Abstract
In this work, the multiple linear regression (MLR) and support vector machine (SVM) methods were applied for modeling and predicting the inhibitory activity of imidazopyridine derivatives with two-dimensional (2D) autocorrelation descriptors calculated from the molecular structure alone for the first time. We define the new objective function as fitness, and it can guide the particle swarm optimization (PSO) method to select important descriptors which are responsible for the inhibitory activity of these compounds. The square of the correlation coefficient (R 2 = 0.897), the square of correlation coefficients of the test set \( \left( {R_{\text{ext}}^{2} = 0.660} \right) \), and the obtained statistical parameter of the calibrating set in the PSO-SVM model was 0.743, which demonstrated the reliability of the model. The PSO-SVM model is superior over the PSO-MLR method in the dataset with imidazopyridine derivatives as Et-PKG inhibitors. Our best quantitative structure–activity relationship model illustrates the importance of an adequate distribution of atomic properties represented in topological frames and reveals atomic masses, van der Waals volumes, Sanderson electronegativities, and polarizabilities to be the most influential atomic properties in the structures of the imidazopyridine derivatives.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Protozoan parasites of the subphylum Apicomplexa are significant threats to both human and animal health worldwide (Diaz et al., 2006). Coccidiosis is caused by related apicomplexans; the Eimeria species of parasites, which invade intestinal epithelial cells, are the causative agents (Liang et al., 2007). Some of the most significant Eimeria species in poultry are E. tenella, E. acervulina, E. necartrix, E. brunetti, and E. maxima (Scribner et al., 2007). During acute infections, these parasites cause significant morbidity and mortality in broiler breeds of chicken. Hence, more than 35 billion chickens are raised annually worldwide and all major poultry operations use anticoccidial agents prophylactically (Scribner et al., 2007). Nearly 30 years ago, polyether ionophore anticoccidials were discovered, which have been successfully used as prophylactic agents to combat coccidiosis (Biftu et al., 2006). Not surprisingly, reports of development of resistance due to the extended and constant chemotherapeutic pressure exerted by this class of compounds are not uncommon (Gurnett et al., 2002). Thus, the need to identify and develop new drugs for the control of coccidiosis is critically important. Recently, some groups (Scribner et al., 2008) reported on some novel anticoccidial agents with potent in vitro and in vivo activity against Eimeria parasites. It was determined that inhibition of parasite growth by these compounds was due to inhibition of the parasite-specific cyclic GMP (cGMP)-dependent protein kinase (PKG). Cyclic GMP (cGMP)-dependent protein kinase (PKG) was biochemically purified from E. tenella (Donald et al., 2002), and it has recently been identified as the likely molecular target of a newly disclosed broad-spectrum coccidiostat (Salowe et al., 2002).
In previous studies, many groups were more interested in synthesis of novel compounds such as E. tenella cGMP-dependent protein kinase (Et-PKG) inhibitors. However, time and cost considerations make it unfeasible to carry out binding bioassays on every molecule. Alternatively, an untested molecule might be evaluated using the information from already obtained bioassays and the ability to build a quantitative structure–activity relationship (QSAR) model. The QSAR model seeks to discover and use mathematical relationships between chemical structure and biological activity. The approach does not depend on any experimental properties; it requires the molecular descriptors that can be calculated from the molecular structure alone. Once the structure of a compound is known, any molecular descriptor can be calculated, whether or not the compound is synthesized. When a reliable model is established, it can predict the inhibitory activity of compounds and evaluate the structural factors responsible for the inhibitory activity. So, in the last decade, computational-based rational design of drugs has increased rapidly. Most of these approaches are focused on using different kinds of molecular descriptors to encode chemical information (Doležal et al., 2009; Li et al., 2009; Mercader et al., 2008). Topological index, geometrical descriptors, and other descriptors are included in QSAR studies. The two-dimensional (2D) autocorrelation descriptor has been successfully used to construct same kinds of QSAR model for modeling biological activities (Saíz-Urra et al., 2007; Sharma et al., 2008; Caballero et al., 2008).
