Abstract
Nephrolithiasis is a costly and prevalent disease that is associated with significant morbidity including pain, infection, and kidney injury. While surgical treatment of kidney stones is generally based on the size and quality of the stones, studies have suggested that specific characteristics of the pyelocalyceal anatomy (i.e. urinary drainage system), such as the infundibulopelvic angle (IPA), can influence the success rate of various treatment modalities. However, the traditional methods of quantifying such anatomic features have typically relied on manual measurements using 2-dimensional (2D) images of a 3-dimensional (3D) system, which can be cumbersome and potentially inaccurate. In this paper, we propose a novel algorithm that automatically identifies and isolates the 3D volume and central frame of the urinary drainage system from computerized tomography (CT) Urograms, which then allows for 3D characterization of the pyelocalyceal anatomy. First, the kidney and pyelocalyceal system were segmented from adjacent soft tissues using an automated algorithm. A centerline tree structure was then generated from the segmented pyelocalyceal anatomy. Finally, the IPA was measured using the derived reconstructions and tree structure. 8 of 11 pyelocalyceal systems were successfully segmented and used to measure the IPA, suggesting that it is technically feasible to use our algorithm to automatically segment the pyelocalyceal anatomy from target images and determine its 3D central frame for anatomic characterization. To the best of our knowledge, this is the first method that allows for an automated characterization of the isolated 3D pyelocalyceal structure from CT images.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The prevalence of kidney stone disease, or nephrolithiasis, has been rising over the last several decades and now affects approximately 1 in 11 individuals in the United States [1]. Most stones that do not spontaneously pass will require surgical treatment with ureteroscopy (retrograde endoscopy through the urethra and bladder), extracorporeal shock wave lithotripsy (stones fragmentation using noninvasive shock waves), percutaneous lithotripsy (endoscopy through 1 cm direct puncture into the kidney), or very rarely laparoscopic or open surgery. An efficient and effective choice of surgical approach is critical given the significant morbidity due to kidney stones, including pain, infection, and renal insufficiency, as well as associated costs, which were estimated to be over $5 billion in 2000 [2]. Currently, more than 40% of patients may not be stone free after surgery [3].
In determining an optimal surgical approach, it is essential to consider anatomic factors and stone features as these affect treatment success rates [4, 5]. However, prior research correlating specific characteristics of the pyelocalyceal anatomy (kidney drainage or urinary collecting system where stones grow), such as the IPA (angle representing the lower pole (i.e. most inferior portion of the drainage system where stones can settle)) and stone-free rates after surgery has often relied on manual measurements of 2D imaging modalities, such as fluoroscopy and intravenous pyelograms, to characterize the 3D urinary collecting system. Data from such studies are conflicting, which may in part be due to the crudeness of the 2D approximations [4, 6]. For example, the range of IPAs in patients using 2D intravenous urograms are not consistent with those measured from 3D resin casts of cadaver kidneys [7, 8]. Furthermore, many of the studies were performed using images taken during surgery, meaning the images were not available pre-operatively to actually aid in treatment planning.
The above indicate a strong need and opportunity for improvement in image-based patient-specific preoperative planning and counseling in the management of stone disease. The high prevalence of CT as a clinical tool provides an ideal avenue to develop algorithms for patient-specific computer-aided treatment guidance. In addition, this type of data at a population level will be highly valuable in the development of novel devices for kidney stone surgery and more general characterization of anatomy.
In this feasibility study, we aimed to automatically segment and isolate the 3D structure of the renal collecting system anatomy in normal CT Urograms that could then be used to measure the IPA, a key feature previously identified as potentially correlating with operative accessibility and thus, success of a given surgical approach.
2 Methods
2.1 Patient Selection and Imaging
The Institutional Review Board approved this study with a waiver of informed consent. Electronic medical records were used to randomly identify patients who had a CT Urogram for evaluation of hematuria (blood in the urine) [9]. Exclusion criteria included any treated or untreated kidney pathology including tumors, presence of kidney stones, anatomic variants, and chronic renal insufficiency as this affects the rate of contrast excretion. Images were manually reviewed to confirm good image quality. All excretory phase sequences (Fig. 1 demonstrates difference between a non-contrast and excretory phase image) in this study were performed in the prone position at an 8 min delay per institutional protocol with 3 mm axial reconstructions.
