Abstract
In the current work, the relationship between the structure and activity of a series of novel thiazolidine-4-carboxylic acid derivatives as potent influenza virus neuraminidase inhibitors was studied using docking, molecular dynamics (MD) simulations, and QSAR analysis. A 7,000 ps MD simulation in a cubic water box were employed to build 3D structure of the 2HU4 in a water environment. After reaching the equilibrium, the inhibitors were docked into the 2HU4 to realize the binding site of the enzyme. The docking analysis showed that the interaction of the inhibitors with residues Arg371, Arg430, Gly429, Ile427, Lys432, Pro431, Trp403, and Tyr347 plays an important role in the activities of the inhibitors. The docked configurations of the inhibitors with the lowest free energy were used to calculate the most feasible descriptors. The selected descriptors were related to the inhibitory activities using stepwise multiple linear regression, classification and regression trees, and least squares support vector regression techniques. The satisfactory results (R 2p = 0.883, Q 2LOO = 0.872, R 2L25%O = 0.835, RMSELOO = 0.310, and RMSEL25%O = 0.352) demonstrate that CART-LS-SVR models present the relationship between influenza virus neuraminidase inhibitors activity and molecular descriptors clearly. An energetic analysis based on MD calculations, revealed that the potency of the most active compound binding is governed by electrostatic and van der Waals contacts. The results provide a set of useful guidelines for the rational design of novel influenza virus neuraminidase inhibitors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Severe public health and economic problems due to influenza can affect millions of people worldwide. Influenza virus is a RNA virus that infects avian and mammalian cells with the assistance of glycoproteins, hemagglutinin, and neuraminidase. Influenza virus neuraminidase has been recognized as a good target for the treatment of influenza because of its critical roles in the life cycle of the virus. It also facilitates not only the virion progeny release but also the general mobility of the virus in the respiratory tract, thereby enhances infection efficiency (Chand et al., 2001).
Structure-based drug design based on the information of original neuraminidase crystal structure has proved valuable in the discovery and development of anti-influenza drugs. Up to date, around 170 neuraminidase X-ray complexes in the protein data bank (PDB) contribute to the development of this drug target. Designing new potent and selective inhibitors have currently emerged as promising therapeutics for influenza since the emergence of viruses resistant to the currently available drugs. Using trial and error approaches for drug design are usually costly and time-consuming, therefore the development of theoretical methods for predicting inhibitory activities of drug-like molecules would be helpful. For this purpose, structure-based method is applied as computer-assisted drug design using molecular docking and molecular dynamics (MD) simulation (Girisha et al., 2009).
In recent years, several drug-like molecules have been reported to possess inhibitory activities against influenza virus neuraminidase. However, little is known about their structure–activity relationship. Mercader and Pomilio (2010) established a QSAR model which suggested the inhibitory activity depending on the electric charges, masses, and polarizabilities of the atoms presented in the molecule as well as its conformation. Murumkar et al. (2011) investigated the interactions between some flavonoids and active site of neuraminidase. However, despite the several inhibitors described in the literature, further researches are required to find potent, promising, and selective inhibitors.
The main aim of this work is to realize the structural basis of the interactions that thiazolidine-4-carboxylic acid derivatives have with the neuraminidase. Xu et al. (2011) synthesized these derivatives as novel influenza virus neuraminidase inhibitors. The crystal structure of neuraminidase (PDB ID: 2HU4) was taken from PDB and MD simulation was performed in a cubic water box, which enhances the similarity of the MD simulation with the cellular environment. The obtained protein structure was used for docking the drugs into the binding site of the receptor. The best docked conformations of inhibitors with the lowest free energies were considered as optimized conformations. Then the descriptor calculation was performed using the best docked conformations of the inhibitors. Furthermore, stepwise multiple linear regression (MLR), classification and regression trees (CART), and least squares support vector regression (LS-SVR) models were constructed to study the relationship between the structures of the inhibitors and the experimental pIC50. The accuracy and robustness of the constructed models was illustrated using (1) leave-one-out and (2) leave-multiple-out cross-validation (LMO-CV) techniques combined with different statistical parameters. Finally, the constructed model and the interaction between the drugs and receptor can not only be used in rapidly and accurately predicting the activities of newly designed inhibitors, but also provide useful guidelines for developing potent influenza virus neuraminidase inhibitors with desired inhibitory activity.
