Abstract
Many pathogenic bacteria require flagella-mediated motility to colonise and persist in their hosts. Helicobacter pylori and Campylobacter jejuni are flagellated epsilonproteobacteria associated with several human pathologies, including gastritis, acute diarrhea, gastric carcinoma and neurological disorders. In both species, glycosylation of flagellin with an unusual sugar pseudaminic acid (Pse) plays a crucial role in the biosynthesis of functional flagella, and thereby in bacterial motility and pathogenesis. Pse is found only in pathogenic bacteria. Its biosynthesis via six consecutive enzymatic steps has been extensively studied in H. pylori and C. jejuni. This review highlights the importance of flagella glycosylation and details structural insights into the enzymes in the Pse pathway obtained via a combination of biochemical, crystallographic, and mutagenesis studies of the enzyme–substrate and –inhibitor complexes. It is anticipated that understanding the underlying structural and molecular basis of the catalytic mechanisms of the Pse-synthesising enzymes will pave the way for the development of novel antimicrobials.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Helicobacter pylori and Campylobacter jejuni are representative members of the epsilon (ε) Proteobacteria. H. pylori and C. jejuni infections cause a broad range of human and animal diseases. H. pylori is found primarily in humans, where it colonises gastric mucosa. H. pylori is the etiological agent of dyspepsia, gastritis, duodenal and gastric ulcers, mucosa-associated lymphoid tissue lymphoma and gastric carcinoma [1,2,3,4,5,6]. In addition, association between H. pylori infection and extra-gastric diseases including iron-deficiency anaemia [7, 8], idiopathic thrombocytopenic purpura [9, 10], cardiovascular diseases [11], chronic liver disorder [12], pancreatic cancer [13], chronic respiratory illness [14], skin diseases [15] and diabetes [16] has been reported. The presence of H. pylori significantly affects the natural microecology of the stomach [17]. It is the first bacterium to be classified as a group I (definite) carcinogen for human gastric cancer by the International Agency for Research on Cancer [18]. Around 75% of gastric cancer and 90% of duodenal ulcer patients had a previous history of H. pylori infection [19, 20]. The clinical outcome of H. pylori infection is determined by a complex interplay between the genetic properties of the bacteria and host genetic factors [21], and may be influenced by co-infection with other bacterial pathogens [22, 23].
The most common mode of transmission is thought to be by person-to-person contact, via gastro-oral, oral–faecal, and oral–oral routes [24, 25], although the full spectrum of H. pylori transmission routes is yet to be determined. Multiple socioeconomic, environmental and behavioural factors contribute to the acquisition and spread of infection. These factors include low socioeconomic status [26, 27], diet [28], use of tobacco [29], a higher number of siblings [30] and a lower educational status of the parents [31, 32]. The prevalence of H. pylori infection increases with age [33] and is significantly higher in developing countries compared to developed countries. About 80% of individuals in developing countries harbour this bacterium, compared with 15–50% of the population in developed ones [34, 35]. For example, a 2011 nationwide study in Australia showed that the prevalence of H. pylori was 15% among adults [35]. The occurrence of H. pylori infection has been decreasing in developed countries [36, 37], largely due to improving hygiene practices and higher standards of living. However, the relative prevalence in developed countries remains significantly higher among people with lower socioeconomic status, such as indigenous and migrant populations [35, 38].
Currently, there is no effective vaccine or single drug available to treat H. pylori infection. The common therapy for the treatment of such infections is a combination of a proton pump inhibitor and two or three antibiotics [39, 40]. However, the effectiveness of this therapy has significantly declined, mainly due to the widespread increase in resistance to clarithromycin and metronidazole [41, 42].
Campylobacter jejuni is a zoonotic pathogen that usually colonises the gut of birds and mammals, but can also infect humans, with variable clinical outcomes including mild, self-limiting, non-inflammatory diarrhea, severe, inflammatory, bloody diarrhea with pyrexia, abdominal cramps, inflammatory bowel disease, Barrett’s oesophagus and irritable bowel syndrome [43]. C. jejuni infections are also associated with acute cholecystitis and celiac disease [43]. The sequelae of the infection include neurological and autoimmune diseases such as Guillain–Barré syndrome [44], Reiter syndrome [45] and reactive arthritis [46].
The prevalence of C. jejuni varies significantly between countries. In developed countries, human infections occur sporadically and at a low frequency [47, 48]. In developing countries, campylobacteriosis is endemic, and asymptomatic infections are common [47, 49]. It is believed that contaminated poultry is the main source of C. jejuni infection [47, 50, 51].
Factors that contribute to the pathogenicity and virulence of H. pylori include the cag pathogenicity island [52], vacuolating cytotoxin A [53], adhesins (including blood-antigen binding protein A [54], sialic acid-binding adhesin [55], heat-shock protein 60 [56], adherence-associated proteins [57], outer membrane protein HopZ [58], N,N′-diacetyllactosediamine-binding adhesin [59] and neutrophil-activating protein [60]), duodenal ulcer promoting gene [61], urease [62], and γ-glutamyl transpeptidase [63]. Furthermore, chemotaxis [64] and flagella-mediated motility [64, 65] have been shown to also play an important role in the development of a robust infection.
In C. jejuni, a diverse group of virulence factors including flagella-mediated motility, chemotaxis, iron acquisition, adhesion, invasion of epithelial cells and quorum sensing contributes to the development of successful infection [64, 66, 67]. The role of motility and chemotaxis in C. jejuni pathogenesis has been the subject of intensive studies, particularly since it has been shown that flagellin biosynthesis and modification of genes are important for colonisation [68, 69].
Bacteria use flagella-driven motility to relocate toward favourable environments and away from toxic chemicals. Flagella-mediated motility allows H. pylori and C. jejuni to penetrate the gastric/intestinal mucus and reach the underlying layer of epithelial cells, where they colonise. In addition, these bacteria rely on their high motility in the viscous layer covering the gastric mucosa of the host to persist despite the natural flow of the gastrointestinal mucus [64]. H. pylori uses motility to colonise the stomach [70] and to establish robust infection [71]. Chemotaxis plays an important role in this, as it enables H. pylori to sense, and responds to, changes in various conditions, including pH, concentration of urea/ammonium and cellular energy status [64]. Deletion of motility-associated genes in C. jejuni and H. pylori was shown to result in attenuated growth in animal models [71, 72]. Furthermore, wild-type H. pylori was shown to have a significantly lower minimum infectious dose in animal models than its non-motile or nonchemotactic mutants [71, 73].
Helicobacter and Campylobacter flagella
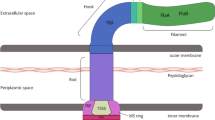
Helicobacter pylori and C. jejuni possess unipolar and bipolar flagella, respectively [66, 74, 75]. The flagellum is assembled from ~ 40 different proteins [76, 77] and has three components: a membrane-embedded basal body, a hook, and a long filament [78, 79]. Interestingly, each H. pylori flagellar filament is covered by a sheath (extension of the outer membrane) which is believed to serve as a protective shield against the low pH of the human stomach [80]. The basal body together with the stator proteins serves as motor that turns the extracellular helical-shaped filament at its base. Cryo-electron tomography studies revealed that the H. pylori basal body possesses a unique periplasmic cage-like structure [75]. This cage is not present in C. jejuni; however, C. jejuni possesses an additional, large periplasmic basal disk at a similar position [81]. It was proposed that these additional structures serve as a robust scaffold for recruitment of stator complexes to the motor, an important feature thought to be linked to the high motility of H. pylori and C. jejuni in viscous environments [75, 81].
The central rod component of the basal body is connected via a hook to a helical filament. The flagellar filament is composed of two proteins, the major flagellin FlaA and the minor flagellin FlaB [82]. The flagellins are exported via the type III secretion apparatus within the basal body following translation and post-translational modification in the cytoplasm [83].
The role of FlaA and FlaB in the biosynthesis of fully functional flagella in C. jejuni and H. pylori has been investigated [84, 85]. In H. pylori, FlaA (53 kDa) and FlaB (54 kDa) show around 56% amino acid sequence identity [85]. A mutagenesis study revealed that isogenic H. pylori flaA mutants were aflagellated and non-motile, while the flaB mutants were flagellated but only partially motile [86, 87]. The C. jejuni FlaA (60 kDa) and FlaB (60 kDa) are more similar to each other (92–95% sequence identity) [82]. The C. jejuni flaA mutant had a truncated flagellum and was unable to colonise the host [88]. Although the C. jejuni flaB gene is not essential for motility [87], its product was reported to play an important role in the defence of C. jejuni against bacteriophage infection [89]. In 1999, it became evident that flagellins of Helicobacter and Campylobacter spp. are glycosylated [87, 90]. C. jejuni flagellins are among the most heavily glycosylated proteins identified to date, with the carbohydrate moieties contributing up to ~ 10% of the total molecular weight [91,92,93].