2.2 Automated Localization and Segmentation of Whole Kidney
Figure 2 demonstrates the workflow of the proposed algorithm. A SIMPLE context learning-based multi-atlas segmentation framework [10] was used to achieve whole kidney segmentation. To achieve the SIMPLE framework, 30 pairs of atlases (anatomical CT scans and corresponding labels) were obtained from MICCAI 2015 MeDiCAL challenges (https://www.synapse.org/#!Synapse:syn3193805/wiki/89480). Two sets of cropped atlases were then formed based on kidney locations (30 pairs each for the left and right kidneys). The atlases were manually cropped by an experienced rater using MIPAV software [11]. Next, the left and right kidneys in target CT Urogram images were automatically localized and cropped using a random forest based localization method [12]. The previously cropped atlases were then registered to the cropped target CT Urogram images using affine and non-rigid registrations by NiftyReg [13]. A SIMPLE based context learning procedure was performed to select the best 10 registered atlases for each target kidney [14]. Finally, the left and right kidney segmentations from the target images were separately derived by performing a joint label fusion (JLF) [15] on the selected atlases.
The workflow of the proposed algorithm. First, the whole kidney was localized and segmented using multi-atlas segmentation. The pyelocalyceal structure was then segmented using a Gaussian Mixture Model and the tree structure was subsequently derived. Key landmarks (yellow dots) were identified from the 3D reconstruction and tree structure to construct an oblique 4 mm thick plane from which the IPA was measured. (Color figure online)
2.3 Automated Segmentation of Pyelocalyceal Anatomy and Validation
Once the kidneys were cropped and segmented from the original excretory phase images, a Gaussian mixture model (GMM) was used to segment the pyelocalyceal anatomy within the kidneys. Empirically, a threshold above 100 Hounsfield Unit (HU) was applied to exclude tissues surrounding the kidney. The GMM with three components was then employed on the histogram of remaining intensities. The two components (from three total) with higher mean HU score were clustered and identified to be the pyelocalyceal anatomy segmentation. The component with smallest mean HU score represented residual kidney organ tissue not completed removed in the initial thresholding step. Finally, a 3D tree structure (center line) was derived from the pyelocalyceal anatomy segmentation using the method described in [16]. Briefly, the method calculated the 3D axis skeleton of the 3D binary volume using a parallel thinning algorithm based on an Euler table.
All pyelocalyceal segmentations were qualitatively evaluated by a radiologist and rated as having excellent, acceptable, or poor accuracy. A random subset of the kidneys that resulted in excellent or acceptable segmentations were then manually segmented by a radiologist and the Dice coefficient was calculated.
2.4 Measurement of Infundibulopelvic Angle in 2D and 3D Images
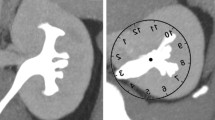
The previously described Elbahnasy method for IPA measurement in 2D images was modified to allow for IPA measurement using 3D images and the above derived 3D tree structure [17]. Key landmarks corresponding to those in the Elbahnasy method were manually identified by a Urologist in 3D slicer software (https://www.slicer.org) using the kidney segmentation, pyelocalyceal anatomy and tree structure derived from above automated algorithm. The landmarks based on the Elbahnasy method were as follows: (1) the center point of the proximal ureter at the lowest plane of the kidney, (2) the center point of the renal pelvis along medial margin of kidney, (3) a point in the inferior branch of the kidney drainage system (i.e. lower pole). The three points were used to create a unique 4 mm thick slice from the 3D volume, and the IPA was measured as the angle between the line connecting points (1) and (2), and the center line through the lowest branch of the kidney drainage system (Fig. 2).
As a comparison, 2D measurements of the IPA were performed on the cropped kidney from an averaged CT image in the coronal direction. This traditional 2D measurement was obtained by estimating the center point (A) of the proximal ureter at the lowest plane of the kidney, estimating the center point (B) of the renal pelvis along medial margin of kidney, approximating the center line (C) through the lower pole branch, and measuring the angle between line C and a line connecting A and B (Fig. 3).
Example results of the segmentation and angle measurements for a single kidney. Top row: 3D reconstruction of the kidney, 3D reconstruction of the pyelocalyceal structure, tree structure. Bottom row: Overlays of reconstructions and tree structure, traditional 2D measurement [14] of IPA (red lines) using an averaged (i.e. flattened) pseudo-2D image generated from CT images to simulate IV pyelogram IPA measurement (horizontal blue line indicates lowest plane of the kidney, sloped vertical blue line indicates medial margin of kidney, (A) center of ureter at lower margin of kidney, (B) center of renal pelvis at medial margin, (C) center line through lower pole branch), and the 3D IPA measurement (red lines) using described method. (Color figure online)
3 Results
3.1 Patients
After exclusion of patients with imaging artifacts or inadequate collecting system distension, CT images of 11 individual kidneys from 8 patients were identified to be appropriate for this feasibility study. Patients ranged in age from 41–80 years old and all had normal kidney function.
3.2 Pyelocalyceal Anatomy Segmentation
The pyelocalyceal anatomy was appropriately segmented in 8 of the 11 kidneys with a rating of excellent or acceptable by the radiologist. Of these, 6 were randomly chosen and manually segmented by the radiologist and Dice coefficients ranged from 0.62 to 0.88.