Materials and methods
Data set
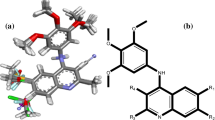
Influenza virus neuraminidase inhibitors of 28 novel thiazolidine-4-carboxylic acid derivatives together with their inhibitory activities (pIC50) were taken from the article recently published by Xu et al. (2011). The 50 % inhibitory concentration (IC50) is defined as the concentration of neuraminidase inhibitor necessary to reduce the activity by 50 % relative to a reaction mixture containing virus but no inhibitor. The chemical structures and experimental activity of these compounds are shown in Fig. 1 and Table 1. The chemical structures of the studied compounds were constructed by HyperChem package (version 7, Hypercube Inc.). Prior to docking study, the energy minimizations for these compounds were performed by AM1 semi-empirical method and Polak-Ribiere algorithm until the root-mean square gradient of 0.1 kcal mol−1.
MD simulation and molecular docking
MD simulations were performed using the GROMACS 3.5.1 package with the standard GROMOS96 43a1 force field (VanGunsterenv et al. 1996. The crystal structure of neuraminidase (PDB ID: 2HU4) consisted of four similar chains. One of the chains was considered for the MD simulations and docking purpose. The system was immersed in a cubic water box and the energy of the complexes was minimized using the steepest descent approach realized in the GROMACS package. MD simulation studies consist of equilibration and production phases. In the first stage of equilibration, the solute (protein and water molecules) was fixed and the position-restrained dynamics simulation of the system was restrained at 310 K. The water permitted to relax about the protein and the relaxation time of water was 30 ps. Finally, the full system was subjected to 7,000 ps MD simulation at 310 K temperature and 1 bar pressure. The particle mesh Ewald (PME) method for long-range electrostatics, a 14 Å cutoff for van der Walls interactions, a 12 Å cutoff for coulomb interaction with updates every 10 steps, and the Lincs algorithm for covalent bond constraints were used in this study.
The molecular docking was carried out using the Molegro Virtual Docker software. All the torsion angles in the small molecules were set free to perform flexible docking. The docking simulations were performed by Molegro virtual docking engine with flexible mode for the inhibitor and the MolDock score with grid resolution of 0.3 Å. The MolDock score optimizer and genetic algorithm search strategy were used for docking with the following settings: an initial population of 200 randomly placed individuals, a maximum number of 250,000 iterations, a crossover rate of 0.80, and scaling factor of 0.5. One Hundred independent docking runs were carried out for each ligand. Results were clustered according to the root-mean-square-deviation (RMSD) criterion. The best docked conformations with the lowest binding free energy for each ligand were used for calculation of the molecular descriptors.
Molecular descriptors calculation
The important step for constructing a QSAR model is to encode the structural features of the inhibitors, named molecular descriptors. A total of 1,142 molecular descriptors were calculated by the Dragon software. Those descriptors that stayed constant for all the molecules were eliminated. Also, pairs of descriptors with a correlation coefficient greater than 0.80 were classified as inter-correlated, and therefore one of them in each correlated pair has been eliminated. A total of 769 descriptors were considered for further investigations (stepwise MLR and CART) after discarding the descriptors with constant and inter-correlated ones. The calculated descriptors were among topological, electronic, geometrics, constitutional, 3D-MoRSE, 2D autocorrelations, BCUT, GETAWAY, and Randic molecular profile categories. In the present work, attempts were made to correlate the activity of influenza virus neuraminidase inhibitors with the best descriptors encoding the steric, hydrophobic, electronic, and structural features of thiazolidine-4-carboxylic acid derivatives.