In the past several years, Ertepinar et al., (1995) applied the QSAR model to study a set of benzimidazole and imidazopyridine derivatives that have previously been tested for their antibacterial activities against Bacillus subtilis. The results revealed that the activity contributions of benzimidazoles and imidazopyridines against B. subtilis depend almost entirely on their relative lipophilic character as defined by their octanol/water partition coefficients, log P. Later, Curtin et al., (1998) discovered and evaluated a series of 3-acylindole imidazopyridine platelet-activating factor (PAF) antagonists. The PAF antagonist, an endogenous phospholipid inflammatory mediator, is a d-glycerol derivative bearing a phosphorylcholine at C3, an acetyl group at C2, and a long-chain alkyl ether moiety at C1. They applied structure–activity relationships (SARs) to study these compounds. The results indicated that modification of the indole and benzoyl spacer of the lead compound 6-(4-fluoro-phenyl)-3-(4-(2-methyl-imidazo[4,5-c]pyridin-3-ylmethyl)-benzoyl)-indole-1-carboxylic acid dimethylamide gave analogs that were more potent, longer-lived, and bioavailable. However, until recently, no authors have investigated a QSAR model for predicting inhibitory activity of imidazopyridine derivatives against Et-PKG.
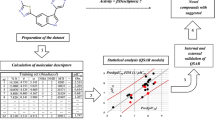
In this context, we calculated 723 descriptors by E-Dragon 1.0 software and present a modeling approach based on SVM to study imidazopyridine derivatives as Et-PKG inhibitors. The particle swarm optimization (PSO) method was used to preselect the proper descriptors from whole descriptor sets. The main purpose of the work is to develop a QSAR model for predicting inhibitory activity of the 107 imidazopyridine derivatives and to better understand the structural features of these compounds and their relation with the inhibitory activity using the 2D autocorrelation descriptor. This study may help us to design new analogs with a better biological profile.
Materials and computational methods
Materials
Experimental data
The studied compounds are 107 imidazopyridine derivatives, which were taken from the literature (Liang et al., 2007; Scribner et al., 2007, 2008; Biftu et al., 2006); their structures are shown in Table 1. These compounds were tested for in vitro efficacy using the Ten_K (Tenella kinase) assay. The Ten_K assay measures inhibition of Et-PKG activity, which is reported as the amount of compound required to inhibit activity by 50% (IC50; nM). And the 50% inhibitory concentration (IC50) values are listed in Table 1. If a compound was tested more than once and the corresponding two IC50 values approximated equality, the average IC50 value is listed in Table 1.
Descriptor calculation
To develop a QSAR model, molecular structures are often represented using molecular descriptors, which encode structural information. The calculation process of the descriptors involved the following steps: the structures of the compounds were drawn using Molinspiration WebME Editor (http://www.molinspiration.com:9080/mi/webme.html) and saved as C.SMI files. Then the C.SMI files were transferred into the software E-Dragon 1.0 (http://www.vcclab.org/lab/edragon/) to calculate different dimension structural descriptors. The software E-Dragon 1.0 can calculate Edge adjacency indexes, GETAWAY, WHIM, 2D autocorrelation indexes, Burden eigenvalues, and 3D MoRSE descriptors. And these descriptors have been successfully used in various QSAR/QSPR studies (Khan et al., 2009; Panek et al., 2005; Davood et al., 2009). In the prereduction step, the calculated descriptors were searched for constant values for all molecules; those detected descriptors were removed, and the other calculated descriptors were as the original variable set.
