Abstract
To observe the diagnostic values of circular RNAs (circRNAs), their expression profiles between gastric cancer patients’ plasma and healthy controls were first assessed by circRNA microarray. Then, circRNA levels were measured by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and RT-droplet digital PCR (RT-ddPCR), respectively. A total of 343 differentially expressed circRNAs were found. The top 10 elevated circRNAs in patients were hsa_circ_0088300, hsa_circ_0075825, hsa_circ_0019172, hsa_circ_0000220, hsa_circ_0035277, hsa_circ_0000301, hsa_circ_0000189, hsa_circ_0090080, hsa_circ_0001888, and hsa_circ_0001874. The top 10 reduced circRNAs were hsa_circ_0004771, hsa_circ_0001190, hsa_circ_0061276, hsa_circ_0092337, hsa_circ_0058495, hsa_circ_0061274, hsa_circ_0075829, hsa_circ_0080845, hsa_circ_0001006, and hsa_circ_0003764. In cancer and dysplasia tissues, hsa_circ_0001017 and hsa_circ_0061276 were downregulated. Their levels were significantly associated with distal metastasis. The area under receiver operating characteristic curve in combinative use was 0.966 with 95.5% sensitivity and 95.7% specificity. Patients with low plasma hsa_circ_0001017 or hsa_circ_0061276 had a much shorter overall survival than those with high levels. Patients whose plasma hsa_circ_0001017 or hsa_circ_0061276 levels recovered to normal after operation had a longer disease-free survival. Finally, the in vitro model indicated gastric cancer cells secreting circRNAs into plasma. In conclusion, RT-ddPCR is a potent non-invasive and absolute quantification method for simultaneous detection of multiple circRNAs. Hsa_circ_0001017 and hsa_circ_0061276 are new potential biomarkers for gastric cancer.
Key message
-
A total of 343 circRNAs are differentially expressed between gastric cancer patients’ plasma and healthy controls.
-
Hsa_circ_0001017 and hsa_circ_0061276 are downregulated in gastric cancer tissues.
-
The RT-ddPCR is a potent method for simultaneous detection of multiple circRNAs in plasma.
-
Hsa_circ_0001017 and hsa_circ_0061276 are potential biomarkers for gastric cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer is the second leading cause of cancer-related deaths worldwide [1, 2]. Though there have been advances in surgical techniques and combined chemotherapy strategies, the 5-year overall survival of this disease in most countries remains less than 30% [3]. The ignored differentially expressed molecular profiles that cause heterogeneity of gastric cancer may be responsible for the difficulty through gastric cancer treatments. As a result, it is necessary to explore the details of molecular characters of gastric cancer.
Meanwhile, considerable efforts have been devoted to the discovery of novel plasma-based biomarkers for the diagnosis of gastric cancer [4,5,6,7]. However, with relatively low sensitivity and specificity, several adaptable gastric cancer-associated plasma biomarkers including carbohydrate antigens 72-4 (CA72-4), CA19-9, and carcinoembryonic antigen (CEA) are hard to replace in the existing endoscopy or x-ray-based examinations [8, 9].
Droplet digital polymerase chain reaction (ddPCR) is a new developed nucleic acid detection method that is based on the dilution of template DNA into independent non-interacting droplets [10]. As the so-called third-generation PCR, ddPCR has single-molecule sensitivity. This novel platform for absolute quantification of DNA (and RNA) holds great promise in clinical diagnostics. It is increasingly popular due to the higher sensitivity and accuracy in quantification than traditional real-time PCR. The ddPCR has been used in the detection of mutations or deletions and copy number variations of cancer-associated genes [11, 12]. According to the detection methods, ddPCR can be divided into two main types: probe- and dye-based methods. The fluorescently quenched oligonucleotide probe-based ddPCR has high specificity but costs too much. By contrast, the application of dyes such as EvaGreen is more accessible [13, 14]. However, both methods performed disability in the multiplex detection of large scale of genes.
Circular RNAs (circRNAs) are recently identified non-coding RNAs that are highly prevalent in eukaryotic transcriptome [15]. CircRNAs are found predominantly located in cytoplasm and can be sorted into exosomes [16]. In consideration of covalently closed loop structure that endows a property of high stability, circRNAs are prized to be novel plasma biomarkers [17]. Importantly, a growing number of studies have demonstrated that circRNAs play a significant role in the pathogenesis of cancers [18, 19]. However, the diagnostic values of peripheral blood circRNAs for gastric cancer remain vague.
In this study, we first used circRNA microarray to detect the differential circRNAs in plasma between patients with gastric cancer and healthy controls. Then we used reverse transcription (RT)-ddPCR to confirm two representative circRNAs, hsa_circ_0001017 and hsa_circ_0061276, in gastric cancer tissues and plasma from patients with gastric cancer. Finally, we analyzed their clinical diagnostic values and possibility as prognosis markers. Our results showed that RT-ddPCR is a potent and costless method of simultaneous detecting two cell-free cancer-associated circRNAs; and the combinative detection of cell-free hsa_circ_0001017 and hsa_circ_0061276 had significant clinical values for gastric cancer diagnosis and its prognosis evaluation.
