Abstract
Prunus species express different ranges and levels of resistance to the root-knot nematodes (RKN) Meloidogyne spp. In Myrobalan plum (Prunus cerasifera), the dominant Ma gene confers a high-level and wide-spectrum resistance to the predominant RKN, Meloidogyne arenaria, Meloidogyne incognita, Meloidogyne javanica and the isolate Meloidogyne sp. Florida which overcomes the resistance of the Amygdalus sources. In Japanese plum (Prunus salicina), a similar wide-spectrum dominant resistance gene, termed R jap , has been hypothesized from an intraspecific segregating cross. In peach, two crosses segregating for resistance to both M. incognita and M. arenaria were used to identify single genes that each control both RKN species in the Shalil (R Mia557 ) and Nemared (R MiaNem ) sources. Localisation of these genes was made possible using the RFLP and SSR- saturated reference Prunus map T×E, combined with a BSA approach applied to some of the genes. The Ma1 allele carried by the Myrobalan plum accession P.2175 was localised on the linkage group 7 at an approximate distance of 2 cM from the SSR marker pchgms6. In the Japanese plum accession J.222, the gene R jap was mapped at the same position in co-segregation with the SSR markers pchgms6 and CPPCT022. The peach genes R Mia557 and R MiaNem , carried by two a priori unrelated resistance sources, were co-localized in a subtelomeric position on linkage group 2. This location was different from the more centromeric position previously proposed by Lu et al. (1999) for the resistance gene Mij to M. incognita and M. javanica in Nemared, near the SSR pchgms1 and the STS EAA/MCAT10. By contrast, R Mia557 and R MiaNem were flanked by STS markers obtained by Yamamoto and Hayashi (2002) for the resistance gene Mia to M. incognita in the Japanese peach source Juseitou. Concordant results for the three independent sources, Shalil, Nemared and Juseitou, suggest that these peach RKN sources share at least one major gene resistance to M. incognita located in this subtelomeric position. We showed that plum and peach genes are independent and, thus, can be pyramided into interspecific hybrid rootstocks based on the plum and peach species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Prunus genus comprises over 400 species, most of them being well adapted to Northern temperate areas and widely distributed in Europe (Rehder 1954). They include fruit-producing species (e.g. peach, almond, plum, apricot and cherry), and several rootstock and ornamental species. Plums and apricots belong to the Prunophora subgenus which is divided into two sections (i.e. Euprunus and Armeniaca, respectively). Peaches and almonds belong to the Amygdalus subgenus. All the Prunus species have an 8-basis chromosome number with various ploidy levels: (2n=2x=16) for peach, almond, Myrobalan and Japanese plums, apricot and sweet cherry, (2n=4x=32) for sour cherry and (2n=6x=48) for European plums (Salesses et al. 1994).
Root-knot nematodes (RKN) (Meloidogyne spp.) are major crop pests all over the world (Sasser 1977; Lamberti 1979). The most economically damaging are the Mediterranean and tropical species, Meloidogyne arenaria, Meloidogyne incognita and Meloidogyne javanica, that are highly polyphagous and reproduce through parthenogenesis (Triantaphyllou 1985) on hundreds of cultivated and wild plant species (de Guiran and Netscher 1970). Genetic resistance of plants has been used to control main RKN species (Minz and Cohn 1962; Kochba and Spiegel-Roy 1975 Kester and Grasselly 1987; Layne 1987; Nyczepir 1991). However, the efficiency of RKN resistance in rootstocks depends on the source of resistance (Scotto La Massese et al. 1984; Esmenjaud et al. 1997). In the subgenus Amygdalus, three types of plant response have been identified. Most of the rootstock material is susceptible to RKN. The peach Shalil and its peach-almond hybrid GF.557 are resistant to M. arenaria and M. incognita but susceptible to M. javanica (Esmenjaud et al. 1994) and to a RKN population from Florida [considered as belonging to a new species and designated as Meloidogyne sp. Florida (Esmenjaud et al. 1997)]. The peach Nemaguard and related material, such as the peach Nemared and the hybrids Garfi almond × Nemared (termed G×N), are also resistant to most M. javanica populations (Ramming and Tanner 1983) but not to Meloidogyne sp. Florida.
