Abstract
The starlet sea anemone Nematostella vectensis has been recently established as a new model system for the study of the evolution of developmental processes, as cnidaria occupy a key evolutionary position at the base of the bilateria. Cnidaria play important roles in estuarine and reef communities, but are exposed to many environmental stressors. Here, I describe the genetic components of a “chemical defensome” in the genome of N. vectensis and review cnidarian molecular toxicology. Gene families that defend against chemical stressors and the transcription factors that regulate these genes have been termed a chemical defensome and include the cytochromes P450 and other oxidases, various conjugating enyzymes, the ATP-dependent efflux transporters, oxidative detoxification proteins, as well as various transcription factors. These genes account for about 1% (266/27,200) of the predicted genes in the sea anemone genome, similar to the proportion observed in tunicates and humans, but lower than that observed in sea urchins. While there are comparable numbers of stress-response genes, the stress sensor genes appear to be reduced in N. vectensis relative to many model protostomes and deuterostomes. Cnidarian toxicology is understudied, especially given the important ecological roles of many cnidarian species. New genomic resources should stimulate the study of chemical stress sensing and response mechanisms in cnidaria and allow us to further illuminate the evolution of chemical defense gene networks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Cnidaria occupy a key basal evolutionary position within Metazoa (Dunn et al. 2008), with recent evidence suggesting that they are early-diverging bilaterians (de Jong et al. 2006; Matus et al. 2006). Cnidaria have important ecological roles as reef structure builders and as predators and prey in planktonic and benthic ecosystems (e.g., Harborne et al. 2006; Sebens 1981). Cnidaria are sensitive to many environmental stressors and have been used as indicators of water quality (Arkhipchuk et al. 2006; Davies and Freeman 1995; Wiger and Stottum 1985). With a better understanding of regulatory processes and development of appropriate endpoints (e.g., Tarrant 2007), cnidaria will become valuable indicators of exposure to disruptive chemicals and other stressors.
The starlet sea anemone Nematostella vectensis has been recently established as a new model system for the study of the evolution of developmental processes (Darling et al. 2005; Putnam et al. 2007) and may act as a model for the basic molecular biology of anthozoans. The remarkable amenability of this species to laboratory manipulation has already made it a productive system for exploring cnidarian development and the origins of bilateral symmetry (Finnerty and Martindale 1999; Finnerty et al. 2004; Fritzenwanker et al. 2004; Kusserow et al. 2005; Magie et al. 2005; Matus et al. 2006; Torras and Gonzalez-Crespo 2005).
N. vectensis is a burrowing estuarine anemone, with populations in the eastern Pacific, northern English Channel, western North Sea, and western Atlantic (Hand and Uhlinger 1994), although it is likely that all but the western Atlantic represent introduced populations (Reitzel et al. 2008a). It can tolerate a remarkably wide ranges of salinities (2–54 ppt), temperatures (−1°C to 28°C), and dissolved oxygen concentrations (Sheader et al. 1997). The facility with which Nematostella populations can be investigated within their natural ecological context (Darling et al. 2005) suggests that this model may also be profitably expanded to address important questions in molecular and evolutionary ecology and toxicology. A mechanistic understanding of stress responses is essential to establishing this model system, as with all model systems.
An important question in biology is how cells and organisms maintain homeostasis in a variable environment. The need to deal with physical, chemical, and biological stressors has driven the evolution of an array of gene families and pathways (also known as “environmental genes” (Ponting 2008)) that afford protection from challenges. The immune system is one such protective mechanism, which responds to biotic stressors such as pathogens (Miller et al. 2007). Another set of genes comprises the “chemical defensome”, encoding a network of defensive proteins that allows the organism to sense, transform, and eliminate potentially toxic chemicals (Goldstone et al. 2006).
The chemical defensome protects against chemically mediated injury by environmental chemicals such as heavy metals, microbial products, and other natural exogenous compounds, as well as anthropogenically derived compounds such as hydrocarbon derivatives and pesticides. These compounds are structurally diverse, requiring either non-specific enzymatic responses or a broad array of specific enzymatic actions. In addition, the maintenance of cellular homeostasis requires the inactivation and elimination of endogenous signaling molecules, such as eicosanoids, and defense against endogenously generated toxicants such as reactive oxygen species (ROS).
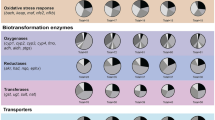
The chemical defensome is comprised of several classes of proteins that function coordinately to protect the cell (Fig. 1). These proteins include enzymes that transform chemicals to less toxic and more readily excretable metabolites, efflux transporters that actively eliminate toxicants and transformed products, antioxidant enzymes protecting against externally and internally generated ROS or other radicals, and soluble receptors and ligand-activated transcription factors that act as sensors of toxicants or cellular damage.
Conceptual organization of the cellular defensome. Organic and inorganic toxicants are actively exported and also subjected to a variety of biotransformative reactions. Modified from Goldstone et al. (2006)
Efflux transporter proteins such as the ATP-binding cassette (ABC) transporters can provide the first line of cellular defense (Dean et al. 2001). Once toxicants enter the cytoplasm, however, biotransformation is often required to inactivate or enhance the elimination of toxicants. Biotransformation enzymes include oxidative enzymes such as the cytochromes P450 (CYPs); reductive enzymes such as aldo-keto reductases (AKR), epoxide hydrolase (EH), and NAD(P)H-quinone oxidoreductase (NQO); and conjugative enzymes including glutathione-S-transferases (GST), sulfotransferases (SULT), UDP-glucuronosyl transferases (UGT), and N-acetyl transferases (NAT). Biotransformation generally results in detoxification, but oxidation, N-acetylation, sulfate, or glutathione conjugation can lead to toxic metabolites in a chemical and cell-specific manner (Gamage et al. 2006; Guengerich et al. 2003; Surh. 1998).
Gene products that protect against injury from chemicals may be especially important in embryos given the complex chemical signaling pathways governing development (Davidson and Erwin. 2006; Hamdoun and Epel 2007), as well as the need to protect the genome of the germ cells (Epel 2003). In adults, some of these proteins also provide protection from environmental factors, such as oxidative stress, that can lead to senescence (Finkel and Holbrook 2000). Many gene products in this network (e.g., CYPs) perform multiple roles, having important endogenous functions (including but not limited to development), as well as functioning in chemical defense.
Here, I show that the major elements of the network of genes and pathways that allow an organism to mount a defense against toxic chemicals appear to be conserved in cnidaria and review relevant aspects of cnidarian molecular toxicology. Almost all of the gene families or superfamilies that are characteristic of the chemical defensive network in deuterostomes (Goldstone et al. 2006) are also represented in the sea anemone (Fig. 1; see also Reitzel et al. 2008b), indicating the presence of this system in the bilaterian ancestor and evolutionary conservation. However, while there is general conformity in the presence of higher order gene groups across taxa, in most cases, gene orthology is more difficult to determine.
Methods
Different types of evidence are available for the genes discussed in this paper. Predicted genes are derived from the US Department of Energy Joint Genome Institute (JGI) predictions of the whole genome shotgun assembly (www.jgi.doe.gov). Many of these predicted genes are supported by expression data from an extensive EST collection (Sullivan et al. 2008). Resources are available online at stellabase.org, cnidbase.bu.edu and nematostella.org. In this study, defensome genes were identified by Hidden Markov Model searches (Hmmer v2.3.2; Eddy 1998) of the JGI gene predictions with conserved domains of known defense genes using the PFAM models. Gene homologies were confirmed by reciprocal BLAST of the predicted genes against Genbank. For this study, the JGI “best models” were used without significant refinement. Alignments were constructed using Muscle v3.6b (Edgar 2004) and are available upon request of the author.
The exact nomenclature of many genes presented in this paper is tentative because of the uncertainty in classifying genes to specific subfamilies within the major superfamilies represented in these analyses. I have attempted to follow the nomenclature guidelines for many of the defined gene superfamilies (Hyndman et al. 2003; Jez and Penning 2001; Mackenzie et al. 2005; Nebert and Vasiliou 2004; Nelson et al. 1993; Vasiliou and Nebert 2005; Vasiliou et al. 2006), but due to evolutionary distances, some of the subfamily assignments are tentative. Thus, new genes here are given names, indicating my understanding of the homologous relationships, but that should not be taken as formal assignments. Formal assignments of new gene names are often reserved by specific nomenclature committees (e.g., the Cytochrome P450 Nomenclature Committee or the Aldo-Keto Reductase Nomenclature Committee). Based on evidence from our previous analysis of the sea urchin genome (Goldstone et al. 2006), gene orthologies may also not be predictive of function.
Phylogenetic trees were constructed by analyzing amino acid sequences using maximum likelihood (RAxML 7.0.3; Stamatakis 2006). Regions of alignment uncertainty were excluded from phylogenetic analysis (Kreil and Ouzounis 2003) by automatic masking using a custom-written script. The WAG-CAT model of amino acid substitution (Whelan and Goldman 2001) with a gamma distribution of substitution rates was used in all likelihood analyses, based on likelihood tests using RAxML.
Defensome gene families
Receptors and signal transduction
Homologs of most important stress receptors are present in the sea anemone genome, including the aryl hydrocarbon receptor (AHR), hypoxia-inducible factor 1 (HIF1α), and the aryl hydrocarbon nuclear translocator (ARNT), metal transcription factor 1 (MTF1), nuclear factor-kappa B (NFkB), and nuclear factor erythroid-derived 2 related 2 (NRF2), detailed below (Fig. 2). Although some of the known components of the vertebrate and invertebrate xenobiotic receptor pathways are missing (e.g., pregnane X receptor, liver X receptor, farnesoid X receptor [PXR, LXR, and FXR]), receptors that are not clearly orthologous to known xenobiotic sensors may substitute, or there may be an increased xenobiotic receptor promiscuity.
Some of the stress-response transcription factor pathways with homologs in N. vectensis. Hypoxia activates both the HIF1α and MTF1 pathway, metal stress activates MTF1, oxidative stress activates the NRF2 pathway (as well as others, not shown), and organic xenobiotics activate AHR or various NRs. These transcription factors have specific response elements (REs) in the regulatory regions of responsive genes, including hypoxida RE (HRE; HIF1α/ARNT), metal RE (MRE; MTF1), antioxidant RE (ARE; NRF2), xenobiotic RE (XRE; AHR/ARNT), and specific NR-REs (e.g., estrogen response elements)
Aryl hydrocarbon receptor (AHR) and related bHLH-PAS proteins
Basic helix-loop-helix PER/ARNT/SIM (bHLH-PAS) family genes encode proteins involved in critical physiological and developmental signaling, including those that mediate responses to certain environmental pollutants (including polynuclear aromatic hydrocarbons) and low oxygen tension (Kewley et al. 2004). bHLH-PAS genes in chordates that have been shown to be important to physiological responses to environmental pollutants include the aryl hydrocarbon receptor (AHR), HIF1α, and the ARNT.