Materials and methods
Patients and specimens
Gastric cancer tissues and fasting plasma samples were obtained from 121 surgical patients who were from two centers for gastroenterology, the Affiliated Hospital of Ningbo University School of Medicine and Yinzhou People’s Hospital (China) between February 2011 and February 2016. The control fasting plasma from 121 healthy volunteers, age and gender matched to patients, were collected from Ningbo No. 2 Hospital (China). All patients received no medical treatment before endoscopy examination or surgery. The diagnosis was confirmed histopathologically. All specimen collection and pre-processing protocol were following previous description [20] and preserved at − 80 °C until RNA isolation.
Tumors were classified according to the Tumor-Node-Metastasis (TNM) staging system (7th ed.); and histological grade was assessed following the National Comprehensive Cancer Network (NCCN) Clinical Practice Guideline of Oncology (V.1.2012).
This study obtained the approval from the Human Research Ethics Committee of Ningbo University (IRB No. 20100303) and all informed consents were obtained. Double-blind manner was used through the entire process of all clinical samples and data collection.
Cell culture
The gastric cancer cell lines BGC-823, MGC-803, and SGC-7901 and normal human gastric epithelial cell line GES-1 were purchased from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 Medium (Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Life Technologies) in a humidified atmosphere at 37 °C with 5% CO2 [21]. When cultured 12, 24, 36, and 48 h, the medium were collected.
Total RNA extraction and reverse transcription
Total RNA was extracted from 200 μl plasma or medium using TRIzol LS reagent (Ambion, Carlsbad, CA, USA), whereas gastric tissue samples were processed using TRIzol reagent (Ambion). Then, the concentrations of total RNA were measured by the DS-11+ Spectrophotometer (DeNovix, Wilmington, DE, USA). The 1% agarose gel electrophoresis was used to assess the integrity of extracted RNA. Finally, total RNA was reverse transcribed to cDNA by GoScript Reverse Transcription (RT) System (Promega, Madison, WI, USA) following the manufacturer’s instruction.
CircRNA microarray analysis
The circRNA expression profiles in three gastric cancer tissues and their adjacent non-tumorous tissues 5 cm away from the edge of tumor, and plasma of the same three patients and three healthy persons were analyzed using Arraystar Human circRNA Array (Arraystar, Rockville, MD, USA). According to Arraystar’s Super RNA Labeling protocol (Arraystar), total extracted RNA was transcribed into fluorescent cRNA by random primer. Then, the labeled cRNAs were hybridized onto the Arraystar Human circRNA Array (6 × 7 K, Arraystar), and incubated for 17 h at 65 °C in an Agilent Hybridization Oven (Agilent Technologies, Santa Clara, CA, USA). The array was scanned by the Axon GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA, USA) after washing the slides.
Scanned images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. R software package was used for quantitative normalization and subsequent data processing. Differentially expressing circRNAs with statistical significance between two groups were identified through fold change filtering or volcano plot filtering. Hierarchical clustering was performed to show the distinguishable circRNAs expression pattern among samples. CircRNAs with fold changes ≥ 2.0 and p values ≤ 0.05 were identified as the differentially expressed ones with significance.
RT-qPCR and RT-ddPCR
The real-time quantitative polymerase chain reaction (qPCR) was performed using GoTaq qPCR Master Mix (Promega) on an Mx3005P Real-Time PCR System (Stratagene, La Jolla, CA, USA) following the manufacturer’s instructions. The ddPCR was constructed on the QX200 Droplet System using ddPCR EvaGreen Supermix (Bio-Rad, Foster City, CA, USA). The divergent primers overlapping the splice junction of hsa_circ_0001017 and hsa_circ_0061276, and the convergent primers of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were designed by Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and synthesized by Sangon Biotech (Shanghai, China). Their relative concentrations were determined by gradient dilution. The suitable concentration and proportion of primers will make the relative droplets population a fitting position that is efficiently distinguished with each other. In general, lower primer concentration and smaller amplicon size are both able to decrease the location of relative droplet population, vice versa. The sequences and optimized primer concentrations were listed in Supplementary Table S1.
The ddPCR was performed using 2× QX200 EvaGreen ddPCR Supermix (Bio-Rad) with 2 μl cDNA. The droplets generation procedure was according to the manufacturer’s protocol. Then, droplets were transferred into PCR tubes; and PCR was performed on a Mastercycler Gradient (Vaudaux-Eppendorf, Deutschland GmbH, Hamburg, Germany). The cycling protocol was as follows: hot-start at 95 °C for 5 min; followed by 45 cycles of 95 °C for 30 s, 60 °C for 1 min, and 72 °C for 30 s; then, a signal stabilization step at 4 °C for 5 min and 90 °C for 5 min, finally holding at 4 °C. Droplets were counted on the QX200 Droplet Reader (Bio-Rad) and analyzed by QuantaSoft software (Bio-Rad).