Within the Prunophora subgenus, plums are the most taxonomically diverse and are adapted to a broad range of climatic and edaphic conditions (Ramming and Cociu 1991; Salesses et al. 1993). Some are used for their fruits (e.g. the Japanese plum, Prunus salicina, or the domestic plums, Prunus domestica and Prunus insititia), the large majority being used as rootstocks for other Prunus species. Among them, the Myrobalan plum (Prunus cerasifera), an outbreeding diploid species, has been recently introduced into selection schemes since some of its clonal selections exhibit beneficial agronomic features (Salesses et al. 1993, 1994) or express resistance to root-knot nematodes (RKN). Clones P.2175, P.1079 and P.2980 of the Myrobalan plum proved to be resistant to the population Meloidogyne sp. Florida (Esmenjaud et al. 1997; Lecouls et al. 1997; Rubio-Cabetas et al. 1999). All three clones carry one dominant allele of a single resistance gene, designated Ma1, Ma2 and Ma3, respectively (Esmenjaud et al. 1996b; Rubio-Cabetas et al. 1998). Each of these Ma resistance alleles confers a high and wide-spectrum resistance to M. arenaria, M. incognita, M. javanica and M. sp. Florida (Lecouls et al. 1997; Rubio-Cabetas et al. 1999) and to the minor species Meloidogyne mayaguensis (Fargette et al. 1996; Rubio-Cabetas et al. 1999). This resistance was not overcome by any of the over-30 RKN species and isolates tested (Esmenjaud et al. 1994, 1997; Fernandez et al. 1994), and was not modified under conditions known as affecting plant defences to RKN such as high temperature and high inoculum pressure (Esmenjaud et al. 1996a). Thus Myrobalan plum appears particularly useful for RKN-resistant rootstock breeding because of the high-level and wide-spectrum RKN resistance of certain accessions. Within perennials, where the genetics of RKN resistance is poorly documented, the Ma gene from Myrobalan plum represent the first genetic system fully characterized. This is also the only system extensively investigated in the Prunus genus for resistance to a plant pest (Lecouls et al. 1997; Lecouls 2000).
Molecular studies have been conducted in order to develop marker-assisted selection (MAS) for Ma. Two reliable SCAR (Sequence Characterized Amplified Region) markers, SCAL19690 and SCAFLP2202, were shown to be linked in coupling to the dominant resistance alleles Ma1 and Ma3 (Lecouls et al. 1999; Bergougnoux et al. 2002). They have been identified by bulked segregant analysis (BSA) (Michelmore et al. 1991) using intraspecific progenies involving P.2175 (Ma1 ma) and several susceptible parents (ma ma). SCAL19 is located less than 1 cM from Ma and SCAFLP2 is co-segregating with Ma, as shown by the analysis of 340 individuals belonging to diverse intra- and inter-specific progenies (M. Claverie, unpublished).
Peach RKN resistance has been first studied in the Nemared rootstock. Markers have been obtained using different F2 progenies such as the intraspecific cross between Lovell and Nemared (Lu et al. 2000) or the almond-peach cross Garfi × Nemared (Jauregui 1998). Recently additional data have been obtained from the Japanese RKN resistant peach Juseitou (Yamamoto and Hayashi 2002).
Here we report results on the precise location of the Myrobalan plum Ma gene in comparison with the putative location of another RKN gene from the Japanese plum on the reference Prunus map. We also give the location of genes for RKN resistance in the two peach sources Nemared and Shalil, in comparison to other available information on peach RKN genes. These data are discussed in the perspective of pyramiding strategies based on marker-assisted selection (MAS) for RKN resistance in Prunus rootstocks.
Materials and methods
Characteristics of plant material and progenies for resistance to RKN species
Various Prunophora and Amygdalus parents (Table 1) showing different RKN spectra for resistance (Esmenjaud et al. 1994, 1997; Lecouls et al. 1997) were used to produce intra- and inter-specific progenies (Table 2). These progenies segregate for several RKN genes carried either by Prunophora or by Amygdalus parents.
Material segregating for Prunophora RKN genes
This material includes intraspecific progenies from Myrobalan or Japanese plums. Myrobalan progenies are crosses of the same resistant clone P.2175 (carrying the heterozygous dominant Ma gene) with each of the three susceptible parents P.2646, P.16.5 and P.2032 (homozygous recessive for Ma) (Esmenjaud et al. 1996b; Lecouls et al. 1997). The segregating progeny of the Japanese plum is a cross between the resistant accession J.222 (resistant to M. arenaria, M. incognita, M. javanica and Meloidogyne sp. Florida) and the susceptible accession J.13. The dominant gene evidenced in this cross has been named R jap and the corresponding resistant and susceptible parental genotypes proposed are (R jap r jap ) and (r jap r jap ), respectively (Lecouls 2000).