Protein glycosylation
Glycosylation (a covalent addition of sugar moieties) of proteins is a ubiquitous co-translational or post-translational modification occurring in all kingdoms of life. More than two-thirds of eukaryotic proteins are believed to be glycosylated [94]. Since the discovery of glycoproteins in archaea Halobacterium sp. and hypothermophilic bacteria Clostridium sp. [95, 96], protein glycosylation in microorganisms has been studied extensively, yielding insights into the structure of the oligosaccharide building blocks of the glycan moieties and the mechanisms of glycosylation [97].
Classification of glycosylation
The three most common types of protein glycosylation, classified according to the nature of the atom of the amino acid to which the sugar moiety is attached, are (1) N-linked, (2) C-linked, and (3) O-linked glycosylation.
N-linked glycosylation
N-linked glycosylation is a common type of post-translational modification of secreted or membrane-embedded proteins, where a sugar molecule is attached to a nitrogen atom (usually the N4 atom of asparagine residues) by oligosaccharyltransferase (OTase) [98]. N-glycosylation contributes to folding, stability, and function of a wide range of proteins that play a role in the regulation of cell differentiation, cell signalling and pathogenesis [99,100,101]. In eukaryotes, the assembly of the building blocks of the glycan occurs at the endoplasmic reticulum (ER) membrane, while in prokaryotes, this process takes place at the plasma membrane. Preassembled blocks of 14 sugars, containing two N-acetylglucosamine, nine mannose, and three glucose residues, are transferred onto the lipid anchor to generate a lipid-linked oligosaccharide (LLO). The LLO is then flipped from the cytosolic side to the luminal face of the eukaryotic ER or to the outer layer of the plasma membrane in prokaryotes, where it serves as a glycosyl donor for the transfer reaction catalysed by OTase [102]. Proteins harbouring the conserved sequence Asn-X-Ser/Thr/Cys in eukaryotes [98, 103] and Asp/Glu-X-Asn-X-Ser/Thr in bacteria [104, 105] act as acceptors in the ER lumen or the bacterial extracytoplasmic space, respectively [102]. Following the transfer, the N-glycan can be modified via terminal glycosylation which gives rise to its structural diversity.
C-linked glycosylation/C-mannosylation
C-linked glycosylation (also known as C-mannosylation) is a relatively rare event that is defined as the covalent attachment of mannose by specific mannosyltransferase to the indole C2 carbon atom of a tryptophan residue on an acceptor protein via a C–C link [106, 107]. This type of glycosylation has been reviewed elsewhere [108, 109]. C-mannosylation is restricted to mammals and related species [110]. The consensus motif for C-linked glycosylation is Trp-X-X-Trp or Trp-X-X-Cys/Phe (where X can be any residue except proline), and the addition of the mannose sugar usually occurs at the first Trp residue [108, 111, 112], although in a motif (Trp-X-X-Trp-X-X-Trp) C-mannosylation can occur on all tryptophan residues [110, 113]. This type of post-translational modification plays a role in protein folding, stability and cell signalling mechanisms [109, 110].
O-linked glycosylation
O-glycosylation is a covalent linkage of a sugar to the side-chain hydroxyl oxygen of serine, threonine, tyrosine, hydroxylysine or hydroxyproline residue [114, 115]. Proline-rich sequences (Thr-Ala-Pro-Pro, Thr-Val-X-Pro, Ser/Thr-Pro-X-Pro and Thr-Ser-Ala-Pro, where X can be any amino acid) are usually preferred for O-glycosylation [114]. However, the consensus sequence of O-glycosylation is yet to be identified.
O-glycosylation in eukaryotes takes place in the Golgi apparatus, ER, and cytoplasm [116,117,118] and involves the sequential addition of nucleotide-activated monosaccharides to acceptor proteins [116]. Sugar units can be of a different chemical nature, which is the source of diversity of O-linked glycan chains. O-linked N-acetylgalactosamine glycans (O-GalNAc) are the most abundant type. They are commonly found in, for example, mucins, fetuin, gonadotropins, and glycophorins [114, 119]. O-linked glycans incorporating N-acetylglucosamine, fucose, xylose, galactose, mannose, glucose, and arabinose have also been found [119, 120].
In prokaryotes, O-glycosylation is believed to occur in the cytoplasm or inner membrane [116]. Two distinct mechanisms of bacterial O-glycosylation have been identified: OTase-dependent and OTase-independent [115]. As the first step of the OTase-dependent O-glycosylation, an initiating glycosyltransferase (GT) catalyses the attachment of the first nucleotide-activated monosaccharide to the membrane-embedded lipid carrier (undecaprenolphosphate) on the cytoplasmic side of the inner membrane of the cell [121]. Further individual monosaccharide groups are subsequently added one by one by different GTs. Then the undecaprenolphosphate-linked glycan is flipped onto the periplasmic side of the membrane and transferred onto the acceptor protein by OTase [121]. The OTase-dependent O-glycosylation pathway appears to exist only in Gram-negative bacteria [122]. A wide range of structurally and functionally diverse membrane-associated proteins, including the components of type IV pili, has been reported to be glycosylated through this mechanism [115, 123]. OTase-independent O-glycosylation, where nucleotide-activated monosaccharides are directly transferred onto acceptor proteins by cytoplasmic GTs, has been observed for proteins that are exported to the outer membrane (e.g. adhesins, autotransporters) or secreted by the basal body [115]. One important function of O-glycosylation is to help pathogenic bacteria to evade host defence mechanisms [92, 93, 115, 124, 125]. The remaining part of this review focuses on the OTase-independent O-glycosylation of flagellins in the representative members of ε-Proteobacteria C. jejuni and H. pylori.
Pseudaminic acid biosynthesis pathway
Flagellin modification via O-linked glycosylation has been characterised in many bacteria, including H. pylori and C. jejuni [91, 115, 126]. C. jejuni FlaA is glycosylated at up to 19 sites [93]; C. jejuni FlaB is also glycosylated, but the exact number of sites remains to be established [127]. FlaA and FlaB in H. pylori are glycosylated at seven and ten sites, respectively [92]. H. pylori decorates its flagellins with glycans that contain only one type of sugar, the sialic acid-like nonulosonate pseudaminic acid (Pse, 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-α-l-manno-2-nonulopyranosonic acid) [91]. In contrast, O-linked glycans attached to C. jejuni flagellins can contain more than one type of sugar, including Pse, legionaminic acid and related derivatives of nonulosonate [93] (Fig. 1). The Pse biosynthesis has been extensively studied in H. pylori and C. jejuni. Nucleotide-activated pseudaminic acid is synthesised via six consecutive enzymatic steps, illustrated in Fig. 2 [128, 129], and then transferred onto flagellins via O-linked glycosylation.
The Pse biosynthesis pathway in H. pylori and C. jejuni [129]
Step one
The first step of the Pse biosynthesis pathway is catalysed by the uridine-5′-diphosphate N-acetylglucosamine (UDP-GlcNAc) 5-inverting 4,6-dehydratase (also known as pseudaminic acid biosynthesis protein B, PseB, EC 4.2.1.115). PseB is a bifunctional enzyme that belongs to the short-chain dehydrogenase/reductase (SDR) superfamily and uses nicotinamide adenine dinucleotide phosphate (NADP+) as a cofactor [130]. PseB catalyses oxidation at the C-4″ atom of the nucleotide sugar UDP-GlcNAc by adding a ketone group, and a subsequent reduction at the C-6″ atom to generate UDP-2-acetamido-2,6-dideoxy-β-l-arabino-4-hexulose (UDP-6-deoxy-4-keto-HexNAc) (Fig. 2) [130,131,132]. PseB from H. pylori and C. jejuni share 63% amino acid sequence identity and follow the same catalytic mechanism [131]. In H. pylori, this enzyme is also involved in the alteration of the O-antigen composition in the lipopolysaccharide (LPS) and the modulation of the urease activity, in addition to its role in the flagellin glycosylation [133]. Inactivation of the pseB gene in H. pylori resulted in an aflagellated non-motile phenotype which suggested that PseB likely plays an important role in bacterial pathogenesis and colonisation [133].
Helicobacter pylori PseB (HpPseB, 333 aa) exists as a hexamer both in solution and in crystal [132]. Each HpPseB monomer consists of two lobes: an N-terminal large lobe (residues 1–174, 208–234, and 265–317) and C-terminal small lobe (residues 175–207, 235–264, and 318–333) (Fig. 3). The N-terminal lobe adopts a Rossmann fold with four additional β-strands, while the C-terminal lobe harbors three α-helices and two β-strands. Detailed structural analysis of the HpPseB/NADP/UDP-N-acetylglucosamine complex revealed that the cofactor binds to the Rossmann fold part of the N-terminal lobe, whereas the sugar substrate binding site is located in the C-terminal lobe (Fig. 3). Biophysical and mutagenesis studies confirmed the presence of three catalytic residues (D132, K133, and Y141) in HpPseB, one of which (K133, equivalent to K127 in C. jejuni PseB) is believed to serve as both a catalytic acid and base during the reaction [131, 132].