The relative quantification of circRNA expression levels in RT-qPCR was calculated by ΔC q method [17, 21]. Lower ΔC q values indicated higher expression levels. The results of RT-ddPCR were showed in two forms: relative expression level and absolute copy number. The former was normalized to GAPDH copy numbers; and the later (copies/μl) was the number exported by the software (Bio-Rad) divided by plasma volume (200 μl).
Cloning and sequencing of RT-ddPCR products
The RT-ddPCR products of circRNAs were purified using the UNIQ-10 PCR Product Purification Kit (Sangon), and then cloned into the pUCm-T vector (Sangon) following the manufacturer’s instructions. DNA sequencing was performed by Sangon Biotech Company, Ltd.
Statistical analysis
Statistical analysis were performed by Statistical Program for Social Sciences (SPSS) 18.0 Software (SPSS, Chicago, IL, USA), R and R-packages (version 3.0.1, R Core Team). K-S nonparametric test and Kruskal-Wallis test were flexibly used according to actual conditions. p < 0.05 was considered to have significant difference.
Results
Plasma circRNA expression profile in patients with gastric cancer
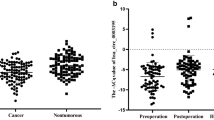
High-throughput human circRNA microarray was conducted to assess the differences of circRNA expression profiles between gastric cancer tissues and their adjacent non-tumorous tissues, and gastric cancer patients’ plasma and healthy controls. A total of 5396 circRNAs were detected (data accessible at NCBI GEO database, accession GSE89143, Guo, 2016, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE89143; accession GSE93541, Guo, 2017, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93541). Differential expressing circRNAs in plasma between cancer and normal control groups were characterized and clustered through fold change filtering and hierarchical clustering, respectively (Fig. 1A, B). In summary, a total of 343 differential expressed circRNAs (twofold change, p < 0.05) were identified. Among them, 171 and 172 circRNAs were upregulated and downregulated, respectively. The top 10 elevated circRNAs in patients were hsa_circ_0088300, hsa_circ_0075825, hsa_circ_0019172, hsa_circ_0000220, hsa_circ_0035277, hsa_circ_0000301, hsa_circ_0000189, hsa_circ_0090080, hsa_circ_0001888, and hsa_circ_0001874. The top 10 reduced ones were hsa_circ_0004771, hsa_circ_0001190, hsa_circ_0061276, hsa_circ_0092337, hsa_circ_0058495, hsa_circ_0061274, hsa_circ_0075829, hsa_circ_0080845, hsa_circ_0001006, and hsa_circ_0003764 (Supplementary Table S2).
Plasma circRNA expression profile in plasma of gastric cancer patients comparing with healthy controls. A Scatter plots are used to evaluate the difference of circRNA expression between plasma from gastric cancer patients and healthy controls. B Hierarchical cluster analysis of all circRNAs that expressed in plasma from three gastric cancer patients. C Overlapped circRNAs between four clusters of differentially expressed circRNAs in tissue and plasma from patients with gastric cancer. Total 17 circRNAs, 3 upregulated (a) and 14 downregulated (b), coordinately expressed between tissue and plasma from patients with gastric cancer
Since blood is the most commonly used sample in laboratory medicine, we performed intersection analysis based on the differential expression patterns of circRNAs between tissues and plasma, i.e., GSE89143 vs. GSE93541. The results showed that 3 and 14 circRNAs were elevated and reduced in patients’ plasma, respectively (Fig. 1C; Table 1).
Accurate quantification of circRNAs in tissues and plasma
The ddPCR is an endpoint PCR, avoiding influences by inhibitors in plasma. In the application of EvaGreen for ddPCR, the distinguishable populations of droplets can be differentiated themselves with different final fluorescent signal intensities (Fig. 2A). As a result, their copy numbers can be determined separately.