Material segregating for Amygdalus RKN genes
The segregating progenies are interspecific crosses between a Myrobalan plum accession and almond-peach hybrids. Two peach resistance sources, Shalil and Nemared, were considered. The Shalil peach was used through its almond-peach hybrid GF.557 that expresses the same RKN resistance to M. arenaria and M. incognita as Shalil. GF.557 is heterozygous for resistance, and segregation was obtained by crossing it (as a male parent) with the susceptible Myrobalan plum P.2032. The dominant gene for resistance to both M. incognita and M. arenaria in GF.557 is designated R Mia557 (= ‘resistance to M. incognita and M. arenaria from GF.557’). The peach Nemared (N) was used through its almond-peach hybrids with the RKN susceptible almond Garfi (G). Accession (G×N)22 (= ‘Felinem’), heterozygous resistant to M. incognita and M. arenaria, was crossed as a male parent with the Myrobalan plum accession P.2175. This Myrobalan × almond-peach progeny was firstly evaluated for its resistance to Meloidogyne sp. Florida, which is not controlled by the Amygdalus resistance sources and thus allows the separation of the resistant individuals carrying the Ma1 resistance allele from P.2175 and the susceptible individuals lacking it. These susceptible individuals (homozygous recessive for Ma) were then evaluated separately for their resistance to each of the M. incognita and M. arenaria species. The same segregation was observed whatever the RKN species, and thus a single dominant gene for resistance to both nematodes, designated R MiaNem (= ‘resistance to M. incognita and M. arenaria from Nemared’), was proposed.
Nematode isolates and RKN resistance evaluation
One isolate representative of each predominant species M. arenaria, M. incognita and M. javanica completed with the isolate Meloidogyne sp. Florida was used (Lecouls et al. 1997). RKN resistance evaluations were performed according to the procedure described by Esmenjaud et al. (1992). All the RKN isolates were maintained on tomato (Lycopersicon esculentum Mill.) cv St Pierre and their identity, at the species level, was verified before inoculation via their isoesterase phenotype (Janati et al. 1982).
DNA extraction and PCR experiments
Genomic DNA of Prunus material was extracted from frozen leaves according to the procedure of Saghai-Maroof et al. (1984) with some modifications. DNA concentrations and quality were evaluated by electrophoresis. For SSR markers, amplifications were performed in a 15-µl final volume containing 40–60 ng of genomic DNA, 0.7 U of Taq polymerase (Life Technologies), 0.2 µM of each primer, 200 µM of each dNTP (Promega Corp., Madison, Wis.), 1.5 mM of MgCl2 and 1× reaction buffer provided with the enzyme. For each SSR, 0.3 pmol of the forward primer was γ33P-ATP end-labeled with polynucleotide kinase (Invitrogen, Cergy-Pontoise, France). PCR conditions were as follows: 94°C for 4 min, then 35 cycles of [94°C for 45 s, annealing temperature provided by the authors (see Tables 3 and 4) for 45 s, 72°C for 45 s], and finally 72°C for 4 min. The labeled PCR products were separated on a 5% denaturing polyacrylamide gel containing 7.5 M urea, in 0.5× TBE running buffer then dried and autoradiographed on X-ray films. For the SCAR or STS (sequence tagged site) markers, PCR amplifications were performed as described by Lecouls et al. (1999) for SCAL19 and SCAN12, Lu et al. (1999) for EAA/MCAT10 STS, and Yamamoto and Hayashi (2002) for OPAP4, OPS14a and OPA11. To recover polymorphism for the SCAL19 marker in the resistant Japanese plum J.222, the multiplex amplification procedure reported by Lecouls (2000) using two forward and one reverse primer was performed. Sequences of those primers are: SCAL19JF1 (5′-TTAGGTGCAGGAATACCA-3′), SCAL19JF2 (5′-CAAATTGATCACCAATGATAC-3′) and SCAL19–2 (5′-CATTGGAGAAGATTGGCCC-3′), respectively.
Elaboration of the resistant and susceptible bulks and evaluation of polymorphism for RFLP and SSR markers in segregating progenies
Segregating crosses, RKN species considered, and the number of individuals used in the different progenies are reported in Table 2. Resistant (R) and susceptible (S) bulks were constituted by 12–15 individuals. Two couples of R and S bulks were constructed for Ma with the two intraspecific progenies P.2175×P.2646 and P.2175×P.16.5. One R and one S bulk was constructed from each of the peach segregating crosses (genes R Mia557 and R MiaNem ). In Japanese plum, no bulks were elaborated from the small-sized cross J.13×J.222 (26 individuals).
Localisation of the Ma gene was initiated using restriction fragment length polymorphism (RFLP) markers from the reference almond-peach map T×E (Joobeur et al. 1998). A set of 46 probes covering the entire genome and separated approximately by a mean distance of 20 cM were chosen. DNAs were digested with EcoRI, HindIII and HpaII, and hybridized with the RFLP probes. The putative location was then more precisely defined by the same BSA approach using SSR markers. Those SSR markers obtained from various teams (Table 3) have been recently placed on the T×E reference map by Aranzana et al. (2002b). The localisation of R Mia557 and R MiaNem was carried out using an equivalent BSA strategy, only based on SSR markers. As preliminary results obtained in Nemared by Jauregui (1998) and Lu et al. (1999) suggest a location of resistance factors in Nemared on linkage group (LG) 2, all ten available SSRs from LG2 (Table 4; Aranzana et al. 2002b) were tested in both segregating progenies for polymorphism or differences in amplification signal intensity between alleles in resistant and susceptible bulks. All parents and grandparents were also deposited in the gels to confirm the origin of the alleles linked to the R genes.