The bHLH gene family has previously been examined in N. vectensis and other species (Simionato et al. 2007). Simionato et al. identified 68 bHLH genes, several of which also contained a PAS domain, including one or two ARNT genes and zero to two HIF genes (the range depends on uncertainty in the phylogenetic clustering; Simionato et al. 2007), but could not identify an AHR in the N. vectensis genome. However, Reitzel et al. (2008b) identify a gene (gi|156394392) as a putative AHR homolog and note that its expression is confirmed through an EST. Both AHR and HIF1α form heterodimers with ARNT to regulate transcription of downstream targets through the recognition of specific DNA response elements. Transcriptional responses to potential activators of AHR and HIF have not been well studied in sea anemones, and no data are available to determine if these response elements are conserved in N. vectensis.
Oxidative and metal stress-response transcription factors
Oxidative stress-response factors in vertebrates include the CNC-bZIP family [nuclear factor erythroid-derived 2 and related factors (NRFs)], the BTB-bZIP proteins BACH1 and BACH2, and the small Maf proteins (MafF, MafG, and MafK in particular). Maf proteins in vertebrates are heterodimeric partners of NF-E2, NRFs, and Bach proteins (Igarashi and Sun 2006). In addition to their roles as heterdimerization partners for various CNC proteins, small Maf proteins have critical roles in vertebrate stress signaling, oncogenesis, and may also have links to the inflammation response (Blank 2008).
N. vectensis has two homologs of the small Maf proteins, an NRE2-like protein homologous to NRF2 and a KEAP1-like protein, which in the absence of oxidative stress in vertebrates, encodes a protein that retains NRF2 in the cytoplasm and enhances its proteasomal degradation (Nguyen et al. 2003). In vertebrates, the NRF2 signaling pathway provides a rapid response to electrophilic or oxidative compounds and has been show to attenuate carcinogenesis and inflammation (Osburn and Kensler 2008).
Other important oxidative stress-responding transcription factors with homologs in sea anemone include MTF1 and NFkB. MTF1 is well known as a metal-responsive transcription factor (Laity and Andrews 2007), but has also been proposed as a generalized sensor of oxidative stress (Murphy 2004; Murphy et al. 1999, 2005). MTF1 may also interact with HIF1α and contribute to HIF1α activation during hypoxia (Murphy et al. 2005).
Nuclear receptors
Ligand-activated nuclear receptors (NRs) function as chemically activated transcription factors, primarily with endogenous functions but also importantly in xenobiotic sensing. Of greatest interest with regards to the chemical defensome are those related to NRs in the NR1H and NR1I subfamilies, which contain vertebrate FXR, LXR, and PXR, constitutively active receptor (CAR), and the vitamin D receptor (VDR), as well as arthropod EcR (ecdysone receptor). Other NRs involved in xenobiotic response in vertebrates include estrogen receptor (ER; NR3A subfamily); the peroxisome proliferator receptors (PPARs; NR1C subfamily), which have target genes involved in lipid metabolism, energy homeostasis, and cell differentiation; and the retinoid X receptor (RXR; NR2B subfamily), which has many target genes involved in xenobiotic metabolism.
N. vectensis and other cnidaria appear to lack many nuclear receptors traditionally studied in response to toxicants (e.g., NR1s, ER; Grasso et al. 2001; Reitzel et al. 2008b). N. vectensis appears to have a modest number of NRs (18), none of which are related to the NR1H (PPAR, LXR, FXR) or NR1I (VDR, PXR, CAR) families. However, there are genes related to hepatocyte nuclear factor 4 (HNF4, NR2A) and to RXR, indicating the presence of ancestral NR2 subfamily members in this cnidarian. An RXR gene has been cloned from a cubazoan, Tripedalia cystophora, and the protein binds 9-cis retinoic acid with high affinity (Kostrouch et al. 1998).
Although there does not appear to be an ER in N. vectensis, the existence of a bilaterian ancestral steroid-binding receptor was inferred based on ancestral protein reconstruction (Thornton et al. 2003). Cnidaria appear to be susceptible to signal disruption by exogenous estrogens (reviewed in Tarrant 2005, 2007), although there appear to be differences between coral and hydra sensitivity (Pascoe et al. 2002; Tarrant et al. 2004). Estrogen signaling may still be important in corals, however, as estrogens have been found in and around spawning corals, and corals have the ability to metabolize estradiol and testosterone (Atkinson and Atkinson 1992; Blomquist et al. 2006; Tarrant et al. 1999, 2003; Twan et al. 2003, 2006).
Efflux transporter proteins
Many toxic compounds are pumped against concentration gradients across membranes in an energy-dependent process. This first line of cellular defense, against amphipathic or slightly lipophilic compounds in particular, is mediated by efflux proteins known as ABC or multidrug efflux transporters, including the p-glycoproteins (PGP/ABCB), mitoxantrone resistance protein (MXR/ABCG2), and multidrug resistance proteins (MRP/ABCC; Dean et al. 2001). Efflux transporters function to export both unmodified substrates and substrates modified by other defensome enzymes (Deeley et al. 2006). In embryos, efflux transporters may provide the primary defense against exogenous toxicants but also play important roles in developmental programs by establishing morphogen gradients (Hamdoun and Epel 2007).
In chordates, the ABC transporters are organized into eight subfamilies designated ABC A through H (Annilo et al. 2006). A subset of these families includes proteins known to export toxicants: the ABCB, ABCC, and ABCG transporters. These proteins are commonly called multidrug resistance transporters after their ability to pump out multiple therapeutic drugs, a major obstacle to the efficacy of the treatment of several pathogens (Dean et al. 2005).
Genome searches revealed that sea anemones have 64 ABC genes organized into six subfamilies, including the three multidrug transporter subfamilies (ABC B, C, and G; Table 1, Supplemental Table S1). There is considerable variation in the total number of ABC genes within eukaryotic genomes, but the relative proportions have tended to stay constant (Annilo et al. 2006; Goldstone et al. 2006). The ABC genes clustered in the ABCA (five genes), ABCD (six genes), and ABCF (four genes) families either do not have known function or do not have known roles in detoxification and will not be considered further here.
N. vectensis has seven ABCB genes, including two related to the ABCB1 (pgp) proteins. The pgp transporters are well known as multidrug resistance proteins involved in the efflux of toxic compounds. Additional genes related to known xenobiotic transporters include six ABCC4 (MRP4)-like genes, found in a sea anemone-specific cluster; six other ABCC-like genes including one ABCC5 (MRP5)-like sequence; and 24 other genes that cluster in a large anemone-specific clade within the ABCC family. Finally, analyses conducted in this study identified six ABCG sequences, including one sequence that clusters closely with the vertebrate ABCG2s. In vertebrates, ABCG2 proteins exhibit broad substrate specificity among xenobiotic compounds and play critical roles in the clearance of certain drugs (Allikmets et al. 1998; Kusuhara and Sugiyama 2007; Miyake et al. 1999).
Other potentially important anemone transporters include the organic anion polypeptides (OATP; solute carrier family 21, SLC21) and organic anion and cation transporters (OAT and OCT; solute carrier family 22, SLC22). Both SLC21 and SLC22 are part of the major facilitator superfamily. OAT substrates examined in vertebrates include estrone sulfate, urate, prostaglandins, heavy metals such as mercury and cadmium, and the herbicide 2,4-dichlorophenoxyacetic acid (Eraly et al. 2004; Kimura et al. 2002; Sweet 2005). OATPs have partially overlapping substrate specificities for steroid conjugates, bile salts, anionic oligopeptides, and anionic xenobiotics including toxins and drugs (Hagenbuch and Meier 2003; Jacobsson et al. 2007). The anemone genome contains 17 OATP genes and 62 SLC22 (OCT and OAT) genes. However, orthology among non-vertebrate SLC families is difficult to assign, precluding any hypotheses regarding substrate specificity.
Oxidative or reductive biotransformation
Cytochromes P450
Oxidative modification of chemicals to more hydrophilic products is often the initial step leading to excretion. In bilaterians, this is carried out by CYP and flavoprotein monooxygenase (FMO) enzymes, especially members of the CYP1, CYP2, CYP3, CYP6, CYP9, and CYP4 families. Toxicant oxidation can, however, also lead to increased toxicity; for example, oxidation of benzo[a]pyrene by CYP1A leads to hepatotoxicity (Uno et al. 2001).
The sea anemone genome contains 82 CYP genes, which are in general not classifiable into established CYP families due to the low (<40%) identity with other known CYPs. A large-scale reclassification of metazoan CYPs taking into account recent genomic data will be required to formally name the N. vectensis CYPs (Nelson, personal communication). However, broader classification into the CYP clan framework (Nelson 1998, 2006) is possible: Clan 2, containing CYP families 1, 2, 17, 18, 21, 33, 34, and 35; Clan 3, containing primarily CYPs 3, 5, 6, 9, 28, 309, 310, and 317; Clan 4, containing CYPs 4, 311, 313, 316, and 318; and the mitochondrial clan, CYPs 11, 12, 24, 27, 44, 49, 302, 314, and 315.
Sea anemone CYPs are principally part of Clan 2 and Clan 3, with 39 and 20 genes in these two Clans, respectively, while there are only three Clan 4 genes (Table 2, Fig. 3, Supplemental Figure S1, Supplemental Table S2). Many of the CYPs in these clans are involved in detoxification of exogenous and endogenous compounds (Lewis et al. 2004), although the functional information is primarily from vertebrates and insects. The anemone Clan 2 CYPs are clustered more closely to the vertebrate CYP17s (and thus the important xenobiotic-detoxifying CYP1s, including aryl hydrocarbon hydroxylases) than to the vertebrate CYP2s. However, it is not clear that CYP1 genes exist outside the deuterostomes (Goldstone et al. 2007), and these sea anemone CYPs cannot be considered early CYP1-like genes. The Clan 3 genes are likewise less closely related to the vertebrate CYP3 or insect CYP6 detoxification genes than to other members of Clan 3, in this case, the CYP5-like genes. CYP5 genes have unusual functionality in that they catalyze a rearrangement of a prostaglandin endoperoxide (Hecker and Ullrich 1989), rather than a substrate oxidation. More generally, the sea anemone Clan 3 CYPs may oxidize prostaglandins, which are potent chemical defenses in marine systems (Paul and Puglisi 2004; Paul et al. 2006).
Distribution of genes in CYP Clans in human, tunicate, sea urchin, and sea anemone. Data for the tunicate and sea urchin assignments were taken from Goldstone et al. (2006)
N. vectensis does not have a CYP19 (aromatase), despite the fact that (low) aromatase activity has been demonstrated in a scleractinian coral, Euphillia ancora (Twan et al. 2003). CYP19 is not present in most invertebrates with a sequenced genome (Goldstone, Nelson, and Stegeman, unpublished data), although it is present in amphioxus (Castro et al. 2005; Mizuta and Kubokawa 2007). It is very possible that a different CYP, not CYP19, possesses aromatase activity.
Other redox enzymes
Other proteins that oxidize or reduce toxicants include the FMO (Ziegler 2002), AKR (Jin and Penning 2007), aldehyde dehydrogenases (ALDH), NQO, and epoxide hydrolase (EPHX). In contrast to the CYPs, much less is known about many of the substrates of these enzymes, even in humans (Krueger and Williams 2005; Penning and Drury 2007).