The relative and absolute quantification of circRNAs. A Detection of plasma circRNA levels by RT-ddPCR with EvaGreen dye. The lowest population (a) of droplets corresponding with the lowest fluorescent amplitude is consisted by the unamplified background template RNA (gray). The second population (b) represents the droplets containing only GAPDH mRNA (65 bp). The third population (c) represents droplets with only hsa_circ_0061276 (46 bp). The fourth population (d) refers to droplets containing both amplified GAPDH mRNA and hsa_circ_0061276. The population with the highest amplitude (e) represents droplets containing hsa_circ_0001017 (175 bp). B Comparing the accuracy between RT-ddPCR and RT-qPCR. Scatter plots with linear regression for measuring hsa_circ_0061276. Scatter plots illustrates the relationship between cDNA volumes (x-axis) and hsa_circ_0061276 levels (y-axis). Y-axis consists of three parts: relative quantification as determined by RT-qPCR (pink), absolute quantification (blue), and relative quantification (gray) by RT-ddPCR. The samples from plasma (a) and tissue (b) were analyzed independently. All error bars indicate 95% CIs calculated by QuantaSoft (Bio-Rad) according to Poisson distribution on scored droplets; p < 0.05. Three independent experiments were performed; and no significant differences were found. C Analysis of GAPDH mRNA levels between patients with gastric cancer and healthy controls. Box-plots for the GAPDH mRNA levels in plasma from pre-operation patients (pink, n = 121), paired post-operation patients (blue, n = 121) with gastric cancer, and healthy controls plasma (gray, n = 121). ***p < 0.001. There is no significant difference in plasma between pre-operation patients and healthy controls
The relative and absolute quantification of DNA by PCR (or RNA by RT-PCR) are two commonly used methods for the detection of nuclear acid-based biomarkers. In recent years, accumulated studies have shown several priorities of ddPCR in detecting free DNA/RNA in multiple body fluids [11, 22]. However, the suitable quantitative approach for detecting circRNAs remains unclear. By optimizing amplicon size and primers’ concentrations (Supplementary Table S1), two circRNAs and GAPDH mRNA could be amplified simultaneously (Fig. 2A). The agarose gel electrophoresis also showed the specificity of amplification (Supplementary Fig. S1). Next, we first explored the accuracy of relative quantification and absolute quantification in our assay by analyzing the expression levels of hsa_circ_0061276, which is the top three lowest circRNAs in plasma of patients group comparing with control group (Supplementary Table S2) and one of the differential circRNAs in intersection analysis (Table 1). As shown in Fig. 2B, among three methods, relative quantification by RT-ddPCR, absolute quantification by RT-ddPCR, and relative quantification by RT-qPCR, the most accurate quantitative method was absolute quantification by RT-ddPCR for both plasma and tissues circRNAs (R 2 = 0.998 and R 2 = 0.972, respectively). For a proper normalization and calculation of relative expression of target genes, it is essential to use a reference gene, which should be stably expressed. Inspired by the instability of relative quantification of cell-free circRNA by RT-ddPCR in plasma (Fig. 2B), we compared the copy numbers of GAPDH mRNA in plasma of gastric cancer patients before operation, post-operation, and healthy controls. As shown in Fig. 2C, its levels were significantly differentiated among three groups. Obviously, GAPDH mRNA may not be a suitable reference gene for the relative detection of RNAs in plasma, at least in our studied patients.
Expression patterns of hsa_circ_0001017 and hsa_circ_0061276 in gastric cancer tissues and paired plasma
It is interesting that fold change of plasma circRNAs between patients and controls in reduced group was bigger than that in elevated one (Supplementary Table S2), and that the number of plasma circRNAs in reduced group was more than that in elevated one (Table 1). So, two reduced circRNAs, hsa_circ_0001017 (30.85-folds changed, ranked middle) and hsa_circ_0061276 (121.54-folds changed, ranked third), were chosen to observe the non-invasion diagnostic values of circRNAs for gastric cancer. To promote the reliability of the verification, we expanded the sample size of gastric cancer patients’ plasma and used their corresponding surgical tissues as comparison.
We found that hsa_circ_0001017 expression levels in gastric cancer tissues were significantly decreased (Fig. 3A). What’s more, its expression levels in gastric dysplasia tissues were even lower than those in gastric cancer tissues. Similar expressing pattern of hsa_circ_0061276 was found (Fig. 3A). The copy numbers of these two circRNAs in plasma from pre-operation patients were reduced, while significantly elevated after 10 days of operation (not received any chemical and biological therapies), to the flat level of healthy controls (Fig. 3B). In details, patients with reduced hsa_circ_0001017 levels (78.5%) were more than those with hsa_circ_0061276 (70.2%) (Fig. 3C). Almost 60% patients’ hsa_circ_0001017 and hsa_circ_0061276 levels diminished in tissues and plasma at the same time. The hsa_circ_0001017 levels in cancer tissues were found associated with age, tumor size, invasion, TNM stages, distal metastasis, and CEA levels (Supplementary Table S3). The hsa_circ_0001017 levels in plasma were found associated with gender, tumor size, differentiation, and distal metastasis (Supplementary Table S3). The hsa_circ_0061276 levels in cancer tissues were found associated with age, tumor size, TNM stages, distal metastasis, and CEA levels (Supplementary Table S4). The hsa_circ_0061276 levels in plasma were found associated with gender, tumor size, differentiation, and CEA levels (Supplementary Table S4). It is worth noting that the association with distal metastasis and tumor size were the common features shared by the hsa_circ_0001017 and hsa_circ_0061276 levels in tissue and plasma.