Theory of CART
CART (Breiman et al., 1984), as a binary tree representation, is able to describe the relationships between the dependent and independent variables with high flexibility and sufficient accuracy. Moreover, CART has the ability to select the most descriptive variables from a large number of descriptors without having the side effect of immunity to outliers, collinearity, and heteroscedasticity. This methodology has the ability to explain and predict both categorical and continuous responses. CART, as a binary recursive splitting procedure, divides the data into mutually exclusive sub-groups, called child nodes, which are more homogeneous with respect to the response than the initial dataset. In general, the configuration of CART consists of two basic steps: (1) growing an over-large tree and (2) tree pruning and optimal tree selection. The tree building process starts by dividing the parent node into two children nodes and searches for the best split. The resulting maximal tree usually provides poor prediction results for new samples. Therefore, the selection of a smaller tree with better predictive ability without losing much accuracy is necessary. The optimal tree size is obtained by pruning, i.e., removing successively branches of the maximal tree. The theories behind the CART have been adequately described elsewhere (Vander Heyden et al., 2005). For CART modeling, the CART toolbox was used with MATLAB Version 7.1.
Theory of LS-SVR
Support vector machine was introduced by Vapnik (1998) and has attracted much research attention in recent years due to its performance over similar techniques in real applications. For a given regression problem, the goal of SVM is to find the optimal hyper-plane, from which the distance to all the data points is minimum. LS-SVR is an alternative method of SVM which fits a linear relation (y = wx + b) between the regression (x) and the dependent variable (y). The best relation is the one that minimizes the cost function (Q) containing a penalized regression error term:
Subjected to
where φ denotes the feature map. The first part of the cost function is a weight decay which is used to regularize weight sizes and penalize large weights. The second part of cost function is the regression error for all training data. The relative weight of this part as compared to the first part was indicated by the parameter γ. The details theory and mathematical equations of LS-SVR has been described in literature (Cheng et al., 2010, 2011). The LS-SVR toolbox (Suykens, Leuven, Belgium) was used with MATLAB Version 7.1 to derive all the LS-SVR models.
Results and discussion
MD simulation on influenza virus neuraminidase
The homology model was clarified using the crystal complex of influenza virus neuraminidase in complex with corresponding ligand Oseltamivir downloaded from PDB database (http://www.rcsb.org/pdb/home/home.do). The PDB ID code is 2HU4. Before MD simulation, the pre-existing ligand was extracted out. A 7,000 ps MD simulation was performed on influenza virus neuraminidase in water box to obtain the protein conformation in water environment. This conformation is more similar to the conformation of neuraminidase in a cell environment. RMSD value of the protein backbone was examined to investigate the stability of the system (protein, water, etc.). Figure 2 shows the time history of RMSD for neuraminidase conformation in a water environment relative to the starting structure. This figure indicates that the RMSD of neuraminidase reaches equilibration and oscillates around in average value after 4,000 ps simulation time. Also, the RMSD value of protein backbone is 0.30 ± 0.02 nm from a 4,000–7,000 ps trajectory. Finally, the influenza neuraminidase structure was used in the docking experiments.