In addition to the 82 CYPs, analyses conducted in this paper identified genes for six FMO enzymes. Although both enzyme families are primarily monooxygenases and have some overlapping substrate specificities (Krueger and Williams 2005; Ziegler 2002), FMOs are generally thought to oxidize soft nucleophiles, while CYPs often catalyze C–H abstraction (Cashman 2005). FMOs are less stable enzymes than CYPs, which has contributed to the relative lack of functional knowledge. The sea anemone FMO enzymes are quite distinct from the known human FMOs, and, as with the anemone CYPs, specific functions cannot be easily guessed at.
The sea anemone genome has at least 12 AKR genes, 21 ALDH, and one EPHX gene. These numbers are comparable to deuterostome gene inventories. In vertebrates, EPHX contributes to the toxicity of benzo[a]pyrene by converting the benzo[a]pyrene epoxides produced by CYP1s to benzo[a]pyrene dihydrodiols (Shimada 2006), which eventually can be oxidized to redox-cycling benzo[a]pyrene quinones by AKR (Palackal et al. 2001; Penning et al. 1999).
One of the most important ALDH reactions in vertebrate development is the irreversible oxidation of retinal to retinoic acid (Lee et al. 1991); retinoids play very important roles in vertebrate patterning and are also likely important in cnidarian development (Bouzaiene et al. 2007; Johnson and Chun 1989; Kostrouch et al. 1998; Muller 1984). ALDH enzymes may also help maintain the cellular redox balance via ROS scavenging and the production of reducing equivalents as NADPH or NADH.
NQO enzymes catalyze the two-electron reduction of quinones to hydroquinones, reducing the formation of semiquinones and the potential for reactive oxygen generation (Vasiliou et al. 2006). Similar to sea urchins (Goldstone et al. 2006), analyses conducted for this paper revealed that sea anemones do not have NQO-like genes. This is in line with the observed lack of NQO genes in the worm, fly, sea squirt, or plants (Vasiliou et al. 2006).
Conjugative biotransformation
Sea anemones possess relatively few proteins with direct homology to xenobiotic-conjugating enzymes, particularly in comparison to the purple sea urchin (Table 2; Goldstone et al. 2006). Sea anemones have genes for 23 GST, including five microsomal GSTs (MAPEG), nine UGT genes, and 22 SULT genes. No NAT genes were found. These numbers are far lower than the large diversification of these gene families seen in sea urchins, but comparable to the numbers observed in mammalian genomes (the human genome contains 13 SULT, 13 UGT, and 21 GST genes; Gamage et al. 2006; Mackenzie et al. 2005; Nebert and Vasiliou, 2004).
Cytosolic GSTs are soluble proteins that catalyze the transfer of glutathione to an electrophilic substrate (Hayes et al. 2005). Microsomal or membrane GSTs (MAPEG) form an evolutionarily distinct class of enzymes that exhibit both glutathione transferase and lipid peroxidase activity (Bresell et al. 2005), thus detoxifying both xenobiotic compounds and ameliorating oxidative stress. The majority (15) of the 18 sea anemone GSTs are readily classifiable, including three mu-class, three omega-class, six sigma-class, one theta-class, one fungal-type, and one zeta-class. The three remaining GSTs appear homologous to the xenobiotic-metabolizing alpha/pi GSTs. This search also found a sequence homologous to the translation elongation factor 1g, which contains a GST domain but does not have glutathione transferase activity. The N. vectensis genome also codes for a total of five MAPEG sequences, including one homologous to vertebrate MAPEG1, one sequence homologous to MAPEG3, and three sequences homologous to prostaglandin E synthase (PTGS). PTGS enzymes are MAPEG superfamily members important to eicosanoid synthesis and involved in the vertebrate inflammation response (Jakobsson et al. 1999). Prostaglandins in corals are very important in chemical defense (reviewed in Paul and Puglisi 2004; Paul et al. 2006) and have been extensively studied in gorgonians. A prostaglandin synthase with 50% identity to mammalian PTGS has been cloned from an Arctic soft coral (Koljak et al. 2001).
SULT and UGT enzymes catalyze the conjugation of sulfuryl groups donated by 3′-phosphoadenosine-5′-phosphosulfate or UDP-glucuronide, respectively, to a wide variety of substrates, including both xenobiotics and endogenous products (Bock and Kohle 2004; Gamage et al. 2006; Runge-Morris and Kocarek 2005). Cytosolic (soluble) SULTs are responsible for the metabolism of xenobiotic and small endogenous substrates (SULT1 and SULT2), while membrane-bound SULTs are involved in endogenous peptide, lipid, and aminosugar sulfonation (Gamage et al. 2006). I found 22 SULT genes in the sea anemone genome, all of which are more closely related to the SULT genes involved in energy metabolism rather than those SULT genes known from vertebrate studies to participate in detoxification reactions. The anemone genes are divided among the SULT3A family (eight genes), SULT3B (two genes), SULT4 (four genes), and carbohydrate keratan/chondroitin SULTs (eight genes). Chondroitin sulfation has been demonstrated in the nematocysts of Hydra magnipapillata (Yamada et al. 2007), and it is possible that the N. vectensis genes are involved in similar functions.
The sea anemone UGT genes are likewise not closely related to the UGT families with known xenobiotic-metabolizing or detoxification roles. UGT1 genes in mammals consist of one gene with as many as 14 different first exons, complicating the assignment of UGT homology (Mackenzie et al. 2005). Based on our previous analysis of the large number of distinct genes in the sea urchin, exon duplication like that observed in the mammalian UGT families is not the only method of UGT diversification. However, the nine anemone UGTs are not classifiable to any of the known vertebrate UGT families, and thus, no function can even be hinted at. This finding is not unique to anemones, as other marine genomes contain what appear to be lineage-specific gene family expansions that are not readily assignable to known UGT functional classes (J. Goldstone, unpublished data).
Antioxidant proteins
ROS, including superoxide, hydrogen peroxide, and hydroxyl radicals, are derived from a variety of cellular processes, including leakage from mitochondrial respiration. Reactive oxygen can also be produced by exposure to toxicants and to ultraviolet radiation. ROS contribute to diseases and pathologies generally deriving from altered gene expression or damage to biomolecules, including proteins, lipids, and DNA (Halliwell and Gutteridge 1999; Lesser 2006). General antioxidant defensive genes include superoxide dismutase (SOD), catalases (CAT), and peroxidases, including glutathione peroxidase (GPX), peroxiredoxin, and thioredoxins (TXNs).
The sea anemone genome has a total of six SOD genes: three Cu/Zn SOD genes, one Mn SOD gene, two Fe SODs, as well as an SOD copper chaperone homolog (which contains an SOD domain but has no dismutase activity). Both EST and cDNA libraries support the expression of all six SOD forms under normal conditions (A. Reitzel, A. Tarrant, J. Goldstone, unpublished data). In addition, there is one CAT, 12 glutathione peroxidase genes, and six other heme peroxidase genes. This abundance of antioxidant defense genes is complemented by a complete glutathione system (glutathione reductase and 4 gamma-glutamyl transferases), as well as TXN and thioredoxin reductase (TXNRD).
Metal detoxification
Heavy metals are important aquatic pollutants resulting from sewage, urban and agricultural runoff, and antifouling paint. Bioconcentration of heavy metals can lead to tissue concentrations that are ten- to 10,000-fold higher than environmental levels, resulting in a variety of toxic effects. Four phytochelatin synthase (PCS) homologs are present in the N. vectensis genome and expressed under normal conditions (A. Reitzel, A. Tarrant, J. Goldstone, unpublished data). Phytochelatins are metal-binding peptides composed primarily of glutathione groups that are important metal detoxifying genes in plants and fungi. Until PCS was discovered in the nematode Caenorhabditis elegans (Clemens et al. 2001), it was believed that phytochelatins were present only in plants and fungi. Now, it is clear that many other lineages contain PCS homologs (Clemens 2006), including the sea urchin (Goldstone et al. 2006). Currently sequenced vertebrate genomes do not contain a gene homologous to PCS, nor do insect genomes, suggesting that phytochelatin synthesis ability was lost independently in some protostome and deuterostome lineages.
No metallothionein (MT) genes, neither plant- nor fungi- nor metazoan-related, were found in the sea anemone genome despite extensive searching, perhaps because of the presence of the alternative metal-complexing phytochelatin system. The absence of MT genes is apparently due to gene loss, as MT proteins are important metal detoxification proteins in plants (Cobbett and Goldsbrough 2002), mollusks (Amiard et al. 2006), sea urchins (Nemer et al. 1985), and vertebrates, and are also present in sponges (Berthet et al. 2005; Philp 1999).
Active efflux of toxic metals is another important route to detoxification. Both OAT and ABC efflux proteins (see above) export metals (Leslie et al. 2001; Sweet 2005), and the anemone contains genes homologous to the transporters (within both the OAT and ABC families) known to facilitate metal export.
Heat shock proteins
Heat shock proteins (HSP) have been implicated in the response to various toxicants, including cadmium, arsenic, and free radicals (Feder and Hofmann 1999). The induction of HSP mRNA and protein by heat shock factor 1 appears to be part of generalized cellular stress response, and HSPs may not only act as chaperones but also assist in refolding of partially denatured proteins (Kim et al 2006). Sea anemones have several families of heat shock proteins, including HSP 90 and 70 and small alpha crystalline HSPs (HSP20s). The largest family of heat shock proteins is the HSP20 family, containing at least 18 genes. Sea anemones also have at least four HSP90 genes and nine HSP70s. Various coral and anemone HSP60, HSP70, and HSP90 proteins and cDNA sequences have been shown to be strongly induced not only by heat or cold shock (Choresh et al. 2007, 2004, 2001; Hashimoto et al. 2004; Robbart et al. 2004; Rossi and Snyder 2001; Rossi et al. 2006; Sharp et al. 1997, 1994; Snyder and Ross 2004) but also by PCB118 (Wiens et al. 2000).
Discussion
The chemical defensome is an integrated network of chemical sensing and response proteins that function as an organized defense against toxic chemicals, both endo- and exogenous (Goldstone et al. 2006). Elucidation of the chemical stress-response repertoire of N. vectensis provides a framework for studies on a number of cnidarian-specific questions as well as on broader evolutionary questions. Characterization of these stress-response genes in N. vectensis facilitates the use of this and other anemones as sentinel species for changing environmental stressors. N. vectensis is a hardy species, tolerating extremes of temperatures unknown to other members of the family Edwardsiidae, which are restricted to temperate and polar coastal seas with less dramatic temperature and salinity variations (Daly 2002). Identification of molecular responses to chemical stress will help us to develop markers that will allow N. vectensis and other anemones to act as sentinels of environmental contamination.