Expression patterns of hsa_circ_0001017 and hsa_circ_0061276 in gastric cancer tissues and the paired plasma. A Relative detection of hsa_circ_0001017 and hsa_circ_0061276 levels in gastric dysplasia tissues (n = 36), gastric cancer tissues (n = 121), and the paired adjacent non-tumorous tissues by RT-qPCR. Larger ΔC q value indicates lower expression. Expression changes between gastric dysplasia tissues, cancer tissues and paired non-tumorous tissues (a, c). ***p < 0.001. There is no significant difference of hsa_circ_0061276 levels between gastric dysplasia tissues and cancer tissues. The purple lines connected the cancer tissue downregulated pairs; and the black lines connected the cancer tissue upregulated pairs (b, d). B Absolutely quantitative detection of hsa_circ_0001017 and hsa_circ_0061276 levels in plasma from paired pre-operation and post-operation patients with gastric cancer (n = 121), and healthy controls (n = 121) by RT-ddPCR. Copy number change between paired pre-operation and post-operation patients with gastric cancer (a, c). ***p < 0.001. There is no significant difference between post-operation patients and healthy control, neither of hsa_circ_0001017 or hsa_circ_0061276 levels. The purple lines connected the post-operation elevated pairs; and the black lines connected the post-operation reduced pairs (b, d). C The proportion of patients with dysregulated hsa_circ_0001017 and hsa_circ_0061276 in gastric cancer tissues and plasma. The 0001017 and 0061276 indicate hsa_circ_0001017 and hsa_circ_0061276, respectively
Potential diagnostic values of hsa_circ_0001017 and hsa_circ_0061276
By receiver operating characteristic (ROC) curve, the diagnostic abilities of hsa_circ_0001017 and hsa_circ_0061276 to effectively differentiate gastric dysplasia from normal controls were shown in Fig. 4A. The area under ROC curve (AUC) of hsa_circ_0001017 reached to 0.871, with 79.4% sensitivity and 81.1% specificity, respectively. For distinguishing gastric cancer patients from healthy controls (Fig. 4B), the AUCs of cell-free hsa_circ_0001017 and hsa_circ_0061276 were found bigger than those of them in cancer tissues (Fig. 4C). It is mentionable that the AUC of the quadruple combination was up to 0.966; and the sensitivity and specificity increased to 95.5% and 95.7%, respectively (Fig. 4B-D). When combinative used of two plasma biomarkers, hsa_circ_0001017 and hsa_circ_0061276, the AUC was 0.912 (Fig. 4C). In Fig. 4C, we listed the all possible combinations with four types of biomarkers to achieve the best effective diagnosis for gastric cancer. The obtained AUC, sensitivity and specificity, relative positive predictive value (PPV), and negative predictive value (NPV) were shown on the heat map (Fig. 4C-E). The NPV could be 96.3% when tissue and plasma hsa_circ_0001017 were combined used; and the PPV was 98.2% when plasma hsa_circ_0061276 was added (Fig. 4D, E).
Potential diagnostic values of hsa_circ_0001017 and hsa_circ_0061276 in gastric cancer. A Diagnostic assessment of hsa_circ_0001017 and hsa_circ_0061276 in gastric dysplasia tissue vs. non-tumorous tissues. The AUC, sensitivity and specificity were inserted. B Diagnostic assessment of plasma (B/circ_0001017) and tissues (T/circ_0001017) hsa_circ_0001017, plasma (B/circ_0061276) and tissues (T/circ_0061276) hsa_circ_0061276, and quadruple combination in gastric cancer cases vs. healthy controls. C Pairwise comparison of the diagnostic values of plasma and tissue hsa_circ_0001017 (B/circ_0001017 and T/circ_0001017) and hsa_circ_0061276 (B/circ_0061276 and T/circ_0061276). The obtained AUC and the corresponding specificity and specificity were shown on the heat map. B, plasma; T, tissue; 1017, hsa_circ_0001017; 61276, hsa_circ_0061276. D The obtained sensitivity (a) and relative negative predictive values (b). E The obtained specificity (a) and relative positive predictive values (b)
Hsa_circ_0001017 and hsa_circ_0061276 can be prognostic indicators of gastric cancer
Next, hsa_circ_0001017 and hsa_circ_0061276 levels in tumor tissues and plasma from patients before operation were categorized as two groups, low or high group, according to the median level. The results showed that patients in the low plasma hsa_circ_0001017 group had a much shorter overall survival (median OS = 60 months) than those in high group (median OS = 84 months; p = 0.009; Fig. 5C, G). Meanwhile, hsa_circ_0061276 levels in plasma from gastric cancer patients were also associated with poor prognosis of patients (Fig. 5D, H). Univariate analyses and further analysis in a multivariate Cox proportional hazard model showed that plasma hsa_circ_0001017 and hsa_circ_0061276 were independent prognostic indicators of OS in patients with gastric cancer (Supplementary Table S5). When the patients were subdivided by tumor stage, plasma hsa_circ_0001017 and hsa_circ_0061276 levels could further distinguish patients with different overall survival times in stages III and IV (Fig. 5C-D).