Linkage analysis
The MAPMAKER software version 3.0 (Lander et al. 1987) was used with a minimum LOD score of 3.0 to construct the local maps around Ma and R MiaNem . Linkage analyses were performed using the Kosambi mapping function (Kosambi 1944) to convert recombination units into genetic distances. The Myrobalan plum local map around the Ma gene was established from SSR and SCAR markers using progenies of the three intraspecific crosses totalizing 288 individuals (Table 2). Concerning the gene R MiaNem , as the BSA strategy confirmed its location on LG2, all the individuals were genotyped to construct the map of this linkage group for the parent G×N. These individuals were also evaluated for resistance to Meloidogyne sp. Florida and, among them, the susceptible individuals were tested for segregation of R MiaNem to both M. arenaria and M. incognita. These latter segregation data were analysed to localise the gene R MiaNem on the aforementioned map of LG2 in G×N. The STS markers OPAP4, OPS14a and OPA11, linked to the Mia gene and obtained by Yamamoto and Hayashi (2002), and the STS EAA/MCAT linked to the Mij gene and obtained by Lu et al. (1999), were also mapped. For the genes R Mia557 and R jap , the number of individuals available in the progenies was limited (Table 2). Nevertheless, an indicative map of the linkage group carrying the gene R Mia557 was constructed from recombination frequencies. In the same way, the local map around R jap was compared with the corresponding T×E reference map (Joobeur et al. 1998; Aranzana et al. 2002b) and with the local map of the Ma gene.
Results
Location of RKN genes in the Prunophora subgenus (Myrobalan and Japanese plums)
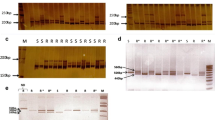
In the Myrobalan plum, only three RFLPs among the 46 probes distributed all over the Prunus genome, revealed polymorphic fragments between the resistant and the susceptible bulks, digested with EcoRI, HindIII and HpaII. These were AG104 (with both EcoRI and HpaII) (Fig. 1), AG63 (with EcoRI) and TSAIII (with HpaII). All three RFLP markers lie on the LG7 of the reference map (Joobeur et al. 1998) and cover 32 cM. This preliminary position of Ma on LG7 was confirmed by the detection of an SSR polymorphism, or the difference in amplification signal intensity between bulks for three SSR markers located on this group, pchgms6, UDP98–405 and CPPCT033 (Table 3). Figure 2 shows an example of the polymorphim observed for pchgms6. Genotyping the individuals of the couples of bulks completed by all other individuals previously characterized for Ma (Table 2), allowed us to locate these markers on the same side of the gene at 2.3, 9.5 and 21.3 cM, respectively. These SSR markers are located on the other side of the gene relative to the SCAR markers SCAL19 and SCAN12 (Bergougnoux et al. 2002) (Fig. 3).
RFLP patterns obtained for the bulks of Ma1-resistant and -susceptible individuals hybridized with the probe AG104. 1R, 1S, 2R and 2S, correspond to resistant (R) and susceptible (S) bulks for the segregating crosses P.2175×P.2646 (1) and P.2175×P.16.5 (2). The arrows indicate the location of the polymorphic bands
Amplification pattern of the SSR pchgms6 in intraspecific Myrobalan progenies segregating for the Ma gene. P.2175 is the resistant (R) parent; P.2646 and P.16.5 are the susceptible (S) parent. 1R, 1S, 2R and 2S correspond to resistant (R) and susceptible (S) bulks for the segregating crosses P.2175×P.2646 (1) and P.2175×P.16.5 (2). The arrow indicates the location of the resistant allele in P.2175. For this allele, the less intense band recovered in lane 1S is due to one recombinant individual in this bulk
Local maps pf SSR (in italics) and SCAR (normal letters) markers linked to the Ma gene in the Myrobalan plum P.2175 (b) and to the R jap gene in the Japanese plum J.222 (c) in comparison with SSR markers located on the LG7 of the almond × peach reference Prunus map Texas × Earlygold (T×E) (a) (Aranzana et al. 2002b). For the Ma gene, distance are expressed in cM using the Kosambi distance given by the MAPMARKER software version 3.0 (Lander et al. 1987) with a minimum LOD score of 3.0. For the R jap gene, distance are expressed in recombination percentages
In the Japanese plum, the SCAR markers linked to Ma and all the SSRs available for this LG7 region were evaluated for their polymorphism in the J.222 and J.13 parents. Polymorphic markers (the SSRs pchgms6, CPPCT033 and CPPCT022), and in particular the new multiplex marker derived from SCAL19, were then directly evaluated on the 26 individuals of the segregating progeny J.13×J.222. On this small-sized cross, the markers pchgms6, CPPCT022 and SCAL19 co-segregated with the R jap gene (Fig. 3), which shows that this gene lies on the LG7 probably in the same position as Ma.