The major components of this defensive gene network are conserved in the sea anemone genome (Fig. 4), indicating that they must have origins prior to the cnidarian–bilaterian split. Interphyla comparison of the components and linkages within the chemical defensome will help us understand the early evolution of the chemical stress response. Despite the fact that the individual genes within the defensome network may vary across organisms, this network may be comprised of evolutionarily conserved modules, which are retained across evolution. Comparing the susceptibility of sea anemone embryos with that of deuterostome and protosome embryos, for a range of chemicals, could lead to fundamental insights into how these defensome “kernels” function to protect embryos from the myriad chemical challenges that could derail development. Predictions of defensome interactions (e.g., the roles of nuclear receptors in simultaneously modulating multiple parts of the defensome) are testable using microarray analysis of gene expression in combination with gene knockdown and protein overexpression.
Gene count comparisons for various classes of defensome genes. The area of each circle is proportional to the total number of genes classified into the defensome. Receptors include bHLH-ZIP, NR, and CNC receptors. Transporters are ABC and OAT transporters; oxidative and reductive modification genes include CYP, FMO, ALDH, EH, and AKR; conjugative genes are GST, MAPEG, UGT, and SULT; antioxidant genes are SOD, CAT, PXR, and GPX. Other genes include PCS, HSP20, HSP70, and HSP90
Signaling network
Ligand-activated transcription factors form a significant component of the defensome, integrating the stress response and potentially activating many different pathways simultaneously. The evolutionary history of both the bZIP (Amoutzias et al. 2007) and bHLH-PAS (Simionato et al. 2007) receptor superfamilies is complicated, but most major clades of these receptors are present in cnidaria. Although some of the known components of the vertebrate and invertebrate xenobiotic receptor pathways are missing, homologs of most important stress receptors are present, including AHR, ARNT, HIF1α, MTF, and NRF2. The deuterostome xenobiotic-responsive NR1H and NR1I subfamilies are missing, however, and it is not currently known whether other NRs are functioning as xenobiotic receptors. Cnidaria have been shown to have a retinoid response, including a functional homolog of the RXR (Bouzaiene et al. 2007; Johnson and Chun 1989; Kostrouch et al. 1998; Muller 1984).
Gene family diversification
Many defense gene families have undergone diversification and expansion in marine invertebrates in comparison to vertebrate genomes. In general, analysis of entire genomes is required to determine that a specific family or superfamily has undergone diversification, and thus current examples of such events are scattered. Class or order level diversification may be presumed, based on the model organisms with sequenced genomes, but caution should be exercised when extrapolating. Although there is general conformity in the presence of higher order gene groups, in many cases, gene orthology is more difficult to determine.
For example, the sea anemone contains 82 CYP genes and those related to CYP gene families 1–4 constitute a large proportion (76%) of the total, suggesting evolutionary pressure to maintain broad functionality in these important defense gene families. Multiple gene duplications in the toxicologically important CYP families appear to have taken place in many different lineages, leading to taxon-specific gene clades that are related to known CYP families yet distinct enough to preclude definitive assignment of names based on current CYP nomenclature guidelines. The extensive birth–death process of CYP diversification is not solely represented by invertebrates—within the vertebrates, there is significant evidence for extensive gene duplication and loss within xenobiotic-metabolizing CYP families (Thomas 2007).
In N. vectensis, particular examples of family diversification relative to known sequences are the CYPs and the ABC transporters. Other defensome gene families do not appear significantly expanded, nor do they have genes distributed into completely novel subfamilies. This observation is interesting in light of the fact that N. vectensis lives in the challenging environment of a temperate estuary and ranges from subtropical to subarctic estuaries (Hand and Uhlinger 1994).
Symbiosis
An important consideration for the study of cnidarian chemical defense genes is that many species are host to photosynthetic endosymbionts (zooxanthellae or zoochorellae). There are unique aspects of both normal physiology and toxicological responses that are related to the presence of endosymbionts. Notably, photosynthesis produces oxygen, and surrounding host tissues require additional protection against ROS to withstand hyperoxygenation, such as additional superoxide dismutase genes. Indeed, Allemand and coworkers have characterized multiple SOD forms in the Mediterranean sea anemone Anemone viridis. They found up to seven SOD activity bands in various tissues and detected several forms of CuZnSOD, MnSOD, and FeSOD in the various compartments (Richier et al. 2003). Two of the CuZnSODs were cloned and encode both extra- and intracellular CuZnSODs with different putative transcription binding sites (Plantivaux et al. 2004).
Although N. vectensis is an apparently asymbiotic anemone, the genome has genes for six different SODs, all of which are expressed under normal conditions. This abundance of SOD genes may be a general pattern, particularly in anthozoans. Interestingly, greater diversity in SOD activities was found in a symbiotic anemone species (A. viridis) than in an asymbiotic species (Actinia schmidti), and the asymbiotic anemone experienced significantly greater oxidative protein damage upon exposure to hyperoxia (Richier et al. 2005). Thus, the presence of photosynthetic endosymbionts and the concomitant possibility of hyperoxia may have driven the evolution of multiple additional SOD forms in cnidaria. Several different catalase forms were also characterized in A. viridis, with tissue-specific distributions and activities (Merle et al. 2007). Inhibition of the host anemone catalase led to symbiont expulsion, suggesting an active response to increased oxidative stress.
A second effect of algal symbiosis is the sensitivity of symbiotic cnidarian species to herbicidal contamination (Jones 2005; Jones and Kerswell 2003). Notably, there is increasing distribution of herbicides such as the s-triazine Irgarol 1051 that have been incorporated into marine antifouling paints along with copper (Carbery et al. 2006; Gardinali et al. 2004), and there is significant runoff of herbicide-containing waste from agricultural regions. Irgarol 1051 is a photosystem II binding agent that inhibits photosynthetic electron transport, resulting in a shortage of NADPH and the formation of singlet oxygen (Fufezan et al. 2002). Acute exposure of coral to Irgarol 1051 resulted in induction of SOD and MXR (ABCG) proteins and decreases of GPX, CAT, and certain CYP proteins (Downs and Downs 2007). While other herbicides have been investigated (e.g., diuron; Harrington et al. 2005; Negri et al. 2005; Raberg et al. 2003), there have been relatively few investigations of the molecular mechanisms of herbicide toxicity in cnidaria, and it is generally thought that damage is primarily a result of the disruption of host–algal symbiosis (Jones 2005).
Reactive oxygen and UV
ROS production can also be an important consequence of UV exposure (Lesser 2006; Mopper and Kieber 2000). While UV responses have been studied in a number of coral species, many coral physiological responses to UV appear to be related to the physiological responses of their algal symbionts (Baruch et al. 2005; Torres et al. 2007; Verde and McCloskey 2002). UV has been shown to interfere with pattern formation in regenerating hydra and promote budding of intact hydra, possibly in response to tissue damage (Ghaskadbi et al. 2005; Znidaric et al. 1992). Both exogenous hydrogen peroxide and UV treatments have been shown to increase DNA strand breaks in cnidaria, demonstrating the potential for genotoxic ROS effects (Baruch et al. 2005; Mitchelmore and Hyatt 2004)
A very important protective mechanism in corals, as well as in diverse other marine organisms, is the accumulation of sunscreening compounds known as mycosporine-like amino acids (MAAs; Shick and Dunlap 2002). MAAs may facilitate larval survival (Wellington and Fitt 2003), as well as adult UV tolerance (Ferrier-Pages et al. 2007; Torres et al. 2007), and may also contribute to antioxidant capacity (Dunlap and Yamamoto 1995; Yakovleva et al. 2004). In contrast to other animals, N. vectensis apparently possesses the shikimic acid pathway thought to be necessary for MAA production (Starcevic et al. 2008), presumably obtained via lateral gene transfer from bacteria. Many cnidaria have been thought to accumulate MAAs from their symbionts (Shick and Dunlap 2002) or from their diet, as is the case for sea urchins (Carroll and Shick 1996). Given the presence of the MAA biosynthetic pathway in the N. vectensis genome and the clustering of sea anemone MAA complement by anemone phylogenetic distribution rather than endosymbiont identity, presence, or other environmental factors (Shick et al. 2002), it is likely that the ability of cnidaria to biosynthesize MAAs is not restricted to N. vectensis.
In contrast to tropical corals and many littoral anemones, N. vectensis is a burrowing anemone, and it is possible that N. vectensis adult may be able to avoid UV damage despite living in shallow ponds. However, larval N. vectensis may require more protection from ROS than adults, leading to the apparent diversification of ROS defenses. Examination of the UV-stress response of N. vectensis will aid in understanding the different roles that antioxidant enzymes and sunscreening compounds may play in protecting sea anemones. Comparisons of N. vectensis, an apparently asymbiotic anemone, with symbiotic anemones (e.g., A. viridis) or symbiotic reef-building corals could elucidate the protective mechanisms required by symbiotic cnidaria and shed light on the role of antioxidant enzymes in thermotolerance and bleaching (Downs et al. 2002; Merle et al. 2007; Richier et al. 2005; Richier et al. 2003).
Molecular toxicology
Many toxicological studies of cnidaria involve metals, particularly copper, cadmium, and zinc. In particular, the acute and structural effects of copper, cadmium, and zinc have been investigated in various hydrozoan species, including both freshwater and marine hydra (Holdway et al. 2001; Karntanut and Pascoe 2000, 2002, 2005) and a variety of anthozoa, including scleractinian corals (Mitchelmore et al. 2007). Other biological responses to heavy metals in cnidaria include coral bleaching (Jones 1997) and effects on coral metabolism (Alutoin et al. 2001; Nystrom et al. 2001), larval mortality, and inhibition of reproduction, including settlement, motility, and fertilization of larvae (Negri and Heyward 2000, 2001; Reichelt-Brushett and Harrison 2000, 2005). Few studies have examined molecular biomarkers or molecular mechanisms of metal stress (Mitchelmore et al. 2002; Morgan et al. 2001).
The availability of the N. vectensis genome will make many mechanistic studies possible and should spur the development of metal stress biomarkers in various species. In particular, the presence of multiple genes for phytochelatin synthase provides obvious markers for metal stress, despite the lack of metalothionein genes.
As with metal contamination, there are few studies of either molecular markers or mechanisms of exposure to organic contaminants other than herbicides in cnidaria (Rougee et al. 2006). A number of biochemical studies of cnidarian CYP biochemistry have been carried out, however. CYP carbon monoxide difference spectra have been observed in six different species of sea anemone (Heffernan and Winston 1998, 2000; Sole and Livingstone 2005) and three different species of scleractinian corals (Favia fragum, Siderastrea sidea, and Montastraea faveolata; Garcia et al. 2005; Gassman and Kennedy 1992; Ramos and Garcia 2007). Furthermore, benzo[a]pyrene hydroxylase activities have been observed in sea anemones, likely due to the action of CYP mixed-function oxygenases (Heffernan et al. 1996; Winston et al. 1998). The presence of inducible (versus constituent) CYP content in corals has also been demonstrated in coral due to benzo[a]pyrene or fuel oil exposure (Ramos and Garcia 2007; Rougee et al. 2006). The same PAH exposures induced components of the reactive oxygen defense systems, including CAT, SOD, and GST. Finally, an important molecular marker of genotoxic damage, the Comet assay of DNA damage, has been assessed in cnidaria in response to benzo[a]pyrene exposures (Mitchelmore and Hyatt 2004). Benzo[a]pyrene was found to increase DNA strand breaks in the temperate anemone Anthopleura elegantissima, suggesting also that cnidaria, like vertebrates, are capable of bioactivating benzo[a]pyrene to genotoxic metabolites, likely by CYPs.