Prognostic significance of hsa_circ_0001017 and hsa_circ_0061276 in gastric cancer patients. A Kaplan–Meier analysis of disease-free survival (DFS) based on hsa_circ_0001017 expression in gastric cancer patients in stages III and IV. B Kaplan–Meier analysis of DFS based on hsa_circ_0061276 in gastric cancer patients in stages III and IV. C Kaplan–Meier analysis of overall survival (OS) based on hsa_circ_0001017 expression in stages III and IV. D Kaplan–Meier analysis of OS based on hsa_circ_0061276 expression in stages III and IV. E Kaplan–Meier analysis of DFS based on hsa_circ_0001017 expression in all patients. F Kaplan–Meier analysis of DFS based on hsa_circ_0061276 in all patients. G Kaplan–Meier analysis of OS based on hsa_circ_0001017 expression in all patients. H Kaplan–Meier analysis of OS based on hsa_circ_0061276 in all patients
Meanwhile, we grouped the patients as “Recovered” or “Not recovered” to monitor the recurrence rate of gastric cancer by the fold changes of plasma hsa_circ_0001017 and hsa_circ_0061276 before and after surgery. The results showed that the gastric cancer patients in the recovered group had a longer disease-free survival (DFS) time, no matter in all clinical stages or in the advanced clinical stages only (stages III and IV; Fig. 5A, B, E, F). Moreover, the recovery situation of hsa_circ_0001017 in stage could be treated as an independent prognostic indicator of DFS in patients with gastric cancer (Supplementary Table S6).
Hsa_circ_0001017 and hsa_circ_0061276 can be secreted from gastric cancer cells
Since hsa_circ_0001017 and hsa_circ_0061276 may be potential biomarkers (Figs. 4 and 5), we next explored the origin of plasma circRNAs. We observed that (1) in tissues, hsa_circ_0061276 levels were lower than GAPDH mRNA levels; (2) in plasma, hsa_circ_0061276 levels were higher than GAPDH mRNA levels. So, we used cultured cell model to investigate the copy number changes of hsa_circ_0001017 and hsa_circ_0061276 in medium. We found that hsa_circ_0001017 and hsa_circ_0061276 levels increased in a time-dependent manner (Fig. 6). This means that gastric cancer cells may secrete circRNAs to the medium and partially explain the detectable circRNAs in patients’ plasma.
Discussion
An ideal biomarker should be non-invasive, accurate, inexpensive, specific, sensitive, reliable, and reproducible [23,24,25]. Consistent with this idea, body fluids are the commonly used materials in the diagnosis of multiple human diseases [24, 25]. Especially, plasma has significant advantages in reflecting gastrointestinal diseases [4, 26].
As so far, microRNAs (miRNAs) have been widely reported to be a kind of biomarkers in body fluids [27, 28]. In recent years, many attentions are attracted by circRNAs, a new member of non-coding RNA family [17, 18]. CircRNAs in tissue samples have been demonstrated to associate with tumor invasion, metastasis, and patient prognosis [29,30,31]. Importantly, the high stability of circRNAs conferred by their circular structure is beneficial for being a novel plasma biomarker [17].
It is declared that ddPCR or RT-ddPCR performed better in the accurate quantification of trace amount of nucleotides in plasma with high resolution, while the disability or large error was showed by qPCR or RT-qPCR [32, 33]. In this study, we compared the different quantification approaches of RT-qPCR and RT-ddPCR. With the highest correlation coefficients, the absolute quantification method by RT-ddPCR was confirmed to be most appropriate calculation method for circRNAs in tissues and plasma (Fig. 2B).
In the economic aspects, the application of dye in ddPCR cost less comparing with probe methods [13, 14], making it more adaptable in laboratory medicine. EvaGreen only has one kind of fluorescence. The ability to multiplex two and more kinds of gene in a single-color ddPCR system is contingent on the ability to distinguish between double-negative, single-positive, and double-positive droplet clusters. In usual, higher primer concentration and larger amplicon size will elevate the position of droplet population, vice versa. Inspired by this, we modulated these parameters to make it a highly flexible platform for simultaneously detection of several circRNAs (Supplementary Table S1). Our strategy is able to efficiently distinguish the droplets population of negative, GAPDH, hsa_circ_0061276, and hsa_circ_0001017 (Fig. 2A).
In this study, we first investigated the global expression profile of cell-free circRNAs in gastric cancer patients’ plasma by a high-throughput microarray. Compared with healthy controls, 343 significantly dysregulated cell-free circRNAs were observed (Fig. 1A, B).