Location of genes in the Amygdalus subgenus (Shalil and Nemared peaches)
Eight SSRs from LG2 in the reference T×E Prunus map (Aranzana et al. 2002b) expressed a clear polymorphism simultaneously in both couples of bulks. In order to localise more precisely R MiaNem , each of the individuals of the two couples of bulks was genotyped together while the other RKN characterized individuals of the progeny. Figure 4 shows an example of the polymorphism of the alleles for the SSR UDP98-025 in segregating individuals from the cross P.2175×(G×N). Data from this cross were integrated in the map of LG2, and the respective positions of the markers were compared in that map and in the T×E map. R MiaNem and R Mia557 are placed on the LG2 in an a priori equivalent subtelomeric position (Fig. 5). In the Japanese peach source Juseitou, Yamamoto and Hayashi (2002) have obtained five STS markers for resistance to M. incognita (gene Mia) and M. javanica (gene Mja), both genes being approximately 3.5-cM apart. Three of these STSs were polymorphic in at least one of our segregating progenies (Fig. 5). The STSs OPA11 and OPS14a, located on one side of Mia, were also located on the same side and in the same order as for R Mia557 and R MiaNem . The STS OPAP4 located on the other side of Mia in Juseitou was also located on the other side of R Mia557 .
Amplification pattern of the SSR UDP98-025 in the interspecific progeny P.2175×(G×N) segregating for the R MiaNem gene. (G×N)22 and Nemared (N) are the resistant (R) parent and grandparent, respectively; Garfi (G) is the susceptible (S) grandparent; P.2175 and Myro2 are Myrobalan controls. The arrows indicate the location of the resistant (low) and susceptible (high) marker alleles in (G×N)22. (+) and (−) indicate the presence of these alleles in coupling with resistance (+) or susceptibility (−) in the individuals of the progeny
Maps of SSR markers (in italics) linked to the genes R MiaNem and R Mia557 for resistance to M. incognita in Nemared (b) and Shalil (c) peaches, respectively, in comparison with markers located on the LG2 of the almond × peach reference Prunus map Texas × Earlygold (T×E) (a) (Aranzana et al. 2002b). For the R MiaNem gene, distance are expressed in cM using the Kosambi distance given by the MAPMARKER software version 3.0 (Lander et al. 1987) with a minimum LOD score of 3.0. For the R Mia557 gene, distances are expressed in recombination percentages. The SCAR markers OPAO4, OPS14a and OPA11, linked to the Mia gene in Juseitou (Yamamoto and Hayashi 2002), and the marker EAA/MCAT10 STS, linked to the Mij gene in Nemared (Lu et al. 1999), are indicated in normal letters
Discussion
Our data illustrate the respective positions of two plum and two peach loci involved in RKN resistance in Prunus species. Thanks to a RFLP approach (for plum) completed with a SSR approach (for plum and peach), plum genes were localised on LG7 and peach genes were shown to reside on LG2. Ma and R jap mapped very close to the SSR marker pchgms6. The location of these genes in a cluster of SSR markers on LG7 makes the localisation of homeologous regions in other Prunus crosses easier, and illustrates the good cross-species transportability of these markers within Prunus species. Nevertheless the amplification of the Myrobalan alleles of pchgms6 in the three-way hybrid crosses [P.2175×(G×N)] was difficult to obtain, presumably because of competition between Prunophora and Amygdalus alleles. A new primer combination, more specific to the Myrobalan alleles, could be defined to solve this problem (M. Claverie, unpublished).