More subtle effects of organic contamination might include in particular disruption of endogenous signaling pathways by exogenous hormones or hormone mimetics. Recent research indicates that cnidaria are susceptible to this sort of signal disruption, although the precise mechanisms are unknown (Fukuhori et al. 2005; Pachura-Bouchet et al. 2006; Pascoe et al. 2002; Tarrant 2005, 2007; Tarrant et al. 2004). In particular, Tarrant et al (2004) observed that spawning and growth rates were reduced in corals exposed to exogenous steroidal estrogens. As noted above, cnidaria do not posses a homolog of the vertebrate estrogen receptor, although there may be other nuclear receptors that function as a steroid receptors (Reitzel et al. 2008b; Tarrant 2007). Cnidaria are steroid-rich organisms (Withers et al. 1982), but the roles these steroids play in normal physiology are not clear (Tarrant 2005). Steroids and secosteroids from gorgonian and soft corals have been show to have antimicrobial and antifouling activity (Qi et al. 2008; Sica and Musumeci 2004), suggesting that many of these compounds may be produced for chemical defense. CYP enzymes often participate in steroid synthesis and modification, and the diversity of N. vectensis CYPs may relate to the diversity of cnidarian steroids, although the steroid content of N. vectensis has not been investigated.
N. vectensis is a physical stress-tolerant organism, tolerating a wide range of environmental conditions (Sheader et al. 1997). With this robustness to physical stress, N. vectensis is in a prime position to act as a sentinel species toward chemical stress in estuaries. N. vectensis is also an excellent laboratory model, with simple maintenance needs, and the generation of clonal stocks by forced regeneration allows great scope for genetic manipulation. Furthermore, this sea anemone is an excellent model for the study of embryonic development (Matus et al. 2006). With the description of these chemical defense genes, we can study the evolution of cellular defense during embryonic development. Development inherently is a robust process; which parts of the process are more susceptible to disruption and from which stressors are not clear (Hamdoun and Epel 2007). The description of this defense gene set will allow us to examine the evolution of generalized cellular stress responses in bilaterian embryos and to understand how these stress responses function in adult cnidaria.
References
Allikmets R, Schriml LM, Hutchinson A, Romano-Spica V, Dean M. A human placenta-specific ATP-binding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Res 1998;58(23):5337–9.
Alutoin S, Boberg J, Nystrom M, Tedengren M. Effects of the multiple stressors copper and reduced salinity on the metabolism of the hermatypic coral Porites lutea. Mar Environ Res 2001;52(3):289–99. doi:10.1016/S0141-1136(01)00105-2.
Amiard JC, Amiard-Triquet C, Barka S, Pellerin J, Rainbow PS. Metallothioneins in aquatic invertebrates: their role in metal detoxification and their use as biomarkers. Aquat Toxicol 2006;76(2):160–202. doi:10.1016/j.aquatox.2005.08.015.
Amoutzias GD, Veron AS, Weiner J 3rd, Robinson-Rechavi M, Bornberg-Bauer E, Oliver SG, et al. One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-binding site specificity. Mol Biol Evol 2007;24(3):827–35. doi:10.1093/molbev/msl211.
Annilo T, Chen ZQ, Shulenin S, Costantino J, Thomas L, Lou H, et al. Evolution of the vertebrate ABC gene family: analysis of gene birth and death. Genomics 2006;88:1–11.
Arkhipchuk VV, Blaise C, Malinovskaya MV. Use of hydra for chronic toxicity assessment of waters intended for human consumption. Environ Pollut 2006;142(2):200–11. doi:10.1016/j.envpol.2005.10.012.
Atkinson S, Atkinson MJ. Detection of estradiol-17-beta during a mass coral spawn. Coral Reefs 1992;11(1):33–5. doi:10.1007/BF00291932.
Baruch R, Avishai N, Rabinowitz C. UV incites diverse levels of DNA breaks in different cellular compartments of a branching coral species. J Exp Biol 2005;208(5):843–8. doi:10.1242/jeb.01496.
Berthet B, Mouneyrac C, Perez T, Amiard-Triquet C. Metallothionein concentration in sponges (Spongia officinalis) as a biomarker of metal contamination. Comp Biochem Physiol C Toxicol Pharmacol 2005;141(3):306–13. doi:10.1016/j.cca.2005.07.008.
Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J Mol Biol 2008;376(4):913–25. doi:10.1016/j.jmb.2007.11.074.
Blomquist CH, Lima PH, Tarrant AM, Atkinson MJ, Atkinson S. 17Beta-hydroxysteroid dehydrogenase (17beta-HSD) in scleractinian corals and zooxanthellae. Comp Biochem Physiol B Biochem Mol Biol 2006;143(4):397–403. doi:10.1016/j.cbpb.2005.12.017.
Bock KW, Kohle C. Coordinate regulation of drug metabolism by xenobiotic nuclear receptors: UGTs acting together with CYPs and glucuronide transporters. Drug Metab Rev 2004;36(3–4):595–615. doi:10.1081/DMR-200033455.
Bouzaiene M, Angers A, Anctil M. Immunohistochemical localization of a retinoic acid-like receptor in nerve cells of two colonial anthozoans (Cnidaria). Tissue Cell 2007;39(2):123–30. doi:10.1016/j.tice.2007.02.001.
Bresell A, Weinander R, Lundqvist G, Raza H, Shimoji M, Sun TH, et al. Bioinformatic and enzymatic characterization of the MAPEG superfamily. FEBS J 2005;272(7):1688–703. doi:10.1111/j.1742-4658.2005.04596.x.
Carbery K, Owen R, Frickers T, Otero E, Readman J. Contamination of Caribbean coastal waters by the antifouling herbicide Irgarol 1051. Mar Pollut Bull 2006;52(6):635–44. doi:10.1016/j.marpolbul.2005.10.013.
Carroll AK, Shick JM. Dietary accumulation of UV-absorbing mycosporine-like amino acids (MAAs) by the green sea urchin (Strongylocentrotus droebachiensis). Mar Biol (Berl) 1996;124(4):561–9. doi:10.1007/BF00351037.
Cashman JR. Some distinctions between flavin-containing and cytochrome P450 monooxygenases. Biochem Biophys Res Commun 2005;338(1):599–604. doi:10.1016/j.bbrc.2005.08.009.
Castro LF, Santos MM, Reis-Henriques MA. The genomic environment around the Aromatase gene: evolutionary insights. BMC Evol Biol 2005;5:43. doi:10.1186/1471-2148-5-43.
Choresh O, Azem A, Loya Y. Over-expression of highly conserved mitochondrial 70-kDa heat-shock protein in the sea anemone Anemonia viridis. J Therm Biol 2007;32(7–8):367–73. doi:10.1016/j.jtherbio.2007.04.006.
Choresh O, Loya Y, Muller WE, Wiedenmann J, Azem A. The mitochondrial 60-kDa heat shock protein in marine invertebrates: biochemical purification and molecular characterization. Cell Stress Chaperones 2004;9(1):38–48.
Choresh O, Ron E, Loya Y. The 60-kDa heat shock protein (HSP60) of the sea anemone Anemonia viridis: a potential early warning system for environmental changes. Mar Biotechnol 2001;3(5):501–8. doi:10.1007/s10126-001-0007-4.
Clemens S. Evolution and function of phytochelatin synthases. J Plant Physiol 2006;163(3):319–32. doi:10.1016/j.jplph.2005.11.010.
Clemens S, Schroeder JI, Degenkolb T. Caenorhabditis elegans expresses a functional phytochelatin synthase. Eur J Biochem 2001;268(13):3640–3. doi:10.1046/j.1432-1327.2001.02293.x.
Cobbett C, Goldsbrough P. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 2002;53:159–82. doi:10.1146/annurev.arplant.53.100301.135154.
Daly M. A systematic revision of Edwardsiidae (Cnidaria, Anthozoa). Invertebr Biol 2002;121(3):212–25.
Darling JA, Reitzel AR, Burton PM, Mazza ME, Ryan JF, Sullivan JC, et al. Rising starlet: the starlet sea anemone, Nematostella vectensis. Bioessays 2005;27(2):211–21. doi:10.1002/bies.20181.
Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science 2006;311(5762):796–800. doi:10.1126/science.1113832.
Davies WJ, Freeman SJ. The Hydra attenuata assay. Methods Mol Biol 1995;43:321–6. Clifton, NJ.
de Jong DM, Hislop NR, Hayward DC, Reece-Hoyes JS, Pontynen PC, Ball EE, et al. Components of both major axial patterning systems of the Bilateria are differentially expressed along the primary axis of a ‘radiate’ animal, the anthozoan cnidarian Acropora millepora. Dev Biol 2006;298(2):632–43. doi:10.1016/j.ydbio.2006.07.034.
Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet 2005;6:123–42. doi:10.1146/annurev.genom.6.080604.162122.
Dean M, Hamon Y, Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res 2001;42(7):1007–17.
Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer 2005;5(4):275–84. doi:10.1038/nrc1590.
Deeley RG, Westlake C, Cole SP. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev 2006;86(3):849–99. doi:10.1152/physrev.00035.2005.
Downs C, Downs A. Preliminary examination of short-term cellular toxicological responses of the coral Madracis mirabilis to acute Irgarol 1051 exposure. Arch Environ Contam Toxicol 2007;52(1):47–57. doi:10.1007/s00244-005-0213-6.
Downs CA, Fauth JE, Halas JC, Dustan P, Bemiss J, Woodley CM. Oxidative stress and seasonal coral bleaching. Free Radic Biol Med 2002;33(4):533–43. doi:10.1016/S0891-5849(02)00907-3.
Dunlap WC, Yamamoto Y. Small-molecule antioxidants in marine organisms: antioxidant activity of mycosporine-glycine. Comp Biochem Physiol B 1995;112(1):105–14. doi:10.1016/0305-0491(95)00086-N.
Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 2008;452(7188):745–9. doi:10.1038/nature06614.
Eddy SR. Profile hidden Markov models. Bioinformatics 1998;14:755–63. doi:10.1093/bioinformatics/14.9.755.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32(5):1792–7. doi:10.1093/nar/gkh340.
Epel D. Protection of DNA during early development: adaptations and evolutionary consequences. Evol Dev 2003;5(1):83–8. doi:10.1046/j.1525-142X.2003.03013.x.
Eraly SA, Monte JC, Nigam SK. Novel slc22 transporter homologs in fly, worm, and human clarify the phylogeny of organic anion and cation transporters. Physiol Genomics 2004;18(1):12–24. doi:10.1152/physiolgenomics.00014.2004.
Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999; 61: 243–82.
Ferrier-Pages C, Richard C, Forcioli D, Allemand D, Pichon M, Shick JM. Effects of temperature and UV radiation increases on the photosynthetic efficiency in four scleractinian coral species. Biol Bull 2007;213(1):76–87.
Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature 2000;408(6809):239–47. doi:10.1038/35041687.
Finnerty JR, Martindale MQ. Ancient origins of axial patterning genes: Hox genes and ParaHox genes in the Cnidaria. Evol Dev 1999;1(1):16–23. doi:10.1046/j.1525-142x.1999.99010.x.
Finnerty JR, Pang K, Burton P, Paulson D, Martindale MQ. Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science 2004;304(5675):1335–7. doi:10.1126/science.1091946.
Fritzenwanker JH, Saina M, Technau U. Analysis of forkhead and snail expression reveals epithelial–mesenchymal transitions during embryonic and larval development of Nematostella vectensis. Dev Biol 2004;275(2):389–402. doi:10.1016/j.ydbio.2004.08.014.
Fufezan C, Rutherford AW, Krieger-Liszkay A. Singlet oxygen production in herbicide-treated photosystem II. FEBS Lett 2002;532(3):407–10. doi:10.1016/S0014-5793(02)03724-9.
Fukuhori N, Kitano M, Kimura H. Toxic effects of bisphenol A on sexual and asexual reproduction in Hydra oligactis. Arch Environ Contam Toxicol 2005;48(4):495–500. doi:10.1007/s00244-004-0032-1.
Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, et al. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci 2006;90(1):5–22. doi:10.1093/toxsci/kfj061.
Garcia E, Ramos R, Bastidas C. Presence of cytochrome P450 in the Carribean corals Siderastrea siderea and Motastaea faveolata. Cinenc 2005;31:1–7.
Gardinali PR, Plasencia MD, Maxey C. Occurrence and transport of Irgarol 1051 and its major metabolite in coastal waters from South Florida. Mar Pollut Bull 2004;49(11–12):1072–83. doi:10.1016/j.marpolbul.2004.08.003.
Gassman NJ, Kennedy CJ. Cytochrome P450 content and xenobiotic metabolizing enzyme activities in the scleractinian coral, Favia fragum (ESPER). Bull Mar Sci 1992;50:320–30.
Ghaskadbi SS, Shetye L, Chiplonkar S, Ghaskadbi S. Ultraviolet irradiation initiates ectopic foot formation in regenerating hydra and promotes budding. J Biosci 2005;30(2):177–82. doi:10.1007/BF02703697.
Goldstone J, Hamdoun AM, Cole B, Ashby MH, Scally M, Dean M, et al. The chemical defensome: environmental sensing and response genes in the Strongylocentrotus purpuratus genome. Dev Biol 2006;300(1):366–84. doi:10.1016/j.ydbio.2006.08.066.
Goldstone JV, Goldstone HM, Morrison AM, Tarrant A, Kern SE, Woodin BR, et al. Cytochrome P450 1 genes in early deuterostomes (tunicates and sea urchins) and vertebrates (chicken and frog): origin and diversification of the CYP1 gene family. Mol Biol Evol 2007;24(12):2619–31. doi:10.1093/molbev/msm200.
Grasso LC, Hayward DC, Trueman JW, Hardie KM, Janssens PA, Ball EE. The evolution of nuclear receptors: evidence from the coral Acropora. Mol Phylogenet Evol 2001;21(1):93–102. doi:10.1006/mpev.2001.0994.
Guengerich FP, McCormick WA, Wheeler JB. Analysis of the kinetic mechanism of haloalkane conjugation by mammalian theta-class glutathione transferases. Chem Res Toxicol 2003;16(11):1493–9. doi:10.1021/tx034157r.
Hagenbuch B, Meier PJ. The superfamily of organic anion transporting polypeptides. Biochim Biophys Acta 2003;1609(1):1–18. doi:10.1016/S0005-2736(02)00633-8.
Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. 3rd ed. Oxford: Oxford University Press; 1999.
Hamdoun A, Epel D. Embryo stability and vulnerability in an always changing world. Proc Natl Acad Sci U S A 2007;104(6):1745–50. doi:10.1073/pnas.0610108104.
Hand C, Uhlinger K. The unique, widely distributed sea anemone, Nematostella vectensis Stephenson: a review, new facts, and questions. Estuaries 1994;17:501–8. doi:10.2307/1352679.
Harborne AR, Mumby PJ, Micheli F, Perry CT, Dahlgren CP, Holmes KE, et al. The functional value of Caribbean coral reef, seagrass and mangrove habitats to ecosystem processes. Adv Mar Biol 2006;50:57–189. doi:10.1016/S0065-2881(05)50002-6.
Harrington L, Fabricius K, Eaglesham G, Negri A. Synergistic effects of diuron and sedimentation on photosynthesis and survival of crustose coralline algae. Mar Pollut Bull 2005;51(1–4):415–27. doi:10.1016/j.marpolbul.2004.10.042.
Hashimoto K, Shibuno T, Murayama-Kayano E, Tanaka H, Kayano T. Isolation and characterization of stress-responsive genes from the scleractinian coral Pocillopora damicornis. Coral Reefs 2004;23(4):485–91.
Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol 2005;45:51–88. doi:10.1146/annurev.pharmtox.45.120403.095857.
Hecker M, Ullrich V. On the mechanism of prostacyclin and thromboxane A2 biosynthesis. J Biol Chem 1989;264(1):141–50.
Heffernan LM, Winston GW. Spectral analysis and catalytic activities of the microsomal mixed-function oxidase system of the sea anemone (phylum: Cnidaria). Comp Biochem Physiol 1998;121(1–3):371–83.
Heffernan LM, Winston GW. Distribution of microsomal CO-binding chromophores and EROD activity in sea anemone tissues. Mar Environ Res 2000;50(1–5):23–7.
Heffernan LM, Zinn RR, Mayeaux MH, Winston GW. Benzo[a]pyrene metabolism in the sea anemone Bunodosoma cavernata. FASEB J 1996;10(6):2868.
Holdway DA, Lok K, Semaan M. The acute and chronic toxicity of cadmium and zinc to two hydra species. Environ Toxicol 2001;16(6):557–65. doi:10.1002/tox.10017.
Hyndman D, Bauman DR, Heredia VV, Penning TM. The aldo-keto reductase superfamily homepage. Chem Biol Interact 2003;143–144:621–31. doi:10.1016/S0009-2797(02)00193-X.
Igarashi K, Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid Redox Signal 2006;8(1–2):107–18. doi:10.1089/ars.2006.8.107.
Jacobsson JA, Haitina T, Lindblom J, Fredriksson R. Identification of six putative human transporters with structural similarity to the drug transporter SLC22 family. Genomics 2007;90(5):595–609. doi:10.1016/j.ygeno.2007.03.017.
Jakobsson PJ, Thoren S, Morgenstern R, Samuelsson B. Identification of human prostaglandin E synthase: a microsomal, glutathione-dependent, inducible enzyme, constituting a potential novel drug target. Proc Natl Acad Sci U S A 1999;96(13):7220–5. doi:10.1073/pnas.96.13.7220.
Jez JM, Penning TM. The aldo-keto reductase (AKR) superfamily: an update. Chem Biol Interact 2001;130–132(1–3):499–525. doi:10.1016/S0009-2797(00)00295-7.
Jin Y, Penning TM. Aldo-keto reductases and bioactivation/detoxication. Annu Rev Pharmacol Toxicol 2007;47:263–92. doi:10.1146/annurev.pharmtox.47.120505.105337.
Johnson EM, Chun YH. In vitro differential developmental toxicity of vitamin A congeners. Teratology 1989;39(4):349–61. doi:10.1002/tera.1420390407.
Jones R. The ecotoxicological effects of Photosystem II herbicides on corals. Mar Pollut Bull 2005;51(5–7):495–506. doi:10.1016/j.marpolbul.2005.06.027.
Jones RJ. Zooxanthellae loss as a bioassay for assessing stress in corals. Mar Ecol Prog Ser 1997;149(1–3):163–71. doi:10.3354/meps149163.
Jones RJ, Kerswell AP. Phytotoxicity of photosystem II (PSII) herbicides to coral. Mar Ecol Prog Ser 2003;261:149–59. doi:10.3354/meps261149.
Karntanut W, Pascoe D. A comparison of methods for measuring acute toxicity to Hydra vulgaris. Chemosphere 2000;41(10):1543–8. doi:10.1016/S0045-6535(00)00068-0.
Karntanut W, Pascoe D. The toxicity of copper, cadmium and zinc to four different Hydra (Cnidaria: Hydrozoa). Chemosphere 2002;47(10):1059–64. doi:10.1016/S0045-6535(02)00050-4.
Karntanut W, Pascoe D. Effects of removing symbiotic green algae on the response of Hydra viridissima (Pallas 1776) to metals. Ecotoxicol Environ Saf 2005;60(3):301–5. doi:10.1016/j.ecoenv.2004.04.001.
Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol 2004;36(2):189–204. doi:10.1016/S1357-2725(03)00211-5.
Kim HP, Morse D, Choi AM. Heat-shock proteins: new keys to the development of cytoprotective therapies. Expert Opin Ther Targets 2006;10(5):759–69. doi:10.1517/14728222.10.5.759.
Kimura H, Takeda M, Narikawa S, Enomoto A, Ichida K, Endou H. Human organic anion transporters and human organic cation transporters mediate renal transport of prostaglandins. J Pharmacol Exp Ther 2002;301(1):293–8. doi:10.1124/jpet.301.1.293.
Koljak R, Jarving I, Kurg R, Boeglin WE, Varvas K, Valmsen K, et al. The basis of prostaglandin synthesis in coral: molecular cloning and expression of a cyclooxygenase from the Arctic soft coral Gersemia fruticosa. J Biol Chem 2001;276(10):7033–40. doi:10.1074/jbc.M009803200.
Kostrouch Z, Kostrouchova M, Love W, Jannini E, Piatigorsky J, Rall JE. Retinoic acid X receptor in the diploblast, Tripedalia cystophora. Proc Natl Acad Sci U S A 1998;95(23):13442–7. doi:10.1073/pnas.95.23.13442.
Kreil DP, Ouzounis CA. Comparison of sequence masking algorithms and the detection of biased protein sequence regions. Bioinformatics 2003;19(13):1672–81. doi:10.1093/bioinformatics/btg212.
Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther 2005;106(3):357–87. doi:10.1016/j.pharmthera.2005.01.001.
Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, et al. Unexpected complexity of the Wnt gene family in a sea anemone. Nature 2005;433(7022):156–60. doi:10.1038/nature03158.
Kusuhara H, Sugiyama Y. ATP-binding cassette, subfamily G (ABCG family). Pflugers Arch 2007;453(5):735–44. doi:10.1007/s00424-006-0134-x.
Laity JH, Andrews GK. Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1). Arch Biochem Biophys 2007;463(2):201–10. doi:10.1016/j.abb.2007.03.019.
Lee MO, Manthey CL, Sladek NE. Identification of mouse liver aldehyde dehydrogenases that catalyze the oxidation of retinaldehyde to retinoic acid. Biochem Pharmacol 1991;42(6):1279–85. doi:10.1016/0006-2952(91)90266-8.