Cartoon representation of the structure of H. pylori PseB in complex with nicotinamide adenine dinucleotide phosphate (NADP+) and uridine-diphosphate-N-acetylglucosamine (PDB ID: 2GN6 [132]). The part of the N-terminal domain that has the Rossmann fold is coloured cyan, and the additional four β-strands are coloured wheat. The NADP molecule is drawn as black sticks, the substrate is drawn using a ball-and-stick representation
Step two
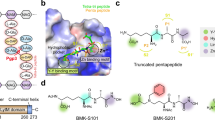
The second step of the Pse biosynthetic pathway (Fig. 2), axial transfer of an amino group onto the C4 atom of UDP-6-deoxy-4-keto-HexNAc to produce UDP-4-amino-4,6-dideoxy-β-l-AltNAc, is catalysed by the UDP-4-amino-4,6-dideoxy-N-acetyl-β-l-altrosamine transaminase, also known as pseudaminic acid biosynthesis protein C, PseC (EC 2.6.1.92). This enzyme belongs to the pyridoxal 5′-phosphate (PLP)-dependent transferase superfamily [128, 134]. PseC uses PLP as a cofactor and l-glutamate as an amino group donor [134, 135]. Insertional inactivation of the H. pylori pseC gene (HP0366) produced a non-motile phenotype that did not produce flagella and lacked O-antigen [133]. In addition, an isogenic H. pylori pseC mutant showed reduced invasion of human gastric epithelial cells. H. pylori PseC (HpPseC) shares no homology with any of the genes for the biosynthesis of the related nine-carbon sugar, sialic acid, in humans and, therefore, it represents a potential target for the design of novel anti-H. pylori therapeutics [136].
Biochemical and structural analyses have shown that HpPseC exists as a homodimer both in solution and in crystal [136]. The HpPseC monomer comprises two domains (Fig. 4). The N-terminal domain (residues 13–245) has a central mixed seven-stranded β-sheet, in which the β-strands are arranged in the topological order ↑β1–↓β7–↑β6–↑β5–↑β4–↑β2–↑β3. The β-sheet is flanked on either side by several α-helices. This domain harbors a β-hairpin structure (residues 211–225 on ↑β8 and ↓β9) that is projected away from one monomer and interacts with the second monomer at the dimer interface (Fig. 4) [136]. The C-terminal domain (residues 1–12 and 246–374) contains an antiparallel β-sheet with the strands arranged in the order ↑β10–↓β11–↑β12. This β-sheet is sandwiched between three α-helices on one side and the edge of the central β-sheet in the N-terminal domain on the other side. Two large and deep active site cavities are located at the dimer interface, and both monomers provide the residues that form the walls of these cavities (Fig. 4) [136]. Detailed crystallographic analysis of the HpPseC/PLP and HpPseC/PLP/UDP-4-amino-4,6-dideoxy-β-l-AltNAc (reaction product) complexes revealed that the cofactor PLP binding site is located at the bottom of the cavity near the C-terminal edge of the central β-sheet. The UDP-sugar binds at a different part of the pocket, connected to the bottom of the PLP binding cavity, and interacts with residues from both monomers. Site-directed mutagenesis has confirmed the crucial role of K183 (equivalent to K181 in C. jejuni PseC) in the catalytic mechanism of HpPseC [135, 136].
Cartoon representation of the structure of the H. pylori PseC dimer (PDB ID: 2FNU [136]). The N- and C-terminal domains of one of the two monomers are coloured green and magenta, respectively. The β-hairpin involved in domain swapping is coloured cyan; the bound pyridoxamine-5′-phosphate (PMP) cofactor is drawn as black sticks; the product UDP-4-amino-4,6-dideoxy-β-l-AltNAc (UD1) is shown using a ball-and-stick representation. The star (*) indicates domains from the second monomer of the PseC homodimer
Step three
The third step of the Pse biosynthesis pathway (Fig. 2) is catalysed by a nucleotide sugar-linked N-acetyltransferase, also known as pseudaminic acid biosynthesis protein H (PseH), or flagellin modification protein H (FlmH) (EC 2.3.1.202). It acetylates at the C4 of the UDP-sugar produced in the previous step, using acetyl-CoA (AcCoA) as an acetyl donor, and produces UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-altrose (UDP-6-deoxy-AltdiNAc) [128]. Mutational inactivation of the C. jejuni pseH gene resulted in the inhibition of the flagellum assembly, thereby rendering bacteria non-motile. This finding suggests that PseH plays a crucial role in bacterial motility and, likely, virulence [124, 137]. PseH is a member of the general control non-repressible 5 (GCN5)-related N-acetyltransferases (GNAT) superfamily [138], representatives of which are present in all kingdoms of life. A common feature of the GNAT enzymes is a V-shaped active site cavity at the central β-sheet and a P-loop that interacts with the pyrophosphate arm of the acetyl-CoA (AcCoA) cofactor [138]. Most members of this superfamily follow the direct acetyl transfer mechanism that proceeds via formation of a tetrahedral intermediate [138].
Helicobacter pylori PseH (HpPseH) (21.4 kDa) is dimeric in solution and in crystal [129, 139]. X-ray crystallographic analysis of the PseH/AcCoA complex revealed that each monomer has a core β-sheet made up of eight β-strands which are arranged in the topological order ↑β0–↓β1–↑β2–↓β3–↑β4–↑β5–↓β7–↑β6 (Fig. 5) [129]. The central β-sheet is flanked by three α-helices on each side. The cofactor AcCoA binds at the V-shaped cavity between strands β4 and β5, so that its pyrophosphate arm interacts with the P-loop between the β4-strand and α4-helix. Analysis of the modelled structure of the PseH/substrate/cofactor complex suggested that the nucleotide- and 4-amino-4,6-dideoxy-β-l-AltNAc-binding pockets are the elements that contribute to the substrate specificity most. Furthermore, a hydrophobic pocket harbouring the 6′-methyl group of the altrose determines preference to the methyl over the hydroxyl group [129]. Examination of the conservation of the amino acid residues in the enzyme active site suggested that PseH follows the common GNAT catalytic mechanism that involves direct acetyl transfer from AcCoA without an acetylated enzyme intermediate, and that S78 and Y138 likely act as a general base and acid in the PseH-catalysed reaction. The crystal structure of a homolog from C. jejuni with a similar fold has also been reported [140].
Cartoon representation of the structure of H. pylori PseH in complex with AcCoA (PDB ID: 4RI1 [129]). The motifs that are conserved across all GNAT enzymes are coloured as follows: motif C—green, motif D—blue, motif A—red, motif B—magenta. Non-conserved N-terminal and C-terminal regions are coloured wheat. The AcCoA cofactor is drawn in black using a stick representation
Step four
The fourth step of the Pse pathway (Fig. 2) is catalysed by an inverting nucleotide sugar hydrolase (UDP-6-deoxy-AltdiNAc hydrolase, PseG, EC 3.6.1.57) that belongs to the glycosyltransferase B (GT-B) family [141]. The enzyme hydrolyses UDP-6-deoxy-AltdiNAc to generate the free sugar 2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose [142]. The enzyme also inverts the stereochemistry at the C-1 atom of the substrate [128, 143]. Insertional inactivation of the H. pylori pseG (HP0326B) gene abolished flagellin production and resulted in a non-motile phenotype [92].
Biochemical and biophysical analyses showed that C. jejuni PseG (CjPseG, 282 aa) exists as a monomer both in solution and in crystal [142]. Structural analysis of CjPseG revealed that it has two domains: N-terminal domain (residues 1–142) and C-terminal domain (residues 153–282), connected by a short α-helix (residues 143–152) [142]. The N-terminal domain harbors a parallel β-sheet made up of seven β-strands (↑β3–↑β2–↑β1–↑β4–↑β5–↑β6–↑β7), sandwiched between two α-helices on one side and three on the other (Fig. 6). The C-terminal domain contains a six-stranded parallel β-sheet, where strands are arranged in the order ↑β10–↑β9–↑β8–↑β11–↑β12–↑β13, and sandwiched between two α-helices on one side and three α-helices on the other. X-ray crystallographic analysis of CjPseG in complex with UDP revealed that the enzyme active site is located in the long cleft at the domain interface, with residues from both domains contributing to the ligand binding pocket [142]. Detailed analysis of the modelled complex with the nucleotide sugar substrate suggested that the UDP-sugar adopts a twist-boat conformation at the active site, thereby exposing the anomeric bond for the nucleophilic attack via an active site water molecule, which facilitates inversion at the C-1 atom [142]. The active site has three highly conserved residues (H17, Y78, and N255) that interact with the sugar moiety of the substrate. A mutagenesis study showed that H17 is crucial for substrate recognition and serves as a catalytic base in the reaction [142].