Data splitting
Rational division of the experimental data set into training, calibration, and test sets is a crucial step in the development and validation of reliable QSAR models. Ideally, this division is performed such that points representing the training, calibration, and test sets are distributed within the whole descriptor space occupied by the entire data set, and each point in the calibration and test set is close to at least one point in the training set. The k-MCA (k-means cluster analysis) may be used in training, calibration, and test series design (Kowalski and Wold, 1982). The idea consists of carrying out partition of the set of compounds under study into several statistically representative classes of chemicals. Thence, one may select from the members of all these classes the training, calibration, and test series. This procedure ensures that any chemical class (as determined by the clusters derived from k-MCA) will be represented in the compound series (training, calibration, and test). It permits the design of training, calibration, and test series which are representative of the entire experimental universe. As a result, k-MCA splits imidazopyridine derivatives into five clusters, with 13, 29, 36, and 45 training-set members and 24 calibration- and test-set members. Selection of the training, calibration, and test sets was carried out by randomly selecting compounds belonging to each cluster. Twenty-five compounds (see Table 1) were selected as the test set, another 25 compounds (see Table 1) were selected as the calibration set, and the remaining 57 compounds formed the training set. The training and calibration set were used to adjust the parameters of the models. And the test set was used to evaluate the predicted ability of the models once they were built.
Computational strategies
Variable selection strategies
The PSO algorithm was proposed by Kennedy and Eberhart, (1995). Similarly to other population-based algorithms, PSO can solve a variety of complicated optimization problems and shows a fast convergence rate in certain problems (Kennedy and Eberhart, 2001). In addition, it is very easy to configure and can be rapidly implemented.
For a population of M particles, particle i can be described as X i = (x i1 , x i2 ,…, x iD ), where i = 1, 2,…, M. Each particle has a velocity along each dimension, represented as V i = (v i1 , v i2 ,…,v iD ). It also memorizes its previous best position P i = (p i1 , p i2 ,…, p iD ) (also known as pbest) and the best position P g of all the particles (also known as gbest) (Pavlidis et al., 2005).
After finding the two best values, P i and P g , the particle updates its velocity according to the following formula:
where c 1 and c 2 are the learning factors that determine how far a particle will move in each iteration, r 1 and r 2 are the elements from two uniform random sequences in the range of [0, 1], and w is the inertia weight. After updating its velocity, the particle’s position is updated:
The velocity V i (t + 1) and position X i (t + 1) are generally confined to an area to prevent the particles from flying out of the solution space.
The iterations are represented by the parameter t in the above formulae. The iterations are terminated when the maximum generation or the designated fitness value is reached.
To select the most significant variables, we define a new objective function, fitness, which can be calculated as fellows:
where a and b are be clamped to the range (0,1); moreover, a+b = 1, R 2 denotes the square of correlation coefficients of the training set, and \( R_{v}^{2} \) denotes the square of correlation coefficients of the calibrating set. Figure 1 shows the flow diagram of the proposed PSO selection variable approach.
The new objective function, fitness, combined the result of the training set and calibration set to guide the PSO-selected important variables, and the values of a and b in the objective function are evaluated on the basis of the square of correlation coefficients \( \left( {R_{\text{ext}}^{2} } \right) \) of the test set. In the present work, to improve the predictive ability of the model, we apply the objective function fitness (\( {\text{fitness}} = a \times R^{2} + b \times R_{v}^{2} \)) to guide the PSO-selected most relevant descriptors from the pool of 96 2D autocorrelation descriptors. Optimized parameters of the objective function fitness are a = 0.4 and b = 0.6 on the basis of maximization of the square of correlation coefficients \( \left( {R_{\text{ext}}^{2} } \right) \) of the test set. Optimized parameters of PSO are as follows: a population of 20 particles, c 1 = c 2 = 1.8, V max = 0.2, X max = 1.0, X min = 0.0, and evolution for 100 generations. And the selected variables are reported in Table 2.
Methods of model construction
As mentioned in the previous section, the PSO method was used to select the most significant descriptors. The selected descriptors were used to construct some models by using multiple linear regression (MLR) and support vector machine (SVM) (Chihchung and Chihjen, 2009) techniques; these models are called PSO-MLR, and PSO-SVM. In each SVM model, the Gaussian radial basis function kernel was used because of its effectiveness and speed in the training process. The following SVM parameters were chosen to be optimized: c, cost (set the parameter C); ν (set the parameter ν of ν-SVC); and g, γ (set γ in kernel function [default 1/k]). The values of optimized SVM parameters are listed in Table 3.