Leslie EM, Deeley RG, Cole SP. Toxicological relevance of the multidrug resistance protein 1, MRP1 (ABCC1) and related transporters. Toxicology 2001;167(1):3–23. doi:10.1016/S0300-483X(01)00454-1.
Lesser MP. Oxidative stress in marine environments: biochemistry and physiological ecology. Annu Rev Physiol 2006;68:253–78. doi:10.1146/annurev.physiol.68.040104.110001.
Lewis DF, Lake BG, Dickins M. Substrates of human cytochromes P450 from families CYP1 and CYP2: analysis of enzyme selectivity and metabolism. Drug Metabol Drug Interact 2004;20(3):111–42.
Mackenzie PI, Walter Bock K, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, et al. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics 2005;15(10):677–85. doi:10.1097/01.fpc.0000173483.13689.56.
Magie CR, Pang K, Martindale MQ. Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis. Dev Genes Evol 2005;215(12):618–30. doi:10.1007/s00427-005-0022-y.
Matus DQ, Pang K, Marlow H, Dunn CW, Thomsen GH, Martindale MQ. Molecular evidence for deep evolutionary roots of bilaterality in animal development. Proc Natl Acad Sci U S A 2006;103(30):11195–200. doi:10.1073/pnas.0601257103.
Merle PL, Sabourault C, Richier S, Allemand D, Furla P. Catalase characterization and implication in bleaching of a symbiotic sea anemone. Free Radic Biol Med 2007;42(2):236–46. doi:10.1016/j.freeradbiomed.2006.10.038.
Miller DJ, Hemmrich G, Ball EE, Hayward DC, Khalturin K, Funayama N, et al. The innate immune repertoire in cnidaria–ancestral complexity and stochastic gene loss. Genome Biol 2007;8(4):R59. doi:10.1186/gb-2007-8-4-r59.
Mitchelmore CL, Hyatt S. Assessing DNA damage in cnidarians using the Comet assay. Mar Environ Res 2004;58(2–5):707–11. doi:10.1016/j.marenvres.2004.03.019.
Mitchelmore CL, Schwarz JA, Weis VM. Development of symbiosis-specific genes as biomarkers for the early detection of cnidarian-algal symbiosis breakdown. Mar Environ Res 2002;54(3–5):345–9. doi:10.1016/S0141-1136(02)00201-5.
Mitchelmore CL, Verde EA, Weis VM. Uptake and partitioning of copper and cadmium in the coral Pocillopora damicornis. Aquat Toxicol 2007;85(1):48–56. doi:10.1016/j.aquatox.2007.07.015.
Miyake K, Mickley L, Litman T, Zhan Z, Robey R, Cristensen B, et al. Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistant cells: demonstration of homology to ABC transport genes. Cancer Res 1999;59(1):8–13.
Mizuta T, Kubokawa K. Presence of sex steroids and cytochrome P450 genes in amphioxus. Endocrinology 2007;148(8):3554–65. doi:10.1210/en.2007-0109.
Mopper K, Kieber DJ. Marine photochemistry and its impacts on carbon cycling. In: De Mora S, Demers S, Vernet M, editors. The effects of UV radiation in the marine environment. Cambridge, UK: Cambridge University Press; 2000. p. 101–30.
Morgan MB, Vogelien DL, Snell TW. Assessing coral stress responses using molecular biomarkers of gene transcription. Environ Toxicol Chem 2001;20(3):537–43. doi:10.1897/1551-5028(2001)020<0537:ACSRUM>2.0.CO;2.
Muller WA. Retinoids and pattern formation in a hydroid. J Embryol Exp Morphol 1984;81:253–71.
Murphy BJ. Regulation of malignant progression by the hypoxia-sensitive transcription factors HIF-1alpha and MTF-1. Comp Biochem Physiol B Biochem Mol Biol 2004;139(3):495–507. doi:10.1016/j.cbpc.2004.04.009.
Murphy BJ, Andrews GK, Bittel D, Discher DJ, McCue J, Green CJ, et al. Activation of metallothionein gene expression by hypoxia involves metal response elements and metal transcription factor-1. Cancer Res 1999;59(6):1315–22.
Murphy BJ, Sato BG, Dalton TP, Laderoute KR. The metal-responsive transcription factor-1 contributes to HIF-1 activation during hypoxic stress. Biochem Biophys Res Commun 2005;337(3):860–7. doi:10.1016/j.bbrc.2005.09.124.
Nebert DW, Vasiliou V. Analysis of the glutathione S-transferase (GST) gene family. Hum Genomics 2004;1(6):460–4.
Negri A, Vollhardt C, Humphrey C, Heyward A, Jones R, Eaglesham G, et al. Effects of the herbicide diuron on the early life history stages of coral. Mar Pollut Bull 2005;51(1–4):370–83. doi:10.1016/j.marpolbul.2004.10.053.
Negri AP, Heyward AJ. Inhibition of fertilization and larval metamorphosis of the coral Acropora millepora (Ehrenberg, 1834) by petroleum products. Mar Pollut Bull 2000;41(7–12):420–7.
Negri AP, Heyward AJ. Inhibition of coral fertilisation and larval metamorphosis by tributyltin and copper. Mar Environ Res 2001;51(1):17–27. doi:10.1016/S0141-1136(00)00029-5.
Nelson DR. Metazoan cytochrome P450 evolution. Comp Biochem Physiol 1998;121(1–3):15–22.
Nelson DR. Cytochrome P450 nomenclature, 2004. Methods Mol Biol 2006;320:1–10. Clifton, NJ.
Nelson DR, Kamataki T, Waxman DJ, Guengerich FP, Estabrook RW, Feyereisen R, et al. The P450 superfamily: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol 1993;12(1):1–51.
Nemer M, Wilkinson DG, Travaglini EC, Sternberg EJ, Butt TR. Sea urchin metallothionein sequence: key to an evolutionary diversity. Proc Natl Acad Sci U S A 1985;82(15):4992–4. doi:10.1073/pnas.82.15.4992.
Nguyen T, Sherratt PJ, Huang HC, Yang CS, Pickett CB. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J Biol Chem 2003;278(7):4536–41. doi:10.1074/jbc.M207293200.
Nystrom M, Nordemar I, Tedengren M. Simultaneous and sequential stress from increased temperature and copper on the metabolism of the hermatypic coral Porites cylindrica. Mar Biol (Berl) 2001;138(6):1225–31. doi:10.1007/s002270100549.
Osburn WO, Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutat Res 2008;659(1–2):31–9. doi:10.1016/j.mrrev.2007.11.006.
Pachura-Bouchet S, Blaise C, Vasseur P. Toxicity of nonylphenol on the cnidarian Hydra attenuata and environmental risk assessment. Environ Toxicol 2006;21(4):388–94. doi:10.1002/tox.20201.
Palackal NT, Burczynski ME, Harvey RG, Penning TM. Metabolic activation of polycyclic aromatic hydrocarbon trans-dihydrodiols by ubiquitously expressed aldehyde reductase (AKR1A1). Chem Biol Interact 2001;130–132(1–3):815–24. doi:10.1016/S0009-2797(00)00237-4.
Pascoe D, Carroll K, Karntanut W, Watts MM. Toxicity of 17alpha-ethinylestradiol and bisphenol A to the freshwater Cnidarian Hydra vulgaris. Arch Environ Contam Toxicol 2002;43(1):56–63. doi:10.1007/s00244-001-0016-3.
Paul VJ, Puglisi MP. Chemical mediation of interactions among marine organisms. Nat Prod Rep 2004;21(1):189–209. doi:10.1039/b302334f.
Paul VJ, Puglisi MP, Ritson-Williams R. Marine chemical ecology. Nat Prod Rep 2006;23(2):153–80. doi:10.1039/b404735b.
Penning TM, Burczynski ME, Hung CF, McCoull KD, Palackal NT, Tsuruda LS. Dihydrodiol dehydrogenases and polycyclic aromatic hydrocarbon activation: generation of reactive and redox active o-quinones. Chem Res Toxicol 1999;12(1):1–18. doi:10.1021/tx980143n.
Penning TM, Drury JE. Human aldo-keto reductases: function, gene regulation, and single nucleotide polymorphisms. Arch Biochem Biophys 2007;464(2):241–50. doi:10.1016/j.abb.2007.04.024.
Philp RB. Cadmium content of the marine sponge Microciona prolifera, other sponges, water and sediment from the eastern Florida panhandle: possible effects on Microciona cell aggregation and potential roles of low pH and low salinity. Comp Biochem Physiol 1999;124(1):41–9. doi:10.1016/S0305-0491(99)00095-4.
Plantivaux A, Furla P, Zoccola D, Garello G, Forcioli D, Richier S, et al. Molecular characterization of two CuZn-superoxide dismutases in a sea anemone. Free Radic Biol Med 2004;37(8):1170–81. doi:10.1016/j.freeradbiomed.2004.06.043.
Ponting CP. The functional repertoires of metazoan genomes. Nat Rev 2008;9(9):689–98.
Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007;317(5834):86–94. doi:10.1126/science.1139158.
Qi SH, Zhang S, Yang LH, Qian PY. Antifouling and antibacterial compounds from the gorgonians Subergorgia suberosa and Scripearia gracillis. Nat Prod Res 2008;22(2):154–66. doi:10.1080/14786410701642441.
Raberg S, Nystrom M, Eros M, Plantman P. Impact of the herbicides 2,4-D and diuron on the metabolism of the coral Porites cylindrica. Mar Environ Res 2003;56(4):503–14. doi:10.1016/S0141-1136(03)00039-4.
Ramos R, Garcia E. Induction of mixed-function oxygenase system and antioxidant enzymes in the coral Montastraea faveolata on acute exposure to benzo(a)pyrene. Comp Biochem Physiol C Toxicol Pharmacol 2007;144(4):348–55. doi:10.1016/j.cbpc.2006.11.006.
Reichelt-Brushett AJ, Harrison PL. The effect of copper on the settlement success of larvae from the scleractinian coral Acropora tenuis. Mar Pollut Bull 2000;41(7–12):385–91.
Reichelt-Brushett AJ, Harrison PL. The effect of selected trace metals on the fertilization success of several scleractinian coral species. Coral Reefs 2005;24(4):524–34. doi:10.1007/s00338-005-0013-5.
Reitzel AM, Darling JA, Sullivan JC, Finnerty JR. Global population genetic structure of the starlet anemone Nematostella vectensis: multiple introductions and implications for conservation policy. Biol Invasions. 2008a (in press).
Reitzel AM, Sullivan JC, Traylor-Knowles N, Finnerty JR. Genomic survey of candidate stress-response genes in the estuarine anemone Nematostella vectensis. Biol Bull 2008b;214(3):233–54.
Richier S, Furla P, Plantivaux A, Merle PL, Allemand D. Symbiosis-induced adaptation to oxidative stress. J Exp Biol 2005;208(Pt 2):277–85. doi:10.1242/jeb.01368.
Richier S, Merle PL, Furla P, Pigozzi D, Sola F, Allemand D. Characterization of superoxide dismutases in anoxia- and hyperoxia-tolerant symbiotic cnidarians. Biochim Biophys Acta 2003;1621(1):84–91.