Cartoon representation of the structure of C. jejuni PseG in complex with uridine-5′-diphosphate (UDP) (PDB ID: 3HBN) [142]. The N-terminal domain (red) is connected with the C-terminal domain (green) by an α-helix (magenta coloured). The UPD molecule is drawn as black sticks
Mechanistic studies showed that the elimination of the nucleotide moiety by CjPseG occurs via a metal-independent C–O bond cleavage mechanism. In the course of the reaction, a catalytic water molecule (hydrogen bonded to H17 and main-chain carbonyl of I13) directly attacks the anomeric carbon (C-1), followed by cleavage of the C–O anomeric bond to remove UDP from the UDP-6-deoxy-AltdiNAc sugar [143].
Step five
The fifth step of the Pse synthesis pathway, a condensation between phosphoenolpyruvate (PEP) and sugar 2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose to generate pseudaminic acid (Pse), is catalysed by Pse synthase (also known as pseudaminic acid biosynthesis protein I, PseI, EC 2.5.1.97) (Fig. 2). Inactivation of the pseI gene H. pylori (HP0178), or of the homologous gene neuB3 in C. jejuni, abolished the synthesis of functional flagella and thereby impaired bacterial motility [92, 144]. Biochemical analysis of C. jejuni NeuB3 (CjNeuB3) showed that a divalent metal ion is required for its activity [145]. Apart from C. jejuni, functional homologues of HpPseI have also been found in many other organisms, where they are involved in the biosynthesis of sialic acid (NeuAc) by catalysing the reaction of a condensation between PEP and N-acetylmannosamine (ManNAc) (or ManNAc-6-P in mammalian cells) [146].
Biophysical and structural analyses of N. meningitidis NeuB (NmNeuB, 349 aa, sharing 30 and 35% sequence identity with CjNeuB3 and HpPseI, respectively) showed that NmNeuB exists as a homodimer both in solution and in crystal [147]. The crystal structure of NmNeuB in complex with PEP and substrate analog N-acetylmannosaminitol (rManNAc, an unreactive, reduced form of ManNAc) [148] revealed that the NmNeuB monomer contains an N-terminal catalytic domain with a (β/α)8 barrel fold (also known as triosephosphate isomerase (TIM)-barrel fold)), connected via a long linker with the C-terminal antifreeze protein-like (AFPL) domain (74 aa) (Fig. 7) [147]. In the homodimer, the AFPL domain from one monomer binds over the active site in the TIM barrel domain of the opposite monomer, capping the active site cavity. This structural architecture is crucial for the activity of the enzyme, as the residues from the linker region and AFPL domain form part of the active site. This feature is common among enzymes with a TIM barrel fold [149]. In the crystal structure of the NmNeuB/PEP/rManNAc complex, the substrate analog forms hydrogen bonds with highly conserved residues D247, Q55, and Y186. Another important conserved residue, R314 on the AFPL domain of the second monomer, interacts with the N-acetyl group of rManNAc via a water molecule. A combination of mutagenesis, biochemical and kinetic analyses have shown that R314 plays an important role in the proper positioning of the sugar substrate at the active site, and thereby contributes to the catalysis [150]. The R314A substitution resulted in the inactivation of NmNeuB [150].
Cartoon representation of the structure of N. meningitidis NeuB in complex with substrate analog N-acetylmannosaminitol (rManNAc), phosphoenolpyruvate (PEP) and Mn2+ (PDB ID: 1XUZ [147]). The linker region and antifreeze protein-like (AFPL) domain are coloured magenta and cyan, respectively. The PEP cofactor is drawn as black sticks, whereas the rManNAc is shown using a ball-and-stick representation; the Mn2+ ion is shown as a blue sphere
Kinetic analysis of C. jejuni NeuB3 showed that the NeuB3-catalysed reaction follows the Michaelis–Menten kinetics, and the condensation reaction occurs by a C–O bond cleavage mechanism that proceeds via an oxocarbenium ion and tetrahedral intermediates [145]. The divalent cation (Mn2+ or Co2+), which is absolutely required for catalysis, acts as an electrophile that activates the carbonyl of the aldehyde for an attack by the C-3 carbon of PEP. Three conserved glutamate residues in the active site were identified as candidates for the role of the catalytic base in the reaction [147].
Step six
The final step of the Pse synthesis pathway (Fig. 2), activation of Pse with cytidine 5′-monophosphate (CMP), is catalysed by the metal-dependent pseudaminic acid cytidyltransferase, also known as CMP-pseudaminic acid synthetase or pseudaminic acid biosynthesis protein F (PseF, EC 2.7.7.81). This enzyme belongs to the nucleotide diphosphate sugar transferases superfamily [128]. Insertional inactivation of the pseF gene in H. pylori (HP0326A) resulted in the non-flagellated, non-motile phenotype [92]. A functional homolog of PseF found in N. meningitidis catalyses the biosynthesis of CMP-N-acetylneuraminic acid (CMP-Neu5Ac), and is termed CMP-5-N-acetylneuraminic acid synthetase (also known as CMP-Neu5Ac synthetase CNS, EC 2.7.7.43) [151].
N. meningitidis CNS (NmCNS) activates 5-N-acetylneuraminic acid (Neu5Ac) by transferring the CMP moiety of CTP to the anomeric OH-group of Neu5Ac in a Mg2+-dependent manner. The Neu5Ac is then transferred and incorporated into the bacterial cell surface components, such as LPS and the polysialic acid capsule, which are important virulence factors. Structural analysis of NmCNS revealed that it exists as a dimer in the crystal [152]. The NmCNS subunit has an α/β-type fold with an ~ 35-residue-long insertion—a β-hairpin (HP) domain—that plays an important role in dimerisation [152]. The core hydrolase domain has an αβα three-layer sandwich architecture which consists of seven β-strands with the topological order (↑β3–↑β2–↑β1–↑β4–↓β8–↑β5–↑β9) (Fig. 8). The central β-sheet is flanked by four α-helices on one side and three on the other. The HP domain protrudes out from the central β-sheet and contains two antiparallel β-strands (β6 and β7) and two α-helices. The active site of NmCNS is located at the interface of the core domain of one monomer and the HP domain of the second (Fig. 8). Analysis of the structure of the NmCNS active site revealed that it contains a hydrophobic pocket formed by Y179, F192, and F193 to aid binding of the methyl group of the N-acetyl moiety of Neu5Ac [153]. This feature is important for substrate recognition, as alanine substitutions at these positions resulted in a significant loss of enzymatic activity [153]. Residue K142 in the HP domain serves to neutralise the negative charge of the carboxylate group of sialic acid by the proper position of R196 via a hydrogen-bonding network. Furthermore, residues D211 and D209 that coordinate the catalytic Mg2+ ions were shown by mutagenesis to play an important role in catalysis. Finally, mutations of Q104 at the active site suggested its role in the metal-binding site of an intermediate complex.
Cartoon representation of the structure of the N. meningitidis CNS homodimer in complex with cytidine-5′-diphosphate (CDP) (PDB ID: 1EYR [151]). The dimerisation hairpin (HP) domain is coloured magenta. The substrate analog CDP is shown using a stick representation
The CNS enzyme follows an ordered sequential kinetic mechanism where the CTP molecule binds to the enzyme first, followed by sialic acid [153]. Horsfall and colleagues proposed a catalytic mechanism for CNS that involves two Mg2+ ions [153]. The role of the Mg2+ ions is to facilitate the correct orientation of the substrates and to activate the α-phosphate moiety of CTP and the sugar hydroxyl group of Neu5Ac. An ordered solvent molecule has been proposed to serve as the general base for the reaction [151]. Kinetic analysis showed that other nucleotides (ATP, GTP, TTP, or UTP) cannot replace CTP as a donor of nucleotide monophosphate in the NmCNS-catalysed reaction.