Molecular docking
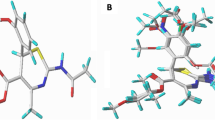
The main aspect for the drug docking was to realize the effective interaction of the drugs with the various amino acid residues in the active sites. The binding sites of the neuraminidase receptor were studied to understand the nature of the residues defining the sites. To realize the binding sites of the influenza neuraminidase receptor, the best potent reported drug in Table 1, compound 27, was docked into the receptor. The applied box size for docking was 70 × 70 × 70 Å and grid resolution was 0.3 Å. The most important binding sites were Arg (118, 371, 428, and 430), Gly (405, 429), Glu433, Ile 427, Lys432, Pro431, Trp403, and Tyr347 (Fig. 3a, b). Polar hydrogen bonds in compound 27 cause to be capable of hydrogen bonding to the polar protein residues (such as Arg, Glu, etc.) in the binding sites. The obtained binding sites show Gly429, Ile427, Lys432, Pro431, Trp403, and three Arginine residues at positions 118, 371, and 428 in their binding pockets, capable of making hydrogen bonding interactions that could potentially interact favorably with O–H, N–H, and COOH groups of the inhibitors. The strongest hydrogen bonds interactions between docked drug and residues are Arg371 (3.11 Å, −2.47 kcal), Ile427 (3.10 Å, −2.49 kcal), Lys432 (2.98 Å, −2.5 kcal), and Pro431 (3.26 Å, −1.67 kcal). However, van der Walls and electrostatic interactions are also important because of presence of both phenyl group and polar atoms, respectively. As a result, substituents which improve hydrogen bonds and electrostatic interactions would prefer in order to enhance the efficiency and potency of the inhibitors. For example compound 27 shows better inhibitory activity compared to compounds 21 and 22 because of the presence of both OH and OCH3 groups. Also, presence of electron withdrawing groups in R1 branch such as –CN (in compound 25) and –NO2 (in compound 26) reduce the pIC50 values. On the other hand, for the same R1 group, the inhibitory activity increase as R2 branch replace with more polar groups. For instance, compound 22 show greater pIC50 than compounds 15, 8, and 1. This pattern is repeated in whole data set, for example compounds 2, 9, 16, 23, and so on. Finally, the optimized conformation for each inhibitor was obtained using Molegro Virtual Docker software. The obtained conformations of the inhibitors were used for descriptor calculation in developing QSAR models.
MD simulation on influenza virus neuraminidase–inhibitor complex
In order to examine conformational variations of inhibitor on the protein conformation, we decided to perform a MD simulation on the neuraminidase–inhibitor complex. MD simulation (7,000 ps) was performed on influenza virus neuraminidase–inhibitor with respect to the compound 27 with the best inhibitory activity in a water box. MD simulation was performed on the receptor–ligand at 310 K. RMSD of the protein backbone was calculated by the MD trajectory to investigate the stability of system including protein, inhibitor and water. As shown in Fig. 2 after approximately 4,000 ps of MD simulation, the structure of the receptor–ligand becomes stable. The average RMSD for the neuraminidase–inhibitor complexes was calculated from a 4,000 to 7,000 ps trajectory, where the data points were fluctuated 0.31 ± 0.02 nm. Therefore, the inhibitor binding to the protein does not affect the conformation of the protein significantly and the stability of the protein conformation in the presence of inhibitor confirmed the docking results.
To investigate the correlation between interaction energy and pIC50, the binding free energy between receptor and the inhibitors within the complex structures were examined using molecular mechanics-generalized Born surface area (MM-GBSA) method (Kollman et al., 2011). The calculations were performed on each complex system using ten snapshots from 4,000 to 7,000 ps MD simulations region. The following equation was used:
where ΔE MM is the change of the gas phase MM energy upon binding, and includes internal (ΔE int), electrostatic (ΔE elect), and van der Waals (ΔE VDW) energies. ΔG solv is the change of the solvation free energy upon binding, and includes the electrostatic solvation free energy ΔG solvGB (polar contribution calculated using generalized Born model), and the non-electrostatic solvation component ΔG solvSA (non-polar contribution estimated by solvent accessible surface area). Finally, TΔS is the change of the conformational entropy upon binding. The binding free energy values obtained from MM-GBSA calculations are reported in Table 1. As shown in Fig. 4, there is a good agreement between binding free energy and pIC50 values (R 2Fit = 0.86). Also, the averaged energy values for two active ligands (compounds 22 and 27) are given in Table 2. The interaction is more favorable for the receptor-27 complex as the difference in the calculated ΔG bind value between two inhibitors was 4.21 kcal/mol. Considering the energy components of the binding free energies, the major favorable contributors to ligand binding are VDW and electrostatic terms, whereas polar and non-polar solvation and entropy terms oppose binding. If we examine the contributions to each binding energy, the most important terms which dictates the difference in the binding affinity are ΔE VDW (4.24 kcal/mol) and ΔE elect (2.05 kcal/mol), which is the key factor for the more favorable ΔG bind and pIC50 values for compound 27.