3.3 Infundibulopelvic Angle
Figure 3 demonstrates the segmentation results, tree structure, as well as 2D and 3D IPA measurements from a single example kidney. The IPA based on the 3D segmentations and tree structures ranged from 14.6° to 81.5° while IPA based on 2D reformatted images ranged from 9.4° to 88.3° (Table 1). Comparisons between the angles based on the 2D and 3D methods demonstrated differences up to 35.6%.
4 Discussion
Kidney stone disease is a chronic condition that often requires many surgeries over a patient’s lifetime. Each surgery is associated with risks and residual stones can have severe consequences so appropriate pre-operative evaluation and intervention are critical [18]. In addition to stone-free rates after surgery, anatomic variation may play a role in stone formation and burden of disease [6]. Thus, accurate characterization of patient anatomy can have both immediate and long-term effects with respect to surgical planning as well as lifelong management, such as the interval between imaging studies.
Augmentation of currently widely available CT Urography with powerful post-processing tools such as 3D modeling and characterization algorithms may aid in advancing patient-tailored medicine in urologic disease. While there is prior work on automated detection of kidney stones and kidney tumors [19, 20] from CT images, this is the first method known to the authors for automated isolation and characterization of the 3D pyelocalyceal frame. There have been some efforts to perform automated identification and segmentation of the pyelocalyceal anatomy using magnetic resonance imaging (MRI). A mean Dice coefficient of 0.72 [21] has been reported, which is within the range achieved in this study, but anatomic characterization was not performed and furthermore, MRIs do not provide adequate visualization of kidney stones and thus are not used for stone patients. The results presented here demonstrate that the proposed algorithm is technically feasible with CT imaging and Dice coefficient calculations indicate that the automated segmentation results compare favorably with manual segmentations for the given geometry.
With respect to the IPA, the relative difference in the measured IPA between the 2D and 3D techniques was noted to be up to 35%. As previously mentioned, prior studies have indicated that anatomic variation may be critical to predicting surgical success, but the data on IPA and other anatomic parameters are inconsistent [4]. As this preliminary data suggests, the discrepancies may be partially attributed to the lower anatomic fidelity of the traditionally utilized 2D images, and the advantages of using 3D techniques are a focus of our future studies. An inherent limitation of such automated algorithms is that the result will only be as reliable as the initial imaging, and imaging quality of CT urograms can be dependent on multiple factors such as kidney function and level of hydration. We aim to further automate our algorithm, assess additional anatomic variables, both novel and previously described, and then correlate these more accurate 3D-based measurements with stone-free rates after stone surgery. Outcomes from such studies may provide valuable tools for patient-specific stone management.
References
Scales, C.D.J., Smith, A.C., Hanley, J.M., Saigal, C.S.: Prevalence of kidney stones in the United States. Eur. Urol. 62, 160–165 (2012)
Saigal, C.S., Joyce, G., Timilsina, A.R.: Direct and indirect costs of nephrolithiasis in an employed population: opportunity for disease management? Kidney Int. 68, 1808–1814 (2005)
Ghani, K.R., Wolf, J.S.: What is the stone-free rate following flexible ureteroscopy for kidney stones? Nat. Rev. Urol. 12, 281–288 (2015)
Danuser, H., Müller, R., Descoeudres, B., Dobry, E., Studer, U.E.: Extracorporeal shock wave lithotripsy of lower calyx calculi: how much is treatment outcome influenced by the anatomy of the collecting system? Eur. Urol. 52, 539–546 (2007)
Geavlete, P., Multescu, R., Geavlete, B.: Influence of pyelocaliceal anatomy on the success of flexible ureteroscopic approach. J. Endourol. 22, 2235–2239 (2008)
Zomorrodi, A., Buhluli, A., Fathi, S.: Anatomy of the collecting system of lower pole of the kidney in patients with a single renal stone: a comparative study with individuals with normal kidneys. Saudi J. Kidney Dis. Transpl. 21, 666–672 (2010)
Marroig, B., Favorito, L.A., Fortes, M.A., Sampaio, F.J.B.: Lower pole anatomy and mid-renal-zone classification applied to flexible ureteroscopy: experimental study using human three-dimensional endocasts. Surg. Radiol. Anat. 37, 1243–1249 (2015)