Current knowledge on inhibitors targeting Pse biosynthesis pathway
The Pse synthesis pathway enzymes are considered promising targets for the development of novel therapeutics since bacterial motility is essential for colonisation and development of persistent infection. This section summaries efforts to identify inhibitors targeting this pathway and investigate their mode of action. Kinetic studies and structure–activity relationship analysis revealed that the activated final product of the Pse pathway, cytidine 5′-monophosphate (CMP)-conjugated Pse, can serve as a natural inhibitor of the first enzyme (PseB) of the Pse biosynthesis pathway [128]. The enzymatic activity of PseB is also inhibited by UDP-α-D-galactose [130]. Recently, Ménard and colleagues have identified five additional PseB inhibitors using a combination of high-throughput screening and in silico approaches [154]. Three out of the five inhibitors were able to penetrate the cell membrane and inhibit flagellin production in C. jejuni with an IC50 (50% inhibitory concentration) of 14 µM. Interestingly, these inhibitors also showed dose-dependent activity against the fourth enzyme of the Pse pathway in H. pylori, PseG [154]. Analysis of the binding mode of the inhibitors, predicted using in silico docking, suggested that they bind in the active site of PseB and PseG, competing with their respective substrates [154]. Since they showed inhibitory activity at concentrations in the micromolar range, it is thought that this class of molecules can be developed into novel anti-infective agents to combat multidrug-resistant bacterial infection.
Although no inhibitors have so far been identified for the remaining four enzymes of the Pse biosynthesis pathway in H. pylori or C. jejuni, the current knowledge on inhibitors of homologous enzymes from other species may guide efforts towards this goal. In a study on the N. meningitidis sialic acid synthase (NmNeuB), a structural and functional homolog of the fifth enzyme (PseI) of the Pse pathway [148], a stable 2-deoxy analog of the putative tetrahedral intermediate of the reaction was identified, which inhibits the NmNeuB activity with an apparent K i of 3.1 μM. The structure of the NmNeuB/inhibitor/Mn2+ complex revealed that the inhibitor binds at the active site in a similar manner to the cognate substrate. A study on the CMP-5-N-acetylneuraminic acid synthetase (NmCNS) from N. meningitides (a structural and functional homolog of PseF) revealed that sulfo-CTP and sulfo-UTP analogs inhibit the NmCNS activity [155], although no structures of the inhibitor complexes are available yet.
Conclusions and therapeutic prospects
Glycoconjugates are widespread in nature and have diverse biological functions. Importantly, most of the characterised bacterial glycoproteins are linked with virulence factors [156]. In fact, many of the glycoproteins produced by bacteria contribute directly to cell adhesion, invasion, immune activation, or evasion of host defence mechanisms [66, 157,158,159,160].
Flagellar glycosylation is essential for the biosynthesis of functional flagella and hence for motility and pathogenesis [156, 161]. Inactivation of the glycosylation genes generated mutants that were either unable to produce flagellins or possessed inactive flagella [69, 127]. The structural similarity between the flagellar glycans of GI tract pathogens and the host’s sialic acids makes a significant contribution to host–pathogen interactions [157]. A recent report showed that Pse residues modulate cytokine interleukin 10 expression, and thereby facilitate bacterial colonisation of the host [162].
The glycosylation sites in flagellins showed variation in terms of their biological functions. In C. jejuni, some glycosylation sites appear to be indispensable for the assembly of the flagellum, while other locations were shown to be crucial for autoagglutination and microcolony formation, which is a prerequisite to the development of biofilms in the host [69, 163]. In H. pylori, Pse biosynthesis is essential not only for the assembly of functional flagella but also for the production of a wide range of virulence factors, including urease and LPS [133]. It has been suggested that glycosylation in H. pylori could also serve to disguise the antigenic epitopes of outer membrane proteins, thereby reducing host immune responses and influencing the clinical outcome of H. pylori infection [133].
The presence of nonulosonates, such as pseudaminic acid and related sugars, on the bacterial cell surface components including pilli, LPS O-antigen, and capsular polysaccharide [164,165,166] has attracted considerable attention over recent years. Prior to its incorporation in biological macromolecules, Pse must be synthesised and activated [128]. Due to the unstable nature of the activated nucleotide sugars and high cost of synthesis by chemical methods, Pse-synthesising enzymes are of great interest in the field of biotechnology, as an alternative route to cost-efficient production of Pse and its analogs. In addition, as highlighted in a recent report, bacterial O-glycosylation systems can be exploited for the production of bioconjugate vaccines targeting a variety of pathogens [167].
Pseudaminic acid is found only in bacterial pathogens. It is not produced or utilised by humans, nor it is present in commensal bacteria that dominate the human gut microbiome [168, 169]. Therefore, the Pse biosynthesis pathway represents an attractive target for selective inhibition, and further detailed studies of the enzymes from this pathway and their validation as targets for novel antimicrobials are well warranted in the view of the global spread of resistance to the existing antibiotics.
References
Warren JR, Marshall B (1983) Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 321:1273–1275
Marshall B, Warren JR (1984) Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 323:1311–1315
Wroblewski LE, Peek RM, Wilson KT (2010) Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev 23:713–739
Kusters JG, van Vliet AH, Kuipers EJ (2006) Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev 19:449–490
Ernst PB, Gold BD (2000) The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu Rev Microbiol 54:615–640
Testerman TL, Morris J (2014) Beyond the stomach: an updated view of Helicobacter pylori pathogenesis, diagnosis, and treatment. World J Gastroenterol 20:12781–12808
Goni E, Franceschi F (2016) Helicobacter pylori and extragastric diseases. Helicobacter 21:45–48
Capurso G, Lahner E, Marcheggiano A, Caruana P, Carnuccio A, Bordi C, Delle Fave G, Annibale B (2001) Involvement of the corporal mucosa and related changes in gastric acid secretion characterize patients with iron deficiency anaemia associated with Helicobacter pylori infection. Aliment Pharmacol Ther 15:1753–1761
Arnold DM, Bernotas A, Nazi I, Stasi R, Kuwana M, Liu Y, Kelton JG, Crowther MA (2009) Platelet count response to H. pylori treatment in patients with immune thrombocytopenic purpura with and without H. pylori infection: a systematic review. Haematologica 94:850–856
Pellicano R, Franceschi F, Saracco G, Fagoonee S, Roccarina D, Gasbarrini A (2009) Helicobacters and extragastric diseases. Helicobacter 14:58–68
Vafaeimanesh J, Hejazi SF, Damanpak V, Vahedian M, Sattari M, Seyyedmajidi M (2014) Association of Helicobacter pylori infection with coronary artery disease: is Helicobacter pylori a risk factor? Sci World J 2014:516354. https://doi.org/10.1155/2014/516354
Pirouz T, Zounubi L, Keivani H, Rakhshani N, Hormazdi M (2009) Detection of Helicobacter pylori in paraffin-embedded specimens from patients with chronic liver diseases, using the amplification method. Dig Dis Sci 54:1456–1459
Schulte A, Pandeya N, Fawcett J, Fritschi L, Risch HA, Webb PM, Whiteman DC, Neale RE (2015) Association between Helicobacter pylori and pancreatic cancer risk: a meta-analysis. Cancer Causes Control 26:1027–1035
Wang L, Guan Y, Li Y, Liu X, Zhang Y, Wang F, Kong L, Guo Q (2015) Association between chronic respiratory diseases and Helicobacter pylori: a meta-analysis. Arch Bronconeumol 51:273–278
Argenziano G, Donnarumma G, Arnese P, Assunta Baldassarre M, Baroni A (2003) Incidence of anti-Helicobacter pylori and anti-CagA antibodies in rosacea patients. Int J Dermatol 42:601–604