All algorithms were written in MATLAB and run on a personal computer [Intel(R) Pentium(R), 4/3.20 GHz, 1.00-GB RAM].
Results and discussion
The studied data set includes selective compounds which are potent inhibitors of E. tenella cGMP-dependent protein kinase (Et-PKG). Et-PKG is essential for survival and represents a desirable therapeutic target (Scribner et al., 2007). High-throughput screening of known kinase inhibitors resulted in the discovery of imidazopyridine derivatives as PKG inhibitors and broad-spectrum anticoccidial agents (Scribner et al., 2008). The imidazopyridine analogs were tested for in vitro efficacy via the Ten_K (Tenella kinase) assay, which measures inhibition of Et-PKG enzyme activity.
QSAR model
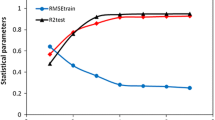
Three 2D autocorrelation MLR and SVM models are reported in this work, respectively. In total, 21 descriptors were employed from the whole 2D autocorrelation pool. Statistical parameters of the two QSAR models are listed in Table 2. As reported in the table, the PSO-SVM model has a higher value of the square of the correlation coefficient (R 2 = 0.897), the adjusted square regression coefficient \( \left( {R_{\text{adj}}^{2} = 0.872} \right) \), the Fisher ratio (F = 35.727), and the Kubinyi, (1994a, b) function (FIT = 2.208) and a lower value of the standard deviation (SD = 0.412) and Akaike’s information criterion (AIC = 84.154) (Kuzmic et al., 2006) than the PSO-MLR model does. So the statistical parameters of the PSO-SVM model are more robust than those of the PSO-MLR model. Comparison of the square of correlation coefficients \( \left( {R_{\text{ext}}^{2} } \right) \) of the test set, the value of PSO-SVM model was higher than another. From the comparison of the calibrating set results, the data indicate that the model is reliable and accurate.
Using the PSO-SVM model, satisfactory results were obtained. The results are shown in Fig. 2. With this model, R 2 for the training set was increased to 0.897 and S was reduced to 0.412. For the calibration set, \( {R_{\rm{v}}^{2} } \) was increased to 0.743, while S was reduced to 0.540. For the test set, R 2 was increased to 0.660, while S was reduced to 0.975, showing the good generalization ability of the SVM model. The smaller scatter of data points in Fig. 2 demonstrates that the SVM model is clearly superior both in fitness and in prediction performance.
The PSO-MLR model gave a squared standard error (s) of 0.872 for the training set, 0.925 for the calibration set, and 1.59 for the test set, and the corresponding correlation coefficients (R 2) were 0.550, 0.304, and 0.166, respectively.
Comparing the PSO-SVM and PSO-MLR models developed above, the PSO-SVM model is expected to be a better predictor for Et-PKG inhibitors than the PSO-MLR model. This indicates the good generalization capability of the PSO-SVM model. This is also demonstrated in Fig. 3, where we compared the predicted results for the test set alone by the PSO-SVM and PSO-MLR models. Clearly, the results using the nonlinear model show a relatively smaller bias than those using the PSO-MLR model.
Figures 4 and 5 show further comparison of the results using the PSO-SVM and PSO-MLR models, which plots the experimental IC50 versus the relative deviation of the corresponding predicted and experimental IC50 values. The plot shows that the MLR model can correctly predict rate constants for 57.89, 60.00, and 56.00% compounds in the training, calibration, and test sets, respectively, within a relative error of between –1.5 and 1.5; for the SVM model, the corresponding proportions were 70.18, 72.00, and 64.00%, respectively. Within a relative error of between –2.0 and 2.0, the SVM model gave a corresponding accuracy of 80.70, 88.00, and 76.00%, respectively, whereas the MLR model can only predict 64.91, 68.00, and 64.00% of the compounds in the respective data set.