QSAR model construction
Hybrid methods of stepwise MLR-LS-SVR and CART-LS-SVR are presented in this work for QSAR study of thiazolidine-4-carboxylic acid derivatives. This means that we have to discuss two stages: (i) descriptor selection (stepwise MLR and CART) and (ii) mapping tool (LS-SVR).
Descriptor selection using stepwise MLR and CART techniques
The objective of descriptor selection is threefold: improving the prediction performance of predictors, providing faster and more cost effective predictors, and providing a better understanding of the underlying process that generated the data. For this purpose, all of the calculated 769 descriptors from previous section were used for stepwise MLR and CART analyses. For stepwise MLR, the calibration set was used to select the most feasible descriptors and to calculate coefficients relating the descriptors to the inhibitory activities of inhibitors. However, the prediction set, consisted of 25 % of molecules, was used to evaluate the generated model. The molecules in each of the calibration and prediction sets were shown in Table 1. Based on the number of molecules in the calibration set (75 % of molecules), we selected three descriptors to construct QSAR models since the number of selected descriptors should keep lower than five times of the number of molecules (Salt et al., 2007). Equation 4 shows the specifications of the obtained MLR model which was made by these descriptors:
where GATS4p, Greay autocorrelation of lag 4/weighted atomic polarizabilities; SP06, shape profile no. 06; More14e, 3D-Morse/weighted atomic Sanderson electronegativities are three descriptors selected in stepwise MLR technique.
In CART technique, tree partitioning will progress until no further split can be occurred. The size of a tree in the CART analysis is an important issue, since an unreasonably big tree can only make the interpretation of results more difficult. In order to reduce the number of variables and obtain the best predictive tree, a fourfold cross-validation has been applied. The number of terminal nodes versus COST function is plotted in Fig. 5. The dashed line in this figure is obtained using the equation of X min +1s, where X min is the value of the minimum COST and s is the standard error of the minimum COST. Figure 6 exhibits the optimal tree with low RMSE for variable selection in CART technique. Three descriptors of VED2 (average eigenvector coefficient sum from distance matrix), BELm3 (lowest eigenvalue no. 3 of burden matrix/weighted atomic masses), and RARS (R matrix average row sum) have been selected using CART technique. As second stage of developing QSAR models, we should use these selected descriptors as inputs for LS-SVR modeling.
Stepwise MLR-LS-SVR and CART-LS-SVR models
In the next step, the selected descriptors in the stepwise MLR and CART techniques were used as inputs for developing the LS-SVR model to predict the value of pIC50 for the influenza virus neuraminidase inhibitors. In order to generate a LS-SVR model, at first the kernel function should be determined, which represents the sample distribution in the mapping space. In this work, the radial basis kernel function (RBF) was used, since it is a non-linear function and could reduce the computational complexity of training procedure. The next step in the construction of LS-SVR model was optimizing of its parameters, including γ and Δ2. The optimized values for the parameters were obtained from grid search method. The optimized values of γ and Δ2 were 29.74 and 162.98 (in stepwise MLR-LS-SVR) and 35.76 and 151.9 (in CART-LS-SVR models), respectively. The predicted values of thiazolidine-4-carboxylic acid derivatives activity using stepwise MLR-LS-SVR and CART-LS-SVR models are listed in Table 1. This table shows that the calculated pIC50 is a good estimate of experimental pIC50. The predicted stepwise MLR-LS-SVR and CART-LS-SVR values of pIC50 were plotted versus their observed values, shown in Figs. 7 and 8, respectively. This figure shows a good agreement between the experimental results and the predicted values.