Ma is the first evidence of the precise localisation of a resistance gene in the Prunus genus. The location of R jap , based on a 26-individual cross, is less fine but seems to be the same as Ma: R jap co-segregates with the two markers flanking Ma in a 2.7-cM interval and with the tightly linked SSR marker (in the T×E map) CPPCT022. Additional segregating individuals should provide recombination events that would precisely define its location. It is likely that the location of Ma and R jap is conserved in cultivated and wild plum species, including diploid to hexaploid species. Locations of R MiaNem and R Mia557 suggest that both genes might be the same: the order of the SSR markers is conserved and, whatever the SSR, the coupling-phase alleles for resistance from Nemared are identical to the coupling-phase alleles for resistance from the Shalil parent (Fig. 5b and c). This could be explained by the limited genetic variability of peach and the almond-peach nature of both segregating parents. Nevertheless, the precise location of peach genes on LG2 in a subtelomeric position appears different from that obtained by Jauregui (1998) who placed one gene or a major QTL in a more centromeric position, in the vicinity of the SSR marker pchgms1. Moreover, our location of R MiaNem was also different from that previously obtained by Lu et al. (1999) in the intraspecific peach cross Lovell × Nemared. These authors have found by a BSA approach that the Mij locus for resistance to M. incognita and M. javanica was located at about 3 cM from the EAA/MCAT10 STS marker (derived from an AFLP marker). We could clearly map this STS marker in a central position on LG2 (Fig. 3b) at more than 45 cM from R MiaNem . Since an equivalent location is observed for R Mia557 , our data based on two independent crosses appear quite reliable and one can be confident for the map position of both peach genes. The differences between Nemared gene locations in different studies could be explained by the presence of a major QTL for M. javanica and M. incognita resistance, acting as a complete-resistance gene depending on either the nematode isolate or the inoculation procedure, or resistant versus susceptible definition or the genetic background. Taken together, these results completed by those of Lu et al. (2000) support the hypothesis of two genes lying on the same linkage group but at an approximate distance of 45 cM.
Since the origin of Nemaguard, the resistant ancestor of Nemared, remains unclear, co-location of R MiaNem and R Mia557 could be explained by a relatively close parentage strongly suggested by the identity of SSRs alleles in coupling with resistance in both Nemared and GF557. It is highly probable that both genes also co-localize with the gene Mia in the peach Juseitou (Yamamoto and Hayashi 2002) since STS markers flanking Mia also flank R MiaNem and R Mia557 in the same order. Concordant results from the three different sources, Shalil, Nemared and Juseitou, suggest that peach RKN sources share at least one major gene (or gene cluster) of resistance to M. incognita located in this subtelomeric position.
In Prunophora, differences in allelism and polymorphism of the genetic markers linked to resistance, associated with co-location of the Ma and R jap genes in Myrobalan and Japanese plums, suggest the conservation of a resistance locus acquired before species separation. This last result, the usual transportability of SSR markers between Prunus species, together with the conservation of locus order and genetic distances around Ma, suggest an even higher level of synteny between Prunus species than previously observed (Joobeur et al. 1998).
Our most beneficial and applied result is that Ma, on the one hand, and the gene(s) specifically controlling M. incognita and M. arenaria in both Nemared and GF.557, on the other hand, are independent, and can be pyramided into new interspecific hybrid rootstock material. Introgression of Ma and peach genes into the genome of new Prunus rootstocks by interspecific hybridisation (e.g. Myrobalan plum × Amygdalus) has been undertaken. These hybrids can cumulate favorable agronomic traits from both origins, together with the complete-spectrum resistance controlled by the Myrobalan Ma gene and the more restricted-spectrum of Amygdalus genes. Indeed, the pyramiding of several genes in the same genotype may limit the risk of resistance breaking (Johnson 1983; Cook and Evans 1987; Roberts 1995), and thus extend the useful life of new rootstocks. For that purpose MAS for Ma is now greatly improved by the availability of the two flanking markers SCAL19 and pchgms6 in a 2.7 cM genetic interval.
References
Aranzana MJ, Garcia-Mas J, Carbo J, Arús P (2002a) Development and variability analysis of microsatellite markers in peach. Plant Breed 121:87–92