As we explained previously, one of the objectives of the current work was to compare the reliability and applicability of the 2D autocorrelation descriptors to describe the property under study compared with those of other different descriptors. Consequently, we developed another six models using the same data set that was included in the 2D autocorrelation QSAR model. The results obtained with Edge adjacency indexes, WHIM, 2D autocorrelation indexes, Burden eigenvalues, 3D MoRSE, and GETAWAY descriptors (Todeschini and Consonni, 2000) are listed in Table 4.
As reported in Table 4, the value of R 2 is <0.670 for all approaches except the 2D autocorrelation, which has an R 2 = 0.897. This approach also yields the best value for other statistical parameters like the standard deviation (S) of the training set and the AIC, which has the lowest value in comparison with the rest of the approaches. In similarly, the Fisher ratio (F) and the Kubinyi function (FIT) are the highest. Additional, the 2D autocorrelation descriptor presents the best R 2v and R 2ext values, which implies that the PSO-SVM model with the 2D autocorrelation descriptor has better predictive ability than the other models. For all these reasons, we consider that the 2D autocorrelation descriptor can be a useful tool for prediction of inhibitory activities taking into account the imidazopyridine derivatives.
Variables’ interpretation of the best model
In the text, the 11 most significant descriptors have been selected by PSO as independent variables of the best model: ATS3m, ATS7m, ATS8m, ATS5v, ATS7v, ATS8e, MATS1v, MATS1e, MATS3e, MATS1p, and GATS5p.
The 2D autocorrelation descriptors represent the topological structure of the compounds but are more complex in nature than the classical topological descriptors. Computation of these descriptors involves the summations of different autocorrelation functions corresponding to different structural lags and leads to different autocorrelation vectors corresponding to the lengths of substructural fragments. Hence, it can distinguish the details of important substructural differences, however, the ‘‘traditional’’ descriptors (for example, log P or pK a ) cannot solve these questions. In the last decades, the 2D autocorrelation descriptor has been proven advantageous for establishing a QSAR model (Saíz-Urra et al., 2007; Caballero et al., 2006; Bauknecht et al. 1994; Moreau and Broto, 1980). Three spatial autocorrelation vectors are employed for modeling inhibitory activities: Broto-Moreau’s autocorrelation coefficients [ATS; Eq. 6] (Moreau and Broto, 1980), Moran’s, (1950) indexes (MATS; Eq. 7]), and Geary’s, (1954) coefficients (GATS; Eq. 8].
where ATS(p k , l), MATS(p k , l), and GATS(p k , l) are Broto-Moreau’s autocorrelation coefficients, Moran’s indexes, and Geary’s coefficients at spatial lag l, respectively; p ki and p kj are the values of physicochemical properties (i.e., atomic masses, atomic van der Waals volumes, atomic Sanderson electronegativities, and atomic polarizabilities) k of atoms i and j, respectively; \( \overline{p}_{k} \) is the average value of property k; and δ(l, d ij ) is a Dirac-delta function defined as
where d ij is the topological distance or spatial lag between atom i and atom j.
The 2D autocorrelation descriptors can be obtained by summing up the products of certain properties of the two atoms located at a given topological distance or spatial lag. There are slight differences among the descriptors of type ATSd, GATSd, and MATSd (Saíz-Urra et al., 2007), but in general, they describe how the considered property is distributed along the topological structure. The most important factor in interpreting them in the model is the topological distance, once weighted equally.
In point of fact, the best model selected an optimum descriptor combination which includes atomic masses, van der Waals volumes, Sanderson electronegativities, and polarizabilities as the most relevant key features. This result illustrates that a certain distribution of these properties is necessarily required to typify the imidazopyridine derivatives.
