-5
Mago R, Tabe L, McIntosh RA, Pretorius Z, Kota R, Paux E, Wicker T, Breen J, Lagudah ES, Ellis JG, Spielmeyer W (2011) A multiple resistance locus on chromosome arm 3BS in wheat confers resistance to stem rust (Sr2), leaf rust (Lr27) and powdery mildew. Theor Appl Genet 123:615–623. doi:10.1007/s00122-011-1611-y
Mcintosh RA, Wellings CR, Park RF (1995) Wheat rusts: an atlas of resistance genes. Csiro Publishing, Clayton
Mcintosh RA, Dubcovsky J, Rogers JM, Morris C, Appels R, Xia X (2013) Catalogue of gene symbols for wheat: 2013-2014. KOMUGI-Integrated Wheat Science Database. http://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2013.pdf
Messmer MM, Seyfarth R, Keller M, Schachermayr G, Winzeler M, Zanetti S, Feuillet C, Keller B (2000) Genetic analysis of durable leaf rust resistance in winter wheat. Theor Appl Genet 100:419–431
Moore JW, Herrera-Foessel S, Lan C, Schnippenkoetter W, Ayliffe M, Huerta-Espino J, Lillemo M, Viccars L, Milne R, Periyannan S, Kong X, Spielmeyer W, Talbot M, Bariana H, Patrick JW, Dodds P, Singh R, Lagudah E (2015) A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat Genet 47:1494–1498. doi:10.1038/ng.3439
Park RF, Goyeau H, Felsenstein FG, Bartoš P, Zeller FJ (2001) Regional phenotypic diversity of Puccinia triticina and wheat host resistance in western Europe, 1995. Euphytica 122:113–127. doi:10.1023/A:1012603500686
Pathan AK, Park RF (2006) Evaluation of seedling and adult plant resistance to leaf rust in European wheat cultivars. Euphytica 149:327–342. doi:10.1007/s10681-005-9081-4
Raats D, Frenkel Z, Krugman T et al (2013) The physical map of wheat chromosome 1BS provides insights into its gene space organization and evolution. Genome Biol 14:R138. doi:10.1186/gb-2013-14-12-r138
Roelfs AP, Singh RP, Saari EE (1992) Rust diseases of wheat: concepts and methods of disease management. CIMMYT, Mexico
Rosewarne GM, Singh RP, Huerta-Espino J, Herrera-Foessel SA, Forrest KL, Hayden MJ, Rebetzke GJ (2012) Analysis of leaf and stripe rust severities reveals pathotype changes and multiple minor QTLs associated with resistance in an Avocet × Pastor wheat population. Theor Appl Genet 124:1283–1294. doi:10.1007/s00122-012-1786-x
Schnurbusch T, Paillard S, Schori A, Messmer M, Schachermayr G, Winzeler M, Keller B (2004) Dissection of quantitative and durable leaf rust resistance in Swiss winter wheat reveals a major resistance QTL in the Lr34 chromosomal region. Theor Appl Genet 108:477–484. doi:10.1007/s00122-003-1444-4
Silva P, Calvo-Salazar V, Condón F, Quincke M, Pritsch C, Gutiérrez L, Castro A, Herrera-Foessel S, von Zitzewitz J, Germán S (2015) Effects and interactions of genes Lr34, Lr68 and Sr2 on wheat leaf rust adult plant resistance in Uruguay. Euphytica 204:599–608. doi:10.1007/s10681-014-1343-6
Singh RP, Rajaram S (1991) Resistance to Puccinia recondita f. sp. tritici in 50 Mexican bread wheat cultivars. Crop Sci 31:1472. doi:10.2135/cropsci1991.0011183X003100060016x
Singh RP, Rajaram S (1992) Genetics of adult-plant resistance of leaf rust in “Frontana” and three CIMMYT wheats. Genome 35:24–31
Singh RP, Mujeeb-Kazi A, Huerta-Espino J (1998) Lr46: a gene conferring slow-rusting resistance to leaf rust in wheat. Phytopathology 88:890–894. doi:10.1094/PHYTO.1998.88.9.890
Singh RP, Huerta-Espino J, Rajaram S (2000) Achieving near-immunity to leaf and stripe rusts in wheat by combining slow rusting resistance genes. Acta Phytopathol Entomol Hungarica 35:133–139
Singh RP, Huerta-Espino J, William HM (2005) Genetics and breeding for durable resistance to leaf and stripe rusts in wheat. Turkish J Agric For 29:121–127
Singh D, Simmonds J, Park RF, Bariana HS, Snape JW (2009) Inheritance and QTL mapping of leaf rust resistance in the European winter wheat cultivar “Beaver”. Euphytica 169:253–261. doi:10.1007/s10681-009-9959-7
Singh A, Pandey MP, Singh AK, Knox RE, Ammar K, Clarke JM, Clarke FR, Singh RP, Pozniak CJ, DePauw RM, McCallum BD, Cuthbert RD, Randhawa HS, Fetch TG (2013a) Identification and mapping of leaf, stem and stripe rust resistance quantitative trait loci and their interactions in durum wheat. Mol Breed 31:405–418. doi:10.1007/s11032-012-9798-4
Singh D, Mohler V, Park RF (2013b) Discovery, characterisation and mapping of wheat leaf rust resistance gene Lr71. Euphytica 190:131–136. doi:10.1007/s10681-012-0786-x
Stein N, Herren G, Keller B (2001) A new DNA extraction method for high-throughput marker analysis in a large-genome species such as Triticum aestivum. Plant Breed 356:354–356
R Core Team (2016) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. doi:10.1093/jhered/93.1.77
William HM, Hoisington D, Singh RP, González-de-León D (1997) Detection of quantitative trait loci associated with leaf rust resistance in bread wheat. Genome 40:253–260
Winzeler M, Mesterhazy A, Park RF et al (2000) Resistance of European winter wheat germplasm to leaf rust. Agronomie 20:783–792. doi:10.1051/agro:2000175
Zadoks J, Chang T, Konzak C (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421
Acknowledgments
We thank J. Raupp, Kansas State University, USA for kindly providing the seeds of the cytogenetic stocks of cv. ‘Chinese Spring’. We thank Bea Senger for her technical assistance with the field experiments as well as in the greenhouse. We also thank Gerhard Herren and Gabriele Buchmann for their technical assistance during the molecular genotyping. We are highly thankful to Dr. Anne Roulin, Department of Plant and Microbial Biology, University of Zurich for helping in statistical data analysis. The financial support for this work was provided by an Advanced Investigator Grant from the European Research Council (ERC-2009-AdG 249996, Durableresistance). SGK is supported by an Ambizione Grant of the Swiss National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by T. Miedaner.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singla, J., Lüthi, L., Wicker, T. et al. Characterization of Lr75: a partial, broad-spectrum leaf rust resistance gene in wheat. Theor Appl Genet 130, 1–12 (2017). https://doi.org/10.1007/s00122-016-2784-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2784-1