Gozen, A.S., Kilic, A.S., Aktoz, T., Akdere, H.: Renal anatomical factors for the lower calyceal stone formation. Int. Urol. Nephrol. 38, 79–85 (2006)
Danciu, I., Cowan, J.D., Basford, M., Wang, X., Saip, A., Osgood, S., Shirey-Rice, J., Kirby, J., Harris, P.A.: Secondary use of clinical data: the Vanderbilt approach. J. Biomed. Inform. 52, 28–35 (2014)
Xu, Z., Burke, R.P., Lee, C.P., Baucom, R.B., Poulose, B.K., Abramson, R.G., Landman, B.A.: Efficient abdominal segmentation on clinically acquired CT with SIMPLE context learning. Proc. SPIE Int. Soc. Opt. Eng. 9413, 94130L (2015)
McAuliffe, M.J., Lalonde, F.M., McGarry, D., Gandler, W., Csaky, K., Trus, B.L.: Medical image processing, analysis and visualization in clinical research. In: 14th IEEE Symposium on Computer-Based Medical Systems (CBMS 2001), Proceedings, pp. 381–386. IEEE (2001)
Criminisi, A., Jamie, S., Konukoglu, E.: Decision forests: a unified framework for classification, regression, density estimation, manifold learning and semi-supervised learning. Found. Trends® Comput. Graph Vis. 7.2(3), 81–227 (2012)
Modat, M., Ridgway, G.R., Taylor, Z.A., Lehmann, M., Barnes, J., Hawkes, D.J., Fox, N.C., Ourselin, S.: Fast free-form deformation using graphics processing units. Comput. Methods Programs Biomed. 98, 278–284 (2010)
Burke, R.P., Xu, Z., Lee, C.P., Baucom, R.B., Poulose, B.K., Abramson, R.G., Landman, B.A.: Multi-atlas segmentation for abdominal organs with gaussian mixture models. Proc. SPIE Int. Soc. Opt. Eng. 9417, 941707 (2015)
Wang, H., Suh, J.W., Das, S.R., Pluta, J.B., Craige, C., Yushkevich, P.A.: Multi-atlas segmentation with joint label fusion. IEEE Trans. Pattern Anal. Mach. Intell. 35, 611–623 (2013)
Kerschnitzki, M., Kollmannsberger, P., Burghammer, M., Duda, G.N., Weinkamer, R., Wagermaier, W., Fratzl, P.: Architecture of the osteocyte network correlates with bone material quality. J. Bone Miner. Res. 28, 1837–1845 (2013)
Elbahnasy, A.M., Shalhav, A.L., Hoenig, D.M., Elashry, O.M., Smith, D.S., McDougall, E.M., Clayman, R.V.: Lower caliceal stone clearance after shock wave lithotripsy or ureteroscopy: the impact of lower pole radiographic anatomy. J. Urol. 159, 676–682 (1998)
Chew, B.H., Brotherhood, H.L., Sur, R.L., Wang, A.Q., Knudsen, B.E., Yong, C., Marien, T., Miller, N.L., Krambeck, A.E., Charchenko, C., Humphreys, M.R.: Natural history, complications and re-intervention rates of asymptomatic residual stone fragments after ureteroscopy: a report from the EDGE research consortium. J. Urol. 195, 982–986 (2016)
Liu, J., Wang, S., Turkbey, E.B., Linguraru, M.G., Yao, J., Summers, R.M.: Computer-aided detection of renal calculi from noncontrast CT images using TV-flow and MSER features. Med. Phys. 42, 144–153 (2015)
Liu, J., Wang, S., Linguraru, M.G., Yao, J., Summers, R.M.: Computer-aided detection of exophytic renal lesions on non-contrast CT images. Med. Image Anal. 19, 15–29 (2015)
Will, S., Martirosian, P., Wurslin, C., Schick, F.: Automated segmentation and volumetric analysis of renal cortex, medulla, and pelvis based on non-contrast-enhanced T1- and T2-weighted MR images. MAGMA 27, 445–454 (2014)
Acknowledgements
This research was supported by the Vanderbilt Institute for Surgery and Engineering (VISE) Fellowship (De), NSF CAREER 1452485 (Landman), NIH grant 5R21EY024036 (Landman), NIH grant 1R21NS064534 (Landman), and NIH grant 1R03EB012461 (Landman). This study was supported in part by VISE/VICTR VR3029 and the National Center for Research Resources, Grant UL1 RR024975-01, and is now the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. The project was also supported in part by using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, TN. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors have no conflict of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Huo, Y., Braxton, V., Herrell, S.D., Landman, B., De, S. (2017). Automated Characterization of Pyelocalyceal Anatomy Using CT Urograms to Aid in Management of Kidney Stones. In: Cardoso, M., et al. Computer Assisted and Robotic Endoscopy and Clinical Image-Based Procedures. CARE CLIP 2017 2017. Lecture Notes in Computer Science(), vol 10550. Springer, Cham. https://doi.org/10.1007/978-3-319-67543-5_9
Download citation
DOI: https://doi.org/10.1007/978-3-319-67543-5_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-67542-8
Online ISBN: 978-3-319-67543-5
eBook Packages: Computer ScienceComputer Science (R0)