Aiming to figure out the impact of each input in the model, we performed a sensibility analysis in two ways (Cherqaoui et al., 1998). In the first, the descriptor under study is removed from the model and the statistical coefficient \( R_{\text{train}}^{2} \) for the training set is analyzed. Comparison between these R 2 values and that calculated when no descriptor is removed gives an idea of the importance of the descriptor removed. In the other method, the mean of the absolute deviation values △mi between the observed and the estimated value for all compounds is calculated. Finally, the contribution factor C i (Cherqaoui et al., 1998) of descriptor i (i = 1–11) is given by
The results of the sensitivity analysis are reported in Table 5. As can be seen, greater decreases in R 2 correspond to higher values of C i . Among the 11 descriptors in the best model, the descriptors with the highest impacts (C i > 9.5%) are GATS5p, ATS8m, ATS5v, and ATS8e. GATS5p indicates that the presence of polarizable atoms at a topological distance of 5 contributes positively to inhibition of Et-PKG enzyme activity. A possible polarizable atom pair at a topological distance of 5 is presented for compound 32, the top inhibitor of Et-PKG enzyme activity in the calibration and test sets. Likewise, ATS8m, ATS5v, and ATS8e indicate that the interaction between each pair of atoms at topological distances of 8, 5, and 8 contributes to the inhibition of Et-PKG enzyme activity, weighted by atomic masses, van der Waals volumes, and Sanderson electronegativities, respectively.
This reflects the fact that the effect of van der Waals volumes on inhibitory activities (IC50) is greater (27.23%) than that of atomic masses (27.03%), Sanderson electronegativities (27.09%), and polarizabilities (18.65%). The vector of ATS7v is weighted by van der Waals volumes representing topological substructures of size 7. The impact of IC50 is similar to that ATS8m. The other descriptors, ATS3m, ATS7m, MATS1v, MATS1e, MATS3e, and MATS1p, influence inhibitory activities to about the same extent that ATS8m does.
This fact may be viewed in terms of association of activity information content with structural fragments of such size and could be related to the great importance of an adequate molecular size and/or shape for proper matching with the receptor. However, further interpretation of the information content of these descriptors is very complex, as their computations involve integration of the structural fragments, and thus it is not possible to traverse backward from a higher state to a lower one (Fernández et al., 2005). These facts make our model interesting mainly as a predictive tool rather than for designing trends that are encoded within the model. Regarding this, our PSO-SVM model related to the 2D autocorrelation descriptor predictor should be useful to predict inhibitory activities of the new synthetic imidazopyridine derivatives against Et-PKG using the Ten_K assay in vitro.
Conclusion
We applied QSAR methodology to model the inhibitory activities of 107 imidazopyridine derivatives using 2D autocorrelation descriptors. To select the pertinent variables, we carried out PSO searches. Satisfactory quantitative models were obtained using the SVM method, achieving good results in calibration-set validations and external predictions. The present work demonstrates that the PSO-SVM approach is a robust and sensitive method to apply to QSAR research of imidazopyridine derivative inhibitory activities. In addition, comparisons with other descriptors such as the 3D MoRSE, Edge adjacency indexes, WHIM, 2D autocorrelation indexes, Burden eigenvalues, and GETAWAY descriptors were also carried out. The statistical parameters of the model and the validation results produced by the methodology that we propose are superior to those produced by the other descriptors. In this sense, the combination of SVM with PSO led to a useful method for the scientific community interested in QSAR development for imidazopyridine derivative inhibitory activities.