Robbart ML, Peckol P, Scordilis SP, Curran HA, Brown-Saracino J. Population recovery and differential heat shock protein expression for the corals Agaricia agaricites and A-tenuifolia in Belize. Mar Ecol Prog Ser 2004;283:151–60. doi:10.3354/meps283151.
Rossi S, Snyder MJ. Competition for space among sessile marine invertebrates: changes in HSP70 expression in two Pacific cnidarians. Biol Bull 2001;201(3):385–93. doi:10.2307/1543616.
Rossi S, Snyder MJ, Gili JM. Protein, carbohydrate, lipid concentrations and HSP70-HSP 90 (stress protein) expression over an annual cycle: useful tools to detect feeding constraints in a benthic suspension feeder. Helgol Mar Res 2006;60(1):7–17. doi:10.1007/s10152-005-0009-0.
Rougee L, Downs CA, Richmond RH, Ostrander GK. Alteration of normal cellular profiles in the scleractinian coral (Pocillopora damicornis) following laboratory exposure to fuel oil. Environ Toxicol Chem 2006;25(12):3181–7. doi:10.1897/05-510R2.1.
Runge-Morris M, Kocarek TA. Regulation of sulfotransferases by xenobiotic receptors. Curr Drug Metab 2005;6(4):299–307. doi:10.2174/1389200054633871.
Sebens KP. Recruitment in a sea anemone population: juvenile substrate becomes adult prey. Science 1981;213(4509):785–7. doi:10.1126/science.213.4509.785.
Sharp VA, Brown BE, Miller D. Heat shock protein (hsp 70) expression in the tropical reef coral Goniopora djiboutiensis. J Therm Biol 1997;22(1):11–9. doi:10.1016/S0306-4565(96)00029-0.
Sharp VA, Miller D, Bythell JC, Brown BE. Expression of low-molecular-weight Hsp-70 related polypeptides from the symbiotic sea-anemone Anemonia-viridis Forskall in response to heat-shock. J Exp Mar Biol Ecol 1994;179(2):179–93. doi:10.1016/0022-0981(94)90113-9.
Sheader M, Suwailem A, Rowe G. The anemone, Nematostella vectensis, in Britain: considerations for conservation management. Aquat Conserv: Mar Freshwat Ecosyst 1997;7:13–25. doi:10.1002/(SICI)1099-0755(199703)7:1<13::AID-AQC210>3.0.CO;2-Y.
Shick JM, Dunlap WC. Mycosporine-like amino acids and related Gadusols: biosynthesis, acumulation, and UV-protective functions in aquatic organisms. Annu Rev Physiol 2002;64:223–62. doi:10.1146/annurev.physiol.64.081501.155802.
Shick JM, Dunlap WC, Pearse JS, Pearse VB. Mycosporine-like amino acid content in four species of sea anemones in the genus Anthopleura reflects phylogenetic but not environmental or symbiotic relationships. Biol Bull 2002;203(3):315–30. doi:10.2307/1543574.
Shimada T. Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab Pharmacokinet 2006;21(4):257–76. doi:10.2133/dmpk.21.257.
Sica D, Musumeci D. Secosteroids of marine origin. Steroids 2004;69(11–12):743–56. doi:10.1016/j.steroids.2004.09.001.
Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, et al. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol 2007;7:33. doi:10.1186/1471-2148-7-33.
Snyder MJ, Ross S. Stress protein (HSP70 family) expression in intertidal bentbic organisms: the example of Anthopleura elegantissima (Cnidaria: Anthozoa). Sci Mar 2004;68:155–62.
Sole M, Livingstone DR. Components of the cytochrome P450-dependent monooxygenase system and ‘NADPH-independent benzo[a]pyrene hydroxylase’ activity in a wide range of marine invertebrate species. Comp Biochem Physiol C Toxicol Pharmacol 2005;141(1):20–31. doi:10.1016/j.cca.2005.04.008.
Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006;22(21):2688–90. doi:10.1093/bioinformatics/btl446.
Starcevic A, Akthar S, Dunlap WC, Shick JM, Hranueli D, Cullum J, et al. Enzymes of the shikimic acid pathway encoded in the genome of a basal metazoan, Nematostella vectensis, have microbial origins. Proc Natl Acad Sci U S A 2008;105:2533–37.
Sullivan JC, Reitzel AM, Finnerty JR. Upgrades to StellaBase facilitate medical and genetic studies on the starlet sea anemone, Nematostella vectensis. Nucleic Acids Res. 2008;36:D607–11, (Database issue) doi:10.1093/nar/gkm941.
Surh YJ. Bioactivation of benzylic and allylic alcohols via sulfo-conjugation. Chem Biol Interact 1998;109(1–3):221–35. doi:10.1016/S0009-2797(97)00134-8.
Sweet DH. Organic anion transporter (Slc22a) family members as mediators of toxicity. Toxicol Appl Pharmacol 2005;204(3):198–215. doi:10.1016/j.taap.2004.10.016.
Tarrant AM. Endocrine-like signaling in cnidarians: current understanding and implications for ecophysiology. Integr Comp Biol 2005;45:201–14. doi:10.1093/icb/45.1.201.
Tarrant AM. Hormonal signaling in cnidarians: do we understand the pathways well enough to know whether they are being disrupted? Ecotoxicology 2007;16(1):5–13. doi:10.1007/s10646-006-0121-1.
Tarrant AM, Atkinson MJ, Atkinson S. Effects of steroidal estrogens on coral growth and reproduction. Mar Ecol Prog Ser 2004;269:121–9. doi:10.3354/meps269121.
Tarrant AM, Atkinson S, Atkinson MJ. Estrone and estradiol-17 beta concentration in tissue of the scleractinian coral, Montipora verrucosa. Comp Biochem Physiol A Mol Integr Physiol 1999;122(1):85–92. doi:10.1016/S1095-6433(98)10155-1.
Tarrant AM, Blomquist CH, Lima PH, Atkinson MJ, Atkinson S. Metabolism of estrogens and androgens by scleractinian corals. Comp Biochem Physiol B Biochem Mol Biol 2003;136(3):473–85. doi:10.1016/S1096-4959(03)00253-7.
Thomas JH. Rapid birth–death evolution specific to xenobiotic cytochrome P450 genes in vertebrates. PLoS Genet 2007;3(5):e67. doi:10.1371/journal.pgen.0030067.
Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science 2003;301(5640):1714–7. doi:10.1126/science.1086185.
Torras R, Gonzalez-Crespo S. Posterior expression of nanos orthologs during embryonic and larval development of the anthozoan Nematostella vectensis. Int J Dev Biol 2005;49(7):895–9. doi:10.1387/ijdb.051980rt.
Torres JL, Armstrong RA, Corredor JE, Gillbes F. Physiological responses of Acropora cervicornis to increased solar irradiance. Photochem Photobiol 2007;83(4):839–50.
Twan WH, Hwang JS, Chang CF. Sex steroids in scleractinian coral, Euphyllia ancora: implication in mass spawning. Biol Reprod 2003;68(6):2255–60. doi:10.1095/biolreprod.102.012450.
Twan WH, Hwang JS, Lee YH, Wu HF, Tung YH, Chang CF. Hormones and reproduction in scleractinian corals. Comp Biochem Physiol A Mol Integr Physiol 2006;144(3):247–53. doi:10.1016/j.cbpa.2006.01.011.
Uno S, Dalton TP, Shertzer HG, Genter MB, Warshawsky D, Talaska G, et al. Benzo[a]pyrene-induced toxicity: paradoxical protection in Cyp1a1(−/−) knockout mice having increased hepatic BaP-DNA adduct levels. Biochem Biophys Res Commun 2001;289(5):1049–56. doi:10.1006/bbrc.2001.6110.
Vasiliou V, Nebert DW. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics 2005;2(2):138–43.
Vasiliou V, Ross D, Nebert DW. Update of the NAD(P)H:Quinone oxidoreductase (NQO) gene family. Hum Genomics 2006;2(5):329–35.
Verde EA, McCloskey LR. A comparative analysis of the photobiology of zooxanthellae and zoochlorellae symbiotic with the temperate clonal anemone Anthopleura elegantissima (Brandt)—II. Effect of light intensity. Mar Biol (Berl) 2002;141(2):225–39. doi:10.1007/s00227-002-0824-7.
Wellington GM, Fitt WK. Influence of UV radiation on the survival of larvae from broadcast-spawning reef corals. Mar Biol (Berl) 2003;143(6):1185–92. doi:10.1007/s00227-003-1150-4.
Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 2001;18(5):691–9.
Wiens M, Ammar MS, Nawar AH, Koziol C, Hassanein HMA, Eisinger M, et al. Induction of heat-shock (stress) protein gene expression by selected natural and anthropogenic disturbances in the octocoral Dendronephthya klunzingeri. J Exp Mar Biol Ecol 2000;245(2):265–76. doi:10.1016/S0022-0981(99)00167-7.
Wiger R, Stottum A. In vitro testing for developmental toxicity using the Hydra attenuata assay. NIPH Ann 1985;8(2):43–7.
Winston GW, Mayeaux MH, Heffernan LM. Benzo[a]pyrene metabolism by the intertidal sea anemone, Bunodosoma cavernata. Mar Environ Res 1998;45(1):89–100. doi:10.1016/S0141-1136(97)00026-3.
Withers NW, Kokke WC, Fenical W, Djerassi C. Sterol patterns of cultured zooxanthellae isolated from marine invertebrates: synthesis of gorgosterol and 23-desmethylgorgosterol by aposymbiotic algae. Proc Natl Acad Sci U S A 1982;79(12):3764–8. doi:10.1073/pnas.79.12.3764.
Yakovleva I, Bhagooli R, Takemura A, Hidaka M. Differential susceptibility to oxidative stress of two scleractinian corals: antioxidant functioning of mycosporine-glycine. Comp Biochem Physiol B Biochem Mol Biol 2004;139(4):721–30. doi:10.1016/j.cbpc.2004.08.016.
Yamada S, Morimoto H, Fujisawa T, Sugahara K. Glycosaminoglycans in Hydra magnipapillata (Hydrozoa, Cnidaria): demonstration of chondroitin in the developing nematocyst, the sting organelle, and structural characterization of glycosaminoglycans. Glycobiology 2007;17(8):886–94. doi:10.1093/glycob/cwm051.
Ziegler DM. An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab Rev 2002;34(3):503–11. doi:10.1081/DMR-120005650.
Znidaric D, Liu A, Kalafatic M. Regeneration and reproduction of irradiated green hydra (Cnidaria). Period Biol 1992;94(2):99–104.
Acknowledgments
Funding was provided by the WHOI Ocean Life Institute and NIH R01-ES015912. This manuscript benefited from discussions with Ann Tarrant, Adam Reitzel, and Cécile Sabourault and from the comments of several anonymous reviewers.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Goldstone, J.V. Environmental sensing and response genes in cnidaria: the chemical defensome in the sea anemone Nematostella vectensis . Cell Biol Toxicol 24, 483–502 (2008). https://doi.org/10.1007/s10565-008-9107-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10565-008-9107-5