Aranzana MJ, Pineda A, Cosson P, Dirlewanger E, Ascasibar J, Cipriani G, Ryder CD, Testolin R, Abbott A, King GJ, Iezzoni AF, Arús P (2002b) A set of simple-sequence repeat (SSR) markers covering the Prunus genome. Theor Appl Genet 106:819–825
Bergougnoux V, Claverie M., Bosselut N, Lecouls AC , Salesses G, Dirlewanger E, Esmenjaud D (2002) Marker-assisted selection of the Ma gene from Myrobalan plum for a complete-spectrum root-knot nematode (RKN) resistance in Prunus rootstocks. Acta Hort 592:223–228
Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R (1999) AC/GT and AG/CT microsatellite repeats in peach [Prunus persica (L.) Batsch]: isolation, characterization and cross-species amplification in Prunus. Theor Appl Genet 99:65–72
Cook R, Evans K (1987) Resistance and tolerance. In: Brown RH, Kerry BR (eds) Principles and practice of nematode control in crops. Academic Press, New York, pp 179–231
Dirlewanger E, Cosson P, Tavaud M, Aranzana MJ, Poizat C, Zanetto A, Arús P, Laigret F (2002) Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor Appl Genet 105:127–138
Downey SL, Iezzoni AF (2000) Polymorphic DNA markers in black cherry (Prunus serotina) are identified using sequences from sweet cherry, peach and sour cherry. J Am Soc Hort Sci 125:76–80
Esmenjaud D, Scotto La Massese C, Salesses G. Minot JC, Voisin R (1992) Method and criteria to evaluate resistance to Meloidogyne arenaria in Prunus cerasifera Ehr. Fundam Appl Nematol 15:385–389
Esmenjaud D, Minot JC, Voisin R, Pinochet J, Salesses G (1994) Inter- and intra-specific resistance variability in Myrobalan plum, peach and peach-almond rootstocks using 22 root-knot nematode populations. J Am Soc Hort Sci 119:94–100
Esmenjaud D, Minot JC, Voisin R (1996a) Effect of durable inoculum pressure and high temperature on root-galling, nematode numbers and survival of Myrobalan plum genotypes (Prunus cerasifera) highly resistant to Meloidogyne spp. Fundam Appl Nematol 19:85–90
Esmenjaud D, Minot JC, Voisin R, Bonnet A, Salesses G (1996b) Inheritance of resistance to the root-knot nematode Meloidogyne arenaria in Myrobalan plum. Theor Appl Genet 92:873–879
Esmenjaud D, Minot JC, Voisin R, Pinochet J, Simard MH, Salesses G (1997) Differential response to root-knot nematodes in Prunus species and correlative genetic implications. J Nematol 29:370–380
Fargette M, Phillips MS, Block VC, Waugh R, Trudgill DL (1996) An RFLP study of relationships between species, populations, and resistance breaking lines of tropical Meloidogyne. Fundam Appl Nematol 19:193–200
Fernandez C, Pinochet J, Esmenjaud D, Salesses G, Felipe A (1994) Resistance among new Prunus rootstocks and selections to the root-knot nematodes in Spain and France. Hortscience 29:1064–1067
Guiran (de) G, Netscher R (1970) Les nématodes du genre Meloidogyne, parasites des cultures tropicales. Cahiers ORSTOM, série Biologie 11:151–185
Janati A, Bergé JB, Triantaphyllou AC, Dalmasso A (1982) Nouvelles données sur l’utilisation des isoestérases pour l’identification des Meloidogyne. Rev Nématol 5:147–154
Jauregui B (1998) Localizacion de marcadores moleculares ligados a caracteres agronomicos en un cruzamiento interespecifico almendro × melocotonero. PhD thesis, University of Barcelona, Spain
Johnson R (1983) Genetic background of durable resistance. In: Lamberti F, Waller JM, Van der Graaff NA (eds) Durable resistance in crops. Plenum, New York, pp 5–26
Joobeur T, Viruel MA, De Vicente MC, Jauregui B, Ballester J, Dettori MT, Verde I, Troco MJ, Messeguer R, Battle I, Quarta R, Dirlewanger E, Arus P (1998) Construction of a saturated linkage map for Prunus using an almond × peach F2 progeny. Theor Appl Genet 97:1034–1041
Kester ED, Grassely C (1987) Almond rootstocks. In: Rom RC, Carlson RF (eds) Rootstocks for fruit crops. John Wiley and sons, New-York, pp 265–293
Kochba J, Spiegel-Roy P (1975) Inheritance to the root-knot nematode (Meloidogyne javanica Chitwood) in bitter almond progenies. Euphytica 24:453–457
Kosambi D (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lamberti F (1979) Economic importance of Meloidogyne spp. in subtropical and Mediterranean climates. In: Lamberti F, Taylor CE (eds) Root-knot nematodes (Meloidogyne spp.): systematic, biology and control. Academic Press, New York, pp 342–357
Lander E, Green P, Abrahamson J, Barlow A, Daley M, Lincoln S, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Layne REC (1987) Peach rootstocks. In: Rom RC, Carlson RF (eds) Rootstocks for fruit crops. John Willey and sons, New-York, pp 185–216
Lecouls AC (2000) Spectre d’activité et marquage moléculaire du gène Ma1 contrôlant la résistance aux nématodes Meloidogyne chez le prunier myrobolan. PhD Thesis, University of Aix-Marseille II, France