Zhou X, Zhang C, Wu J, Zhang G (2013) Association between Helicobacter pylori infection and diabetes mellitus: a meta-analysis of observational studies. Diabetes Res Clin Pract 99:200–208
Blaser MJ (2006) Who are we? Indigenous microbes and the ecology of human diseases. EMBO Rep 7:956–960
IARC (1994) Infection with Helicobacter pylori. IARC monographs on the evaluation of carcinogenic risks to humans/World Health Organization. Int Agency Res Cancer 61:177–240
De Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M (2012) Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol 13:607–615
Blaser MJ (1998) Science, medicine, and the future: Helicobacter pylori and gastric diseases. BMJ 316:1507–1510
Mbulaiteye SM, Hisada M, El-Omar EM (2009) Helicobacter pylori associated global gastric cancer burden. Front Biosci (Landmark Ed) 14:1490–1504
Du Y, Agnew A, Xp Ye, Robinson PA, Forman D, Crabtree JE (2006) Helicobacter pylori and Schistosoma japonicum co-infection in a Chinese population: helminth infection alters humoral responses to H. pylori and serum pepsinogen I/II ratio. Microbes Infect 8:52–60
Cárdenas-Mondragón MG, Carreon-Talavera R, Camorlinga-Ponce M, Gomez-Delgado A, Torres J, Fuentes-Panana EM (2013) Epstein Barr virus and Helicobacter pylori co-infection are positively associated with severe gastritis in pediatric patients. PLoS One 8:e62850
Vale F, Vítor J (2010) Transmission pathway of Helicobacter pylori: does food play a role in rural and urban areas? Int J Food Microbiol 138:1–12
Bastos J, Carreira H, La Vecchia C, Lunet N (2013) Childcare attendance and Helicobacter pylori infection: systematic review and meta-analysis. Eur J Cancer Prev 22:311–319
Hollander WJ, Holster IL, Hoed CM, Deurzen F, Vuuren AJ, Jaddoe VW, Hofman A, Perez Perez GI, Blaser MJ, Moll HA (2013) Ethnicity is a strong predictor for Helicobacter pylori infection in young women in a multi-ethnic European city. J Gastroenterol Hepatol 28:1705–1711
Zhu Y, Zhou X, Wu J, Su J, Zhang G (2014) Risk factors and prevalence of Helicobacter pylori infection in persistent high incidence area of gastric carcinoma in Yangzhong city. Gastroenterol Res Pract 2014:481365
Zhang XY, Zhang P-Y, Aboul-Soud MA (1899) From inflammation to gastric cancer: role of Helicobacterá pylori. Oncology Lett 13:543–548
Brenner H, Arndt V, Bode G, Stegmaier C, Ziegler H, Stümer T (2002) Risk of gastric cancer among smokers infected with Helicobacter pylori. Int J Cancer 98:446–449
Rothenbacher D, Bode G, Brenner H (2000) Helicobacter pylori among siblings. Lancet 355:1998–1999
Ford AC, Forman D, Bailey AG, Goodman KJ, Axon AT, Moayyedi P (2007) Effect of sibling number in the household and birth order on prevalence of Helicobacter pylori: a cross-sectional study. Int J Epidemiol 36:1327–1333
Jeon CY, Haan MN, Cheng C, Clayton ER, Mayeda ER, Miller JW, Aiello AE (2012) Helicobacter pylori infection is associated with an increased rate of diabetes. Diabetes Care 35:520–525
Inoue M (2017) Changing epidemiology of Helicobacter pylori in Japan. Gastric Cancer 20:3–7
Dunn B, Cohen H, Blaser MJ (1997) Helicobacter pylori. Clin Microbiol Rev 10:720–741
Pandeya N, Whiteman DC (2011) Prevalence and determinants of Helicobacter pylori sero-positivity in the Australian adult community. J Gastroenterol Hepatol 26:1283–1289
Leja M, Axon A, Brenner H (2016) Epidemiology of Helicobacter pylori infection. Helicobacter 21:3–7
Eusebi LH, Zagari RM, Bazzoli F (2014) Epidemiology of Helicobacter pylori infection. Helicobacter 19:1–5
Cherian S, Forbes D, Sanfilippo F, Cook A, Burgner D (2008) The epidemiology of Helicobacter pylori infection in African refugee children resettled in Australia. Med J Aust 189:438–441
Mitchell H, Katelaris P (2016) Epidemiology, clinical impacts and current clinical management of Helicobacter pylori infection. Med J Aust 204:376–380
Graham D, Rimbara E (2015) Helicobacter pylori update: gastric cancer, reliable therapy, and possible benefits. Gastroenterology 148:719–731. https://doi.org/10.1053/j.gastro.2015.01.040
Gatta L, Vakil N, Vaira D, Scarpignato C (2013) Global eradication rates for Helicobacter pylori infection: systematic review and meta-analysis of sequential therapy. BMJ 347:f4587. https://doi.org/10.1136/bmj.f4587
Ierardi E, Giorgio F, Losurdo G, Di Leo A, Principi M (2013) How antibiotic resistances could change Helicobacter pylori treatment: a matter of geography. World J Gastroenterol 19:8168–8180
Man SM (2011) The clinical importance of emerging Campylobacter species. Nat Rev Gastroenterol Hepatol 8:669–685
Brooks WA (2009) Campylobacter jejuni HS: 23 and Guillain–Barre syndrome, Bangladesh. Emerg Infect Dis 15:1313–1315
Leung FYK, Littlejohn GO, Bombardifr C (1980) Reiter’s syndrome after Campylobacter jejuni enteritis. Arthritis Rheumatol 23:948–950
Pope JE, Krizova A, Garg AX, Thiessen-Philbrook H, Ouimet JM (2007) Campylobacter reactive arthritis: a systematic review. Semin Arthritis Rheum 37:48–55
Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM (2015) Global epidemiology of Campylobacter infection. Clin Microbiol Rev 28:687–720
Platts-Mills JA, Kosek M (2014) Update on the burden of Campylobacter in developing countries. Curr Opin Infect Dis 27:444
Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case–control study. Lancet 382:209–222
Amour C, Gratz J, Mduma E, Svensen E, Rogawski ET, McGrath M, Seidman JC, McCormick BJ, Shrestha S, Samie A (2016) Epidemiology and impact of campylobacter infection in children in 8 low-resource settings: results from the MAL-ED study. Clin Infect Dis 63:1171–1179
Hudson J, Nicol C, Wright J, Whyte R, Hasell S (1999) Seasonal variation of Campylobacter types from human cases, veterinary cases, raw chicken, milk and water. J Appl Microbiol 87:115–124
Backert S, Schwarz T, Miehlke S, Kirsch C, Sommer C, Kwok T, Gerhard M, Goebel UB, Lehn N, Koenig W (2004) Functional analysis of the cag pathogenicity island in Helicobacter pylori isolates from patients with gastritis, peptic ulcer, and gastric cancer. Infect Immun 72:1043
Cover TL (1996) The vacuolating cytotoxin of Helicobacter pylori. Mol Microbiol 20:241–246
Ilver D, Arnqvist A, Ögren J, Frick I-M, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Borén T (1998) Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373–377
Mahdavi J, Sondén B, Hurtig M, Olfat FO, Forsberg L, Roche N, Ångström J, Larsson T, Teneberg S, Karlsson KA (2002) Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 297:573–578
Yamaguchi H, Osaki T, Kurihara N, Taguchi H, Hanawa T, Yamamoto T, Kamiya S (1997) Heat-shock protein 60 homologue of Helicobacter pylori is associated with adhesion of H. pylori to human gastric epithelial cells. J Med Microbiol 46:825–831
de Jonge R, Durrani Z, Rijpkema SG, Kuipers EJ, van Vliet AH, Kusters JG (2004) Role of the Helicobacter pylori outer-membrane proteins AlpA and AlpB in colonization of the guinea pig stomach. J Med Microbiol 53:375–379
Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B (1999) Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res 27:3325–3333
Rossez Y, Gosset P, Boneca IG, Magalhães A, Ecobichon C, Reis CA, Cieniewski-Bernard C, Curt MJC, Léonard R, Maes E (2014) The LacdiNAc specific adhesin LabA mediates adhesion of Helicobacter pylori to human gastric mucosa. J Infect Dis 210:1286–1295
Evans D, Evans DG, Takemura T, Nakano H, Lampert HC, Graham DY, Granger DN, Kvietys PR (1995) Characterization of a Helicobacter pylori neutrophil-activating protein. Infect Immun 63:2213–2220
Lu H, Hsu PI, Graham DY, Yamaoka Y (2005) Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology 128:833–848
Eaton KA, Brooks C, Morgan D, Krakowka S (1991) Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun 59:2470–2475
Chevalier C, Thiberge JM, Ferrero RL, Labigne A (1999) Essential role of Helicobacter pylori γ-glutamyltranspeptidase for the colonization of the gastric mucosa of mice. Mol Microbiol 31:1359–1372
Lertsethtakarn P, Ottemann KM, Hendrixson DR (2011) Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 65:389–410
Josenhans C, Suerbaum S (2002) The role of motility as a virulence factor in bacteria. Int J Med Microbiol 291:605–614
Guerry P (2007) Campylobacter flagella: not just for motility. Trends Microbiol 15:456–461
Zhu J, Hua X, Hou J, Zhao W (2008) The virulence determinants of Campylobacter jejuni and its ability to colonize hosts. Rev Med Microbiol 19:13–18