References
Bauknecht H, Zell A, Bayer H, Levi P, Wagener M, Sadowski J, Gasteiger J (1994) Locating biologically active compounds in medium-sized heterogeneous datasets by topological autocorrelation vectors: dopamine and benzodiazepine agonists. J Chem Inf Comput Sci 36:1205–1213
Biftu T, Feng D, Fishe M, Liang GB, Qian XX, Scribne A, Dennis R, Lee S, Liberato PA, Brown C, Gurnett A, Leavitt PS, Thompson D, Mathew J, Misura A, Samaras S, Tamas T, Sina JF, McNulty KA, McKnight CG, Schmatz DM, Wyvratt M (2006) Synthesis and SAR studies of very potent imidazopyridine antiprotozoal agents. Bioorg Med Chem Lett 16:2479–2483
Caballero J, Garriga M, Fernández M (2006) 2D Autocorrelation modeling of the negative inotropic activity of calcium entry blockers using Bayesian-regularized genetic neural networks. Bioorg Med Chem 14:3330–3340
Caballero J, Fernández M, González-Nil FD (2008) Structural requirements of pyrido[2,3-d]pyrimidin-7-one as CDK4/D inhibitors: 2D autocorrelation, CoMFA and CoMSIA analyses. Bioorg Med Chem 16:6103–6115
Cherqaoui D, Esseffar M, Villemin D, Cense JM, Chastrette M, Zakarya D (1998) Structure-musk odour relationship studies of tetralin and indan compounds using neural networks. New J Chem 22:839–843
Chihchung C, Chihjen L (2009) LIBSVM—a library for support vector machines. Available at: http://www.csie.ntu.edu.tw/~cjlin/libsvm
Curtin ML, Davidsen SK, Heyman HR, Garland RB, Sheppard GS, Florjancic AS, Xu LH, Carrera GM, Steinman DH, Trautmann JA, Albert DH, Magoc TJ, Tapang P, Rhein DA, Conway RG, Luo GJ, Denissen JF, Marsh KC, Morgan DW, Summers JB (1998) Discovery and evaluation of a series of 3-acylindole imidazopyridine platelet-activating factor antagonists. J Med Chem 41:74–95
Davood A, Nematollahi A, Iman M, Shafiee A (2009) Computational studies of new 1,4-dihydropyridines containing 4-(5)-chloro-2-ethyl-5-(4)-imidazolyl substituent: QSAR and docking, Med Chem Res. doi:10.1007/s00044-009-9171-2
Diaz CA, Allocco J, Powles MA, Yeung L, Donald RGK, Anderson JW, Liberator PA (2006) Characterization of Plasmodium falciparum cGMP-dependent protein kinase (PfPKG): antiparasitic activity of a PKG inhibitor. Mol Biochem Parasitol 146:78–88
Doležal R, Damme SV, Bultinck P, Waisser K (2009) QSAR analysis of salicylamide isosteres with the use of quantum chemical molecular descriptors. Eur J Med Chem 44:869–876
Donald RGK, Allocco J, Singh SB, Nare B, Salowe SP, Wiltsie J, Liberator PA (2002) Toxoplasma gondii cyclic GMP-dependent kinase: chemotherapeutic targeting of an essential parasite protein kinase. Eukaryot Cell 1:317–328
Ertepinar H, Gök Y, Geban Ö, Zden SÖ (1995) A QSAR study of the biological activities of some benzimidazoles and imidazopyridines against Bacillus subtilis. Eur J Med Chem 30:171–175
Fernández M, Caballero J, Morales AH, Castro EA, González MP (2005) Quantitative structure–activity relationship to predict differential inhibition of aldose reductase by flavonoid compounds. Bioorg Med Chem 13:3269–3277
Geary RF (1954) The contiguity ratio and statistical mapping. Incorp Stat 5:115–145
Gurnett AM, Liberator PA, Dulski PM, Salowe SP, Donald RGK, Anderson JW, Wiltsie J, Diaz CA, Harris G, Chang B, Darkin-Rattray SJ, Nare B, Crumley T, Blum PS, Misura AS, Tamas T, Sardana MK, Yuan J, Biftu T, Schmatz DM (2002) Purification and molecular characterization of cGMP-dependent protein kinase from Apicomplexan paras. J Biol Chem 277:15913–15922
Kennedy J, Eberhart RC (1995) Particle swarm optimization. In: Proceedings of the IEEE International joint conference on neural networks. Vol 4, pp 1942–1948