Lecouls AC, Salesses G, Minot JC, Voisin R, Bonnet A, Esmenjaud D (1997) Spectrum of the Ma genes for resistance to Meloidogyne spp. in Myrobalan plum. Theor Appl Genet 85:1325–2334
Lecouls AC, Rubio-Cabetas MJ, Minot JC, Voisin R, Bonnet A, Salesses G, Dirlewanger E, Esmenjaud D (1999) RAPD and SCAR markers linked to the Ma1 root-knot nematode resistance gene in Myrobalan plum (Prunus cerasifera Ehr.). Theor Appl Genet 99:328–336
Lu ZX, Sossey-Alaoui K, Reighard GL, Baird WV, Abbott AG (1999) Development and characterization of a co-dominant marker linked to root-knot nematode resistance, and its application to peach rootstocks breeding. Theor Appl Genet 99:115–123
Lu ZX, Reighard GL, Nyczepir AP, Beckman TG, Ramming DW (2000) Inheritance of resistance to root-knot nematodes in Prunus rootstocks. HortScience 35:1344–1346
Michelmore RW, Paran I, Kesseli V (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Minz G, Cohn E (1962) Susceptibility of peach rootstocks to root-knot nematodes. Plant Dis Rep 46:531–534
Nyczepir AP (1991) Nematode management strategies in stone fruits in the United States. J Nematol 23:334–341
Ramming DW, Tanner O (1983) Nemared peach rootstock. HortScience 18:376
Ramming DW, Cociu V (1991) Plum (Prunus). In: Moore JV, Ballington JR (eds) Genetic resources of temperate fruit and nut crops. Acta Hort 290:239–288
Rehder A (1954) Manual of cultivated trees and shrubs, 2nd edn. Dioscorides Press, Portland
Roberts PA (1995) Conceptual and practical aspects of variability in root-knot nematodes related to host plant resistance. Annu Rev Phytopathol 33:199–221
Rubio-Cabetas MJ, Lecouls AC, Salesses G, Bonnet A, Minot JC, Voisin R, Esmenjaud D (1998) Evidence of a new gene for high resistance to Meloidogyne spp. in Myrobalan plum (Prunus cerasifera). Plant Breed 117:567–571
Rubio-Cabetas MJ, Minot JC, Voisin R, Esmenjaud D, Salesses G, Bonnet A (1999) Response of the Ma genes from Myrobalan plum to Meloidogyne hapla and M. mayaguensis. HortScience 34:1266–1268
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location and population dynamics. Proc Natl Acad Sci USA 88:8014–8018
Salesses G, Grasselly C, Renaud R, Claverie J (1993) Les porte-greffe des espèces fruitières à noyau du genre Prunus. In: Gallais A, Bannerot H (eds) Amélioration des espèces cultivées. INRA, Paris, pp 605–619
Salesses G, Grasselly C, Bernhard R (1994) Utilisation des espèces indigènes et exotiques pour l’amélioration des Prunus cultivés, variétés et porte-greffe. C R Acad Agric France 80:77–88
Sasser JN (1977) Worldwide dissemination and importance of the root-knot nematodes Meloidogyne spp. J Nematol 22:585–589
Scotto La Massese C, Grasselly C, Minot JC, Voisin R (1984) Différence de comportement de 23 clones et hybrides de Prunus à l’égard de quatre espèces de Meloidogyne. Rev Nématol 7:265–270
Sosinski B, Gannavarapu M, Hager LD, Beck LE, King GJ, Ryder CD, Rajapakse S, Baird WV, Ballard RE, Abbott AG (2000) Characterization of microsatellite markers in peach [Prunus persica (L.) Batsch]. Theor Appl Genet 97:1034–1041
Testolin R, Marrazzo T, Cipriani G, Quarta R, Verde I, Dettori MT, Pancaldi M, Sansavini S (2000) Microsatellite DNA in peach (Prunus persica L. Batch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome 43:512–520
Triantaphyllou AC (1985) Cytogenetics, cytotaxonomy and phylogeny of root-knot nematodes. In: Sasser JN, Carter CC (eds) An advanced treatise on Meloidogyne, Vol I. North Carolina State University Graphics, Raleigh, pp 113–126
Yamamoto T, Hayashi T (2002) New root-knot nematode resistance genes and their STS markers in peach. Sci Hort 96:81–90
Acknowledgements
This work was partly funded by the Commission of the European Union via the FAIR Programme of Research and Technological Development (Research project no. FAIR6-CT 984139; 1999–2003) and by the Conseil Regional d’Aquitaine (2000–2002). The participation of Anne-Claire Lecouls in this work was supported by a Research Training Grant (no. BTH 00535) from INRA and ‘Region Provence-Alpes-Côte d’Azur’, France (1997–2000). The authors also thank the technical staff of the INRA ‘Domaine des Jarres’ experimental farm for producing the Myrobalan plum intra-and inter-specific material, and of the ‘Domaine de l’Amarine’ experimental farm for providing the Japanese-plum cuttings used for the genetic and marker studies. The authors are grateful to H. Duval who created the segregating cross for the Japanese plum.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H.C. Becker
Rights and permissions
About this article
Cite this article
Claverie, M., Bosselut, N., Lecouls, A.C. et al. Location of independent root-knot nematode resistance genes in plum and peach. Theor Appl Genet 108, 765–773 (2004). https://doi.org/10.1007/s00122-003-1463-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-003-1463-1