Our data indicated that the expression patterns of hsa_circ_0001017 and hsa_circ_0061276 in expanded sample sizes of plasma and tissue samples were consistent with the results of microarray analysis. The levels of hsa_circ_0001017 and hsa_circ_0061276 in pre-operation plasma and gastric cancer tissues were found lower than those in the controls (Fig. 3A, B). The average copy numbers of cell-free hsa_circ_0001017 and hsa_circ_0061276 increased to normal levels after 10 days of operation (Fig. 3B). In the following analysis, the copy number changes of those circRNAs in plasma from post-operation patients were confirmed to be independent monitoring indicators for the recurrence of gastric cancer (Fig. 5A, B, E, F). Furthermore, the reduced hsa_circ_0001017 and hsa_circ_0061276 in plasma of gastric cancer patients were significantly correlated with higher mortality in the overall gastric cancer patients and TNM III–IV stage cohort (Fig. 5C, D, G, H). Finally, the in vitro model indicated that gastric cancer cells may secrete circRNAs into plasma (Fig. 6).
Gastric epithelial dysplasia represents a recognized precursor lesion of gastric cancer [5, 34]. That the significantly reduced hsa_circ_0001017 expression in gastric epithelial dysplasia tissues compared to non-tumorous tissues (Fig. 3A) and the effective diagnosis (with AUC up to 0.871, Fig. 4A) indicated that hsa_circ_0001017 may be a predictor for early diagnosis of gastric cancer.
The gastric cancer tissue and plasma hsa_circ_0001017 and hsa_circ_0061276 levels were closely related with major clinicopathological factors (Supplementary Table S3 and Table S4). All these results strongly suggested that hsa_circ_0001017 and hsa_circ_0061276 may be novel potential gastric cancer-associated biomarkers.
Due to the low sensitivity or specificity of the existing blood biomarkers of gastric cancer, a panel approach of combinations will allow us to pursue more precise diagnosis. As shown in Fig. 4C, we evaluated the pairwise combination of four independent indexes from hsa_circ_0001017 and hsa_circ_0061276. Overall, in all the datasets, the AUC of combinative use of hsa_circ_0001017 and hsa_circ_0061276 was up to 0.966, with PPV = 95.7% and NPV = 95.5% (Fig. 4C-E). A combination of plasma hsa_circ_0001017 and hsa_circ_0061276 for non-invasion diagnosis exhibited 0.912 of AUC, with a high sensitivity and PPV up to 97.6% and 96.1%, respectively. These mean that diagnosis in combination with hsa_circ_0001017 and hsa_circ_0061276 will be in satisfactory specificity and sensitivity. Nevertheless, to confirm the specificity of this method, larger cohort of patients should be analyzed in the future.
In conclusion, our study first revealed the cell-free circRNAs profile in plasma of patients with gastric cancer. Further, we found that hsa_circ_0001017 and hsa_circ_0061276 both decreased in gastric cancer tissues and patients’ plasma. Moreover, their levels were tightly associated with main clinicopathological features of patients with gastric cancer. As a result, hsa_circ_0001017 and hsa_circ_0061276 may be valuable blood-based biomarkers, either alone or in combination, to screen gastric cancer and estimate prognosis.
References
Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y (2009) Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol 24:652–657
Li T, Mo X, Fu L, Xiao B, Guo J (2016) Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 7:8601–8612
Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A et al (2015) Global surveillance of cancer survival 1995-2009: analysis of individual data for 25, 676, 887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 385:977–1010
Hashad D, Elbanna A, Ibrahim A, Khedr G (2016) Evaluation of the role of circulating long non-coding RNA H19 as a promising novel biomarker in plasma of patients with gastric cancer. J Clin Lab Anal 30:1100–1105
Li Q, Shao Y, Zhang X, Zheng T, Miao M, Wang B, Ye G, Xiao B, Guo J (2015) Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol 36:2007–2012
Cui L, Lou Y, Zhang X, Zhou H, Deng H, Song H, Yu X, Xiao B, Wang W, Guo J (2011) Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin Biochem 44:1050–1057
Zhou H, Xiao B, Zhou F, Deng H, Zhang X, Lou Y, Gong Z, Du C, Guo J (2012) MiR-421 is a functional marker of circulating tumor cells in gastric cancer patients. Biomarkers 17:104–110
Schneider J, Schulze G (2003) Comparison of tumor M2-pyruvate kinase (tumor M2-PK), carcinoembryonic antigen (CEA), carbohydrate antigens CA 19-9 and CA 72-4 in the diagnosis of gastrointestinal cancer. Anticancer Res 23:5089–5093
Carpelan-Holmstrom M, Louhimo J, Stenman UH, Alfthan H, Haglund C (2002) CEA, CA 19-9 and CA 72-4 improve the diagnostic accuracy in gastrointestinal cancers. Anticancer Res 22:2311–2316
Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA (1992) Quantitation of targets for PCR by use of limiting dilution. BioTechniques 13:444–449