Model validation is nowadays recognized as a compulsory stage in QSAR model development. Golbraikh and Tropsha (2002)considered a QSAR model to be predictive, if all of the following conditions are satisfied: (1) Q 2LOO > 0.5, (2) R 2p > 0.6, r 20 (i.e., predicted vs. observed activities imply regressions through the origin) is close to R 2p such that (R 2p − r 20 )/R 2p < 0.1. In addition, according to the recommendation of Roy and Roy (2002), an additional statistic for external validation (r 2m ) was calculated using Eq. (5):
For a model with good external predictability, r 2m value should be greater than 0.5. The MLR-LS-SVR and CART-LS-SVR models were further evaluated by applying the LMO-CV technique. In the L25 %O procedure, a group of 25 % of molecules was randomly selected from the training set. Then each group was left out and was predicted by the model developed from the remaining observations. This procedure was carried out 200 times. The theory behind these validation methods has been sufficiently described elsewhere (Jalali-Heravi et al., 2009; Shahbazikhah et al., 2011). The statistical results are given in Table 3 shows the stepwise MLR-LS-SVR and CART-LS-SVR models possessed all the criteria to be considered as a predictive model. It is clear from Table 3 that the results of Q 2LOO , R 2L25%O and their corresponding RMSEs for the CART-LS-SVR model are superior compared with those of the stepwise MLR-LS-SVR. The RMSEs of both LOO and LMO have been reduced more than 40 % using CART-LS-SVR technique. The strong correlation between the calculated activity and the experimental activity demonstrated the robustness of the models, and the feasibility and advantage of the computational approach in this study.
Descriptors appeared in the QSAR models
The correlation matrix between selected descriptors in stepwise MLR and CART techniques together with pIC50 is listed in Table 4. The methods for calculations of these descriptors and the meaning of them are explained in the Handbook of Molecular Descriptors by Todeschini and Consonni (2000). As can be seen in the Table 4, the effective descriptors for inhibitory activity were GATS4p, More14e, BELm3, and VED2; therefore, we will only explain these descriptors. The appearance of average Geary autocorrelation of lag 4/weighted atomic polarizabilities (GATS4p) among other descriptors in the model shows the importance of polarizability and surface area of the inhibitor. Both these factors are very important for the inhibition mechanism. For instance, for compounds 6, 13, 20, and 27 which they have the same rigid substituents of R1, the values of pIC50 increase when R2 is replaced with more polarizable groups of PhCH2CO– and NH2CH2CO–. Therefore, the value of pIC50 increase as the GATS4p value increases. Also, compound 27 (the most active compound) has the highest value of GATS4p. This pattern is also repeated for other sets of inhibitors, for example, compounds 1, 8, 15, and 22 and so on. BELm3 (lowest eigenvalue no. 3 of burden matrix/weighted atomic masses) is among BCUT descriptors which is related to molecular graph theory Bonchev (1983) and they influence transport phenomena as well as entropy contributions. BCUT descriptors were proposed as molecular descriptor with high discrimination power, to be used in the recognition and ordering of molecular structures. The basic assumption was that the lowest eigenvalues contain contributions from all atoms and thus reflect the topology of the whole molecule.
The next descriptor related to inhibitory activity is 3D-Morse/weighted atomic Sanderson electronegativities (Mor14e), which is the 3D-molecule representation of structures based on electron diffraction (MoRSE) descriptor. 3D-MoRSE codes 3D structure of a molecule. By increasing the value of Mor14e, inhibitory activity increases. For instance, for the compound 2, the value of Mor14e was 0.241 with pIC50 of 4.695, and for the compound 9, the value of Mor14e was 0.448 with pIC50 of 5.123. Therefore, the substituent with a higher value of Mor14e would prefer as a potent influenza virus neuraminidase inhibitor. VED2 (average eigenvector coefficient sum from distance matrix, topological descriptor) can be calculated from the information theory, and it measures the complexity of the molecule in terms of the diversity of elements that includes in its chemical structure, such as the type of atoms, bonds, cycles, etc.