Kennedy J, Eberhart RC (2001) Swarm intelligence. Morgan Kaufmann Publishers, San Francisco
Khan AKR, Sahu VK, Singh RK, Khan SA (2009) Comparative QSTR study of saturated alcohols based on topological, constitutional, geometrical, and getaway descriptors. Med Chem Res. doi: 10.1007/s00044-009-9166-z
Kowalski RB, Wold S (1982) Pattern recognition in chemistry. In: Krishnaiah PR, Kanal LN (eds) Handbook of statistics. North-Holland, Amsterdam, pp 673–677
Kubinyi H (1994a) Variable selection in QSAR studies. I. An evolutionary algorithm. Quant Struct Act Relat 13:285–294
Kubinyi H (1994b) Variable selection in QSAR studies. II. A highly efficient combination of systematic search and evolution. Quant Struct Act Relat 13:393–401
Kuzmic P, Cregar L, Millis SZ, Goldman M (2006) Mixed-type noncompetitive inhibition of anthrax lethal factor protease by aminoglycosides. FEBS J 273:3054–3062
Li F, Chen JW, Wang ZJ, Li J, Qiao XL (2009) Determination and prediction of xenoestrogens by recombinant yeast-based assay and QSAR. Chemosphere 74:1152–1157
Liang GB, Qian XX, Feng D, Fishe M, Brown CM, Gurnett A, Leavitt PS, Liberato PA, Misura AS, Tamas T, Schmatz DM, Wyvratt M, Biftu T (2007) Synthesis and SAR studies of potent imidazopyridine anticoccidial agents. Bioorg Med Chem Lett 17:3558–3561
Mercader AG, Duchowicz PR, Fernández FM, Castro EA (2008) Modified and enhanced replacement method for the selection of molecular descriptors in QSAR and QSPR theories. Chemometr Intell Lab 92:138–144
Moran PAP (1950) Notes on continuous stochastic phenomena. Biometrika 37:17–23
Moreau G, Broto P (1980) The autocorrelation of a topological structure: a new molecular descriptor. Nouv J Chim 4:359–360
Panek JJ, Jezierska A, Vračko M (2005) Kohonen network study of aromatic compounds based on electronic and nonelectronic structure descriptors. J Chem Inf Model 45:264–272
Pavlidis NG, Parsopoulos KE, Vrahatis MN (2005) Computing Nash equilibria through computational intelligence methods. J Comput Appl Math 175:113–136
Saíz-Urra L, González MP, Teijeira M (2007) 2D-autocorrelation descriptors for predicting cytotoxicity of naphthoquinone ester derivatives against oral human epidermoid carcinoma. Bioorg Med Chem 15:3565–3571
Salowe SP, Wiltsie J, Liberator PA, Donald RGK (2002) The role of a parasite-specific allosteric site in the distinctive activation behavior of Eimeria tenella cGMP-dependent protein kinase. Biochemistry 41:4385–4391
Scribner A, Dennis R, Hong J, Lee S, McIntyre D, Perrey D, Feng D, Fisher M, Wyvratt M, Leavitt P, Liberator P, Gurnett A, Brown C, Mathew J, Thompson D, Schmatz D, Biftu T (2007) Synthesis and biological activity of imidazopyridine anticoccidial agents: part I. Eur J Med Chem 42:1334–1357
Scribner A, Dennis R, Lee S, Ouvry G, Perrey D, Fisher M, Wyvratt M, Leavitt P, Liberator P, Gurnett A, Brown C, Mathew J, Thompson D, Schmatz D, Biftu T (2008) Synthesis and biological activity of imidazopyridine anticoccidial agents: part II. Eur J Med Chem 43:1123–1151
Sharma S, Prabhakar YS, Singh P, Sharma BK (2008) QSAR study about ATP-sensitive potassium channel activation of cromakalim analogues using CP-MLR approach. Eur J Med Chem 43:2354–2360
Todeschini R, Consonni V (2000) Handbook of molecular descriptors. Wiley, Weinheim
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cheng, Z., Zhang, Y. & Zhang, W. QSAR studies of imidazopyridine derivatives as Et-PKG inhibitors using the PSO-SVM approach. Med Chem Res 19, 1307–1325 (2010). https://doi.org/10.1007/s00044-009-9272-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-009-9272-y