Riva F, Bidard FC, Houy A, Saliou A, Madic J, Rampanou A, Hego C, Milder M, Cottu P, Sablin MP et al (2017) Patient-specific circulating tumor DNA detection during neoadjuvant chemotherapy in triple-negative breast cancer. Clin Chem 6:691–699
Guttery DS, Page K, Hills A, Woodley L, Marchese SD, Rghebi B, Hastings RK, Luo J, Pringle JH, Stebbing J et al (2015) Noninvasive detection of activating estrogen receptor 1 (ESR1) mutations in estrogen receptor-positive metastatic breast cancer. Clin Chem 61:974–982
Shen F, Du W, Kreutz JE, Fok A, Ismagilov RF (2010) Digital PCR on a SlipChip. Lab Chip 10:2666–2672
Shen F, Sun B, Kreutz JE, Davydova EK, Du W, Reddy PL, Joseph LJ, Ismagilov RF (2011) Multiplexed quantification of nucleic acids with large dynamic range using multivolume digital RT-PCR on a rotational SlipChip tested with HIV and hepatitis C viral load. J Am Chem Soc 133:17705–17712
Yao T, Chen Q, Fu L, Guo J (2017) Circular RNAs: biogenesis, properties, roles, and their relationships with liver diseases. Hepatol Res 47:497–504
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S (2015) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25:981–984
Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J (2015) Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 444:132–136
Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y (2015) Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 6:6001–6013
Yang W, Du WW, Li X, Yee AJ, Yang BB (2016) Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene 35:3919–3931
Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, Ye G, Zhang X, Xiao B, Guo J (2014) Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer 120:3320–3328
Li P, Chen H, Chen S, Mo X, Li T, Xiao B, Yu R, Guo J (2017) Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer 116:626–633
Janku F, Huang HJ, Fujii T, Shelton DN, Madwani K, Fu S, Tsimberidou AM, Piha-Paul SA, Wheler JJ, Zinner RG et al (2017) Multiplex KRASG12/G13 mutation testing of unamplified cell-free DNA from the plasma of patients with advanced cancers using droplet digital polymerase chain reaction. Ann Oncol 28:642–650
Yu D, Li HX, Liu Y, Ying ZW, Guo JJ, Cao CY, Wang J, Li YF, Yang HR (2017) The reference intervals for serum C-terminal agrin fragment in healthy individuals and as a biomarker for renal function in kidney transplant recipients. J Clin Lab Anal 31:e22059
Klapkova E, Fortova M, Prusa R, Moravcova L, Kotaska K (2016) Determination of urine albumin by new simple high-performance liquid chromatography method. J Clin Lab Anal 30:1226–1231
Ay E, Marakoğlu K, Kizmaz M, Ünlü A (2016) Evaluation of Netrin-1 levels and albuminuria in patients with diabetes. J Clin Lab Anal 30:972–977
Capello M, Bantis LE, Scelo G, Zhao Y, Li P, Dhillon DS, Patel NJ, Kundnani DL, Wang H, Abbruzzese JL et al (2017) Sequential validation of blood-based protein biomarker candidates for early-stage pancreatic cancer. J Natl Cancer Inst 109. https://doi.org/10.1093/jnci/djw266
Min QH, Chen XM, Zou YQ, Zhang J, Li J, Wang Y, Li SQ, Gao QF, Sun F, Liu J et al (2017) Differential expression of urinary exosomal microRNAs in IgA nephropathy. J Clin Lab Anal. https://doi.org/10.1002/jcla.22226
Raitoharju E, Seppala I, Lyytikainen LP, Viikari J, Ala-Korpela M, Soininen P, Kangas AJ, Waldenberger M, Klopp N, Illig T et al (2016) Blood hsa-miR-122-5p and hsa-miR-885-5p levels associate with fatty liver and related lipoprotein metabolism—the young Finns study. Sci Rep 6:38262
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C (2016) Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular carcinoma development. Medicine (Baltimore) 95:e3811
Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W (2016) Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res 44:e87
Zhao ZJ, Shen J (2017) Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol 14:514–521
Sillence KA, Roberts LA, Hollands HJ, Thompson HP, Kiernan M, Madgett TE, Welch CR, Avent ND (2015) Fetal sex and RHD genotyping with digital PCR demonstrates greater sensitivity than real-time PCR. Clin Chem 61:1399–1407
Huggett JF, Cowen S, Foy CA (2015) Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem 61:79–88
Yang Y, Shao Y, Zhu M, Li Q, Yang F, Lu X, Xu C, Xiao B, Sun Y, Guo J (2016) Using gastric juice lncRNA-ABHD11-AS1 as a novel type of biomarker in the screening of gastric cancer. Tumour Biol 37:1183–1188
Acknowledgments
This study was supported by grants from the Applied Research Project on Nonprofit Technology of Zhejiang Province (no. 2016C33177), the Scientific Innovation Team Project of Ningbo (no. 2017C110019), National Natural Science Foundation of China (no. 81772279), and the K.C. Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
ESM 1
(PDF 291 kb)
Rights and permissions
About this article
Cite this article
Li, T., Shao, Y., Fu, L. et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med 96, 85–96 (2018). https://doi.org/10.1007/s00109-017-1600-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-017-1600-y