Qualitative energy analysis
To get a detailed view of the effects of individual residues on inhibitory activities of the receptor–ligand systems, a residue decomposition of the total energy was performed to evaluate the energetic influences of critical residues on the binding. For this purpose, energy of interaction between individual residues and three active ligands (compounds 22, 23, and 27) was calculated with the calculate interaction energy protocol encoded in Molegro Virtual Docker for the three receptor–ligand complexes. The residue-based decomposition of interaction energies in three systems identified several critical residues of 2HU4. Table 5 lists the average energy contributions of these key residues of interest for three systems. As can be seen, the interaction between ligands and the residues Arg371, Arg430, Gly429, Ile427, Lys432, Pro431, Trp403, and Tyr347 are the most favorable dominant contributions to the binding of three ligands to 2HU4. Also, the energy interaction for Ile427, Lys432, and Pro431 residues (the strongest interactions) are about 50 % greater for the most active drug (compound 27) than compounds 22 and 23. These key residues are well coincided with the molecular docking studies and QSAR results. Thus, these critical residues together with the obtained descriptors and CART-LS-SVR model may provide guidance for the rational design to discover more potent influenza virus neuraminidase inhibitors.
Design of new potent inhibitors
As shown in the previous sections, molecular docking, MD, and QSAR analysis provided detailed insight into the structural requirements for potent activity of the inhibitors. We have employed this information to design several inhibitors with improved activity. The most potent molecule (compound 27) was used as a reference structure to design new molecules. To show the practical values of developed models, we designed a series of new inhibitors and predicted their pIC50 values by the established QSAR models. Modifications were made by placing (1) strongly hydrophilic and electronegative groups in R1 and (2) more polarizable groups in R2. Table 6 shows the structure and predicted pIC50 values of newly designed compounds. There is a good agreement between binding free energy and pIC50 values (R 2Fit = 0.76) as shown in Fig. 4 which show the predicted pIC50 values are acceptable. It can be seen that some designed derivatives (for example compounds new 2 and new 6) showed higher activities than compound 27 which were the most active in the database. In addition, examining the contributions to each binding energy for compounds 27 and new 6 in Table 2, the most important terms which dictate the difference in the binding affinity are ΔE VDW and ΔE elect. These results obtained from the developed models serve as computational predictions which can be used to guide the design of new potent inhibitors.
Conclusions
In this work, molecular docking, MD simulation and QSAR analysis were performed to explore structural features and binding mechanism of thiazolidine-4-carboxylic acid derivatives as potent influenza virus neuraminidase inhibitors. MD simulation followed by docking studies was used to find the best conformation of 2HU4–inhibitor complex with lowest binding free energy for each inhibitor. The combination of stepwise MLR, CART, and LS-SVR techniques was successfully applied for selecting the best molecular descriptors and predicting the inhibitor activity of thiazolidine-4-carboxylic acid derivatives against influenza virus neuraminidase. Various methods were used to validate these studies, including LOO cross-validation, LMO cross-validation, R 2p , r 20 , r 2m and calibration–prediction set methods. It is shown that the four parameters of GATS4p, More14e, BELm3, and VED2 chosen by QSAR analysis affect significantly the inhibition process of the drug-like molecules. The selected descriptors indicated that steric parameters and electronic interactions such as electronegativity, polarizability, and polar surface area affected the inhibition activity of these inhibitors. Furthermore, the residue-based decomposition of interaction energies in three receptor–ligand systems identified several critical residues for ligand binding. A group of residues have been found, namely Ile427, Lys432, and Pro431, which are important in receptor–ligand interactions. Moreover, the analysis of the energetic binding components using MM-GBSA method reveals that while the VDW energy drives binding of the inhibitors, electrostatic energy alone does not completely explain the affinity differential. In summary, the models built in this study provide detailed and deep insights for understanding the different chemical parameters affecting ligand–receptor interactions and provide valuable suggestions for novel inhibitor design and further chemical optimization.