Wassenaar TM, van der Zeijst BA, Ayling R, Newell DG (1993) Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. Microbiology 139:1171–1175
Ewing CP, Andreishcheva E, Guerry P (2009) Functional characterization of flagellin glycosylation in Campylobacter jejuni 81-176. J Bacteriol 191:7086–7093. https://doi.org/10.1128/jb.00378-09
Eaton K, Morgan D, Krakowka S (1992) Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J Med Microbiol 37:123–127
Ottemann KM, Lowenthal AC (2002) Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect Immun 70:1984–1990
Hendrixson DR, DiRita VJ (2004) Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol 52:471–484
Terry K, Williams SM, Connolly L, Ottemann KM (2005) Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect Immun 73:803–811. https://doi.org/10.1128/iai.73.2.803-811.2005
Suerbaum S (1995) The complex flagella of gastric Helicobacter species. Trends Microbiol 3:168–170
Qin Z, W-t Lin, Zhu S, Franco AT, Liu J (2017) Imaging the motility and chemotaxis machineries in Helicobacter pylori by cryo-electron tomography. J Bacteriol 199:e00695–e00716
Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547
Parkhill J, Wren B, Mungall K, Ketley J, Churcher C, Basham D, Chillingworth T, Davies R, Feltwell T, Holroyd S (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668
Chevance FF, Hughes KT (2008) Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465
Evans LD, Hughes C, Fraser GM (2014) Building a flagellum outside the bacterial cell. Trends Microbiol 22:566–572
Geis G, Suerbaum S, Forsthoff B, Leying H, Opferkuch W (1993) Ultrastructure and biochemical studies of the flagellar sheath of Helicobacter pylori. J Med Microbiol 38:371–377
Beeby M, Ribardo DA, Brennan CA, Ruby EG, Jensen GJ, Hendrixson DR (2016) Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc Natl Acad Sci USA 113:E1917–E1926
Nuijten P, Van Asten F, Gaastra W, Van Der Zeijst B (1990) Structural and functional analysis of two Campylobacter jejuni flagellin genes. J Biol Chem 265:17798–17804
Minamino T, Imada K, Namba K (2008) Mechanisms of type III protein export for bacterial flagellar assembly. Mol BioSyst 4:1105–1115
Guerry P, Alm R, Power M, Logan S (1991) Role of two flagellin genes in Campylobacter motility. J Bacteriol 173:4757–4764
Josenhans C, Labigne A, Suerbaum S (1995) Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J Bacteriol 177:3010–3020
Suerbaum S, Josenhans C, Labigne A (1993) Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA-and flaB-negative mutants by electroporation-mediated allelic exchange. J Bacteriol 175:3278–3288
Josenhans C, Ferrero RL, Labigne A, Suerbaum S (1999) Cloning and allelic exchange mutagenesis of two flagellin genes of Helicobacter felis. Mol Microbiol 33:350–362
Wassenaar TM, Bleumink-Pluym N, Van der Zeijst B (1991) Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J 10:2055–2061
Lis L, Connerton IF (2016) The minor flagellin of Campylobacter jejuni (FlaB) confers defensive properties against bacteriophage infection. Front Microbiol 7:1908. https://doi.org/10.3389/fmicb.2016.01908
Szymanski CM, Yao R, Ewing CP, Trust TJ, Guerry P (1999) Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol 32:1022–1030
Schirm M, Schoenhofen IC, Logan SM, Waldron KC, Thibault P (2005) Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal Chem 77:7774–7782
Schirm M, Soo E, Aubry A, Austin J, Thibault P, Logan S (2003) Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol Microbiol 48:1579–1592
Thibault P, Logan SM, Kelly JF, Brisson JR, Ewing CP, Guerry P (2001) Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J Biol Chem 276:34862–34870
Apweiler R, Hermjakob H, Sharon N (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta 1473:4–8
Mescher MF, Strominger JL (1976) Purification and characterization of a prokaryotic glucoprotein from the cell envelope of Halobacterium salinarium. J Biol Chem 251:2005–2014
Sleytr UB (1975) Heterologous reattachment of regular arrays of glycoproteins on bacterial surfaces. Nature 257:400–402. https://doi.org/10.1038/257400a0
Nothaft H, Szymanski CM (2010) Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol 8:765–778
Gavel Y, von Heijne G (1990) Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng Des Sel 3:433–442
Wiederschain GY (2009) Essentials of glycobiology. Biochemistry 74:1056–1056. https://doi.org/10.1134/s0006297909090156
Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J (1999) Essentials of glycobiology. Cold Spring Harbor, New York
Dennis JW, Nabi IR, Demetriou M (2009) Metabolism, cell surface organization, and disease. Cell 139:1229–1241
Schwarz F, Aebi M (2011) Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol 21:576–582
Zielinska DF, Gnad F, Wiśniewski JR, Mann M (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141:897–907
Kowarik M, Young NM, Numao S, Schulz BL, Hug I, Callewaert N, Mills DC, Watson DC, Hernandez M, Kelly JF (2006) Definition of the bacterial N-glycosylation site consensus sequence. EMBO J 25:1957–1966
Chen MM, Glover KJ, Imperiali B (2007) From peptide to protein: comparative analysis of the substrate specificity of N-linked glycosylation in C. jejuni. Biochemistry 46:5579–5585
Hofsteenge J, Mueller DR, de Beer T, Loeffler A, Richter WJ, Vliegenthart JF (1994) New type of linkage between a carbohydrate and a protein: C-glycosylation of a specific tryptophan residue in human RNase Us. Biochemistry 33:13524–13530
de Beer T, Vliegenthart JF, Loeffler A, Hofsteenge J (1995) The hexopyranosyl residue that is C-glycosidically linked to the side chain of tryptophan-7 in human RNase Us is α-Mannopyranose. Biochemistry 34:11785–11789
Furmanek A, Hofsteenge J (2000) Protein C-mannosylation: facts and questions. Acta Biochim Pol 47:781–789
Ihara Y, Inai Y, Ikezaki M, Matsui I-SL, Manabe S, Ito Y (2015) C-mannosylation: modification on tryptophan in cellular proteins. In: Taniguchi N, Endo T, Hart GW, Seeberger PH, Wong CH (eds) Glycoscience: biology and medicine. Springer, New York, pp 1091–1099
Shcherbakova A, Tiemann B, Buettner FF, Bakker H (2017) Distinct C-mannosylation of netrin receptor thrombospondin type 1 repeats by mammalian DPY19L1 and DPY19L3. Proc Natl Acad Sci USA 114:2574–2579
Krieg J, Hartmann S, Vicentini A, Gläsner W, Hess D, Hofsteenge J (1998) Recognition signal for C-mannosylation of Trp-7 in RNase 2 consists of sequence Trp-xx-Trp. Mol Biol Cell 9:301–309
Julenius K (2007) NetCGlyc 1.0: prediction of mammalian C-mannosylation sites. Glycobiology 17:868–876
Hofsteenge J, Blommers M, Hess D, Furmanek A, Miroshnichenko O (1999) The four terminal components of the complement system AreC-mannosylated on multiple tryptophan residues. J Biol Chem 274:32786–32794
Christlet THT, Veluraja K (2001) Database analysis of O-glycosylation sites in proteins. Biophys J 80:952–960
Iwashkiw JA, Vozza NF, Kinsella RL, Feldman MF (2013) Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol Microbiol 89:14–28
Szymanski CM, Wren BW (2005) Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol 3:225–237
Gemmill TR, Trimble RB (1999) Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim Biophys Acta 1426:227–237
Luo Y, Haltiwanger R (2005) O-fucosylation of notch occurs in the endoplasmic reticulum. J Biol Chem 280:11289–11294. https://doi.org/10.1074/jbc.M414574200
Spiro RG (2002) Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12:43R–56R
Dell A, Morris HR (2001) Glycoprotein structure determination by mass spectrometry. Science 291:2351–2356
Hug I, Feldman MF (2011) Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology 21:138–151
Dell A, Galadari A, Sastre F, Hitchen P (2011) Similarities and differences in the glycosylation mechanisms in prokaryotes and eukaryotes. Int J Microbiol 2010:148178. https://doi.org/10.1155/2010/148178
Vik Å, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobsen W, Koomey M (2009) Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc Natl Acad Sci USA 106:4447–4452
Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, Pattarini D, Majam G, Thibault P, Logan S (2006) Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol 60:299–311
Lloyd DH, Viac J, Werling D, Rème CA, Gatto H (2007) Role of sugars in surface microbe–host interactions and immune reaction modulation. Vet Dermatol 18:197–204