References
Bonchev D (1983) Information theoretic indices for characterization of chemical structures. Wiley, Chichester
Breiman L, Friedman JH, Olshen RA, Stone CG (1984) Classification and regression trees. Wadsworth International Group, Belmont
Chand P, Kotian PL, Dehghani A (2001) Systematic structure-based design and stereoselective synthesis of novel multisubstituted cyclopentane derivatives with potent antiinfluenza activity. J Med Chem 44:4379–4392
Cheng Z, Zhang Y, Zhang W (2010) QSAR studies of imidazopyridine derivatives as Et-PKG inhibitors using the PSO-SVM approach. Med Chem Res 19:1307–1325
Cheng Z, Zhang Y, Fu W (2011) Predictive QSAR models of 3-acylamino-2-aminopropionic acid derivatives as partial agonists of the glycine site on the NMDA receptor. Med Chem Res 20:1235–1246
Girisha HR, Chandra JN, Boppana S, Malviya M, Sadashiva CT, Rangappa KS (2009) Active site directed docking studies: synthesis and pharmacological evaluation on cis- 2,6-dimethyl piperidine sulfonamides as inhibitors of acetylcholinesterase. Eur J Med Chem 44:4057–4062
Golbraikh A, Tropsha A (2002) Beware of q2. J Mol Graph Model 20:269–276
Jalali-Heravi M, Asadollahi-Baboli M, Mani-Varnosfaderani A (2009) Shuffling multivariate adaptive regression splines and adaptive neuro-fuzzy inference system as tools for QSAR study of SARS inhibitors. J Pharm Biomed Anal 50:853–860
Kollman PA, Massova I, Reyes C, Kuhn B et al (2011) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897
Mercader AG, Pomilio AB (2010) QSAR study of flavonoids and biflavonoids as influenza H1N1 virus neuraminidase inhibitors. Eur J Med Chem 45:1724–1730
Murumkar PR, Le L, Truong TN, Yadav MR (2011) Determination of structural requirements of influenza neuraminidase type A inhibitors and binding interaction analysis with the active site of A/H1N1 by 3D-QSAR CoMFA and CoMSIA modeling. Med Chem Commun 2:710–719
Roy P, Roy K (2002) On some aspects of variable selection for partial least squares regression models. QSAR Comb Sci 27:302–313
Salt DW, Ajmani S, Crichton R, Livingstone DJ (2007) An improved approximation to the estimation of the critical F values in best subset regression. J Chem Inf Model 47:143–149
Shahbazikhah P, Asadollahi-Baboli M, Khaksar R, Alamdaria RF, Zare-Shahabadi V (2011) Predicting partition coefficients of migrants in food simulant/polymer systems using adaptive neuro-fuzzy inference system. J Braz Chem Soc 22:1446–1451
Todeschini R, Consonni V (2000) Handbook of molecular descriptors. Wiley, Weinheim
Vander Heyden Y, Deconinck E, Hancock T, Coomans D, Massart DL (2005) Classification of drugs in absorption classes using the classification and regression trees (CART) methodology. J Pharm Biomed Anal 39:91–103
VanGunsterenv SR, Eising AA, Huenberger PH, Kruger P, Mark AE (1996) Biomolecular simulation: the GROMOS 96 manual and user guide. Verlag der Fachvereine, Zürich
Vapnik V (1998) Statistical learning theory. Wiley, New York
Xu Y, Liu Y, Jing F, Xie Y, Shi F, Fang H, Li M, Xu W (2011) Design, synthesis and biological activity of thiazolidine-4-carboxylic acid derivatives as novel influenza neuraminidase inhibitors. Bioorg Med Chem 19:2342–2348
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Asadollahi-Baboli, M., Mani-Varnosfaderani, A. Molecular docking, molecular dynamics simulation, and QSAR model on potent thiazolidine-4-carboxylic acid inhibitors of influenza neuraminidase. Med Chem Res 22, 1700–1710 (2013). https://doi.org/10.1007/s00044-012-0175-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-012-0175-y