Twine SM, Paul CJ, Vinogradov E, McNally DJ, Brisson JR, Mullen JA, McMullin DR, Jarrell HC, Austin JW, Kelly JF (2008) Flagellar glycosylation in Clostridium botulinum. FEBS J 275:4428–4444
Logan SM (2006) Flagellar glycosylation—a new component of the motility repertoire? Microbiology 152:1249–1262
Schoenhofen IC, McNally DJ, Brisson JR, Logan SM (2006) Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology 16:8C–14C
Salah Ud-Din AIM, Liu YC, Roujeinikova A (2015) Crystal structure of Helicobacter pylori pseudaminic acid biosynthesis N-acetyltransferase PseH: implications for substrate specificity and catalysis. PLoS One 10:e0115634. https://doi.org/10.1371/journal.pone.0115634
Creuzenet C, Schur MJ, Li J, Wakarchuk WW, Lam JS (2000) FlaA1, a new bifunctional UDP-GlcNAc C6 dehydratase/C4 reductase from Helicobacter pylori. J Biol Chem 275:34873–34880
Morrison JP, Schoenhofen IC, Tanner ME (2008) Mechanistic studies on PseB of pseudaminic acid biosynthesis: a UDP-N-acetylglucosamine 5-inverting 4, 6-dehydratase. Bioorg Chem 36:312–320
Ishiyama N, Creuzenet C, Miller WL, Demendi M, Anderson EM, Harauz G, Lam JS, Berghuis AM (2006) Structural studies of FlaA1 from Helicobacter pylori reveal the mechanism for inverting 4, 6-dehydratase activity. J Biol Chem 281:24489–24495
Hopf PS, Ford RS, Zebian N, Merkx-Jacques A, Vijayakumar S, Ratnayake D, Hayworth J, Creuzenet C (2011) Protein glycosylation in Helicobacter pylori: beyond the flagellins? PLoS One 6:e25722
Schoenhofen IC, McNally DJ, Vinogradov E, Whitfield D, Young NM, Dick S, Wakarchuk WW, Brisson J-R, Logan SM (2006) Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J Biol Chem 281:723–732
Obhi RK, Creuzenet C (2005) Biochemical characterization of the Campylobacter jejuni Cj1294, a novel UDP-4-keto-6-deoxy-GlcNAc aminotransferase that generates UDP-4-amino-4, 6-dideoxy-GalNAc. J Biol Chem 280:20902–20908
Schoenhofen IC, Lunin VV, Julien JP, Li Y, Ajamian E, Matte A, Cygler M, Brisson JR, Aubry A, Logan SM (2006) Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J Biol Chem 281:8907–8916
Javed MA, Alphen LB, Sacher J, Ding W, Kelly J, Nargang C, Smith DF, Cummings RD, Szymanski CM (2015) A receptor-binding protein of Campylobacter jejuni bacteriophage NCTC 12673 recognizes flagellin glycosylated with acetamidino-modified pseudaminic acid. Mol Microbiol 95:101–115. https://doi.org/10.3390/ijms17071018
Salah Ud-Din AIM, Tikhomirova A, Roujeinikova A (2016) Structure and functional diversity of GCN5-related N-acetyltransferases (GNAT). Int J Mol Sci 17:1018. https://doi.org/10.3390/ijms17071018
Liu YC, Ud-Din AI, Roujeinikova A (2014) Cloning, purification and preliminary crystallographic analysis of the Helicobacter pylori pseudaminic acid biosynthesis N-acetyltransferase PseH. Acta Crystallogr Sect F Struct Biol Cryst Commun 70:1276–1279
Song WS, Nam MS, Namgung B, Yoon SI (2015) Structural analysis of PseH, the Campylobacter jejuni N-acetyltransferase involved in bacterial O-linked glycosylation. Biochem Biophys Res Commun 458:843–848
Hu Y, Chen L, Ha S, Gross B, Falcone B, Walker D, Mokhtarzadeh M, Walker S (2003) Crystal structure of the MurG: UDP–GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc Natl Acad Sci USA 100:845–849
Rangarajan ES, Proteau A, Cui Q, Logan SM, Potetinova Z, Whitfield D, Purisima EO, Cygler M, Matte A, Sulea T (2009) Structural and functional analysis of Campylobacter jejuni PseG a UDP-sugar hydrolase from the pseudaminic acid biosynthetic pathway. J Biol Chem 284:20989–21000
Liu F, Tanner ME (2006) PseG of pseudaminic acid biosynthesis a UDP-sugar hydrolase as a masked glycosyltransferase. J Biol Chem 281:20902–20909
Linton D, Karlyshev AV, Hitchen PG, Morris HR, Dell A, Gregson NA, Wren BW (2000) Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol Microbiol 35:1120–1134
Chou WK, Dick S, Wakarchuk WW, Tanner ME (2005) Identification and characterization of NeuB3 from Campylobacter jejuni as a pseudaminic acid synthase. J Biol Chem 280:35922–35928
Cotton T, Parker E, Joseph D (2014) The role of sialic acid synthases in sialic acid biosynthesis. Chem N Z 78:69–74
Gunawan J, Simard D, Gilbert M, Lovering AL, Wakarchuk WW, Tanner ME, Strynadka NC (2005) Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol. J Biol Chem 280:3555–3563
Liu F, Lee HJ, Strynadka NC, Tanner ME (2009) Inhibition of Neisseria meningitidis sialic acid synthase by a tetrahedral intermediate analogue. Biochemistry 48:9194–9201
Silverman J, Balakrishnan R, Harbury P (2001) Reverse engineering the (β/α)8 barrel fold. Proc Natl Acad Sci USA 98:3092–3097
Joseph DD, Jiao W, Parker EJ (2013) Arg314 is essential for catalysis by N-acetyl neuraminic acid synthase from Neisseria meningitidis. Biochemistry 52:2609–2619
Mosimann SC, Gilbert M, Dombroswki D, To R, Wakarchuk W, Strynadka NC (2001) Structure of a sialic acid-activating synthetase, CMP-acylneuraminate synthetase in the presence and absence of CDP. J Biol Chem 276:8190–8196
Krapp S, Münster-Kühnel AK, Kaiser JT, Huber R, Tiralongo J, Gerardy-Schahn R, Jacob U (2003) The crystal structure of murine CMP-5-N-acetylneuraminic acid synthetase. J Med Microbiol 334:625–637
Horsfall LE, Nelson A, Berry A (2010) Identification and characterization of important residues in the catalytic mechanism of CMP-Neu5Ac synthetase from Neisseria meningitidis. FEBS J 277:2779–2790
Ménard R, Schoenhofen IC, Tao L, Aubry A, Bouchard P, Reid CW, Lachance P, Twine SM, Fulton KM, Cui Q (2014) Small-molecule inhibitors of the pseudaminic acid biosynthetic pathway: targeting motility as a key bacterial virulence factor. Antimicrob Agents Chemother 58:7430–7440
Wong JH, Sahni U, Li Y, Chen X, Gervay-Hague J (2009) Synthesis of sulfone-based nucleotide isosteres: identification of CMP-sialic acid synthetase inhibitors. Org Biomol Chem 7:27–29
Schmidt MA, Riley LW, Benz I (2003) Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol 11:554–561
Rubin EJ, Trent MS (2013) Colonize, evade, flourish: how glyco-conjugates promote virulence of Helicobacter pylori. Gut Microbes 4:439–453
Varki A (2008) Sialic acids in human health and disease. Trends Mol Med 14:351–360
Schauer R (2009) Sialic acids as regulators of molecular and cellular interactions. Curr Opin Struct Biol 19:507–514
Ohtsubo K, Marth JD (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126:855–867
Goon S, Kelly JF, Logan SM, Ewing CP, Guerry P (2003) Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol Microbiol 50:659–671
Stephenson HN, Mills DC, Jones H, Milioris E, Copland A, Dorrell N, Wren BW, Crocker PR, Escors D, Bajaj-Elliott M (2014) Pseudaminic acid on Campylobacter jejuni flagella modulates dendritic cell IL-10 expression via Siglec-10 receptor: a novel flagellin-host interaction. J Infect Dis 210:1487–1498
McNally DJ, Hui JP, Aubry AJ, Mui KK, Guerry P, Brisson JR, Logan SM, Soo EC (2006) Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81–176 using a focused metabolomics approach. J Biol Chem 281:18489–18498. https://doi.org/10.1074/jbc.M603777200
Knirel YA, Vinogradov EV, L’vov VL, Kocharova NA, Shashkov AS, Dmitriev BA, Kochetkov NK (1984) Sialic acids of a new type from the lipopolysaccharides of Pseudomonas aeruginosa and Shigella boydii. Carbohydr Res 133:C5–C8
Kenyon JJ, Marzaioli A, Hall RM, De Castro C (2014) Structure of the K2 capsule associated with the KL2 gene cluster of Acinetobacter baumannii. Glycobiology 24:554–563
Smedley JG, Jewell E, Roguskie J, Horzempa J, Syboldt A, Stolz DB, Castric P (2005) Influence of pilin glycosylation on Pseudomonas aeruginosa 1244 pilus function. Infect Immun 73:7922–7931
Pan C, Sun P, Liu B, Liang H, Peng Z, Dong Y, Wang D, Liu X, Wang B, Zeng M (2016) Biosynthesis of conjugate vaccines using an O-linked glycosylation system. MBio 7:e00443–00416
Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230
Tra VN, Dube DH (2014) Glycans in pathogenic bacteria–potential for targeted covalent therapeutics and imaging agents. Chem Commun (Camb) 50:4659–4673
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict interest.
Rights and permissions
About this article
Cite this article
Salah Ud-Din, A.I.M., Roujeinikova, A. Flagellin glycosylation with pseudaminic acid in Campylobacter and Helicobacter: prospects for development of novel therapeutics. Cell. Mol. Life Sci. 75, 1163–1178 (2018). https://doi.org/10.1007/s00018-017-2696-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